Abstract
Increasing ocean temperatures severely affects marine species and ecosystems. Among other things, rising temperatures cause coral bleaching and loss of breeding grounds for marine fish and mammals. Motivated by the need to understand better these global problems, researchers from all over the world generated huge amounts of oceanographic data during the last years. However, most of this data remain isolated in their own silos. One approach to provide safe accessibility to these silos is to map local, often database-specific identifiers, to shared global identifiers. This mapping can then be used to build interoperable knowledge graphs (KGs), where entities such as publications, people, places, specimens, environmental variables and institutions are all part of a single, shared knowledge space. This short paper describes one such effort, the OceanGraph KG, including the modeling and publication processes, and the current and prospective uses of the dataset.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction and Motivation
We are transitioning from the era of Big Data to Big Knowledge, and semantic knowledge bases such as KGs play an important role in this transition. This is evident from the expanding investments in KG research and development by major corporate players, resulting in widely used systems such as IBM Watson, Google entity search, Apple Siri, and Amazon product graph. KGs are an increasingly critical component of the Semantic Web (SW) [1] and serve as information hubs for general use as well as for domain-specific applications. There is no common definition about what a KG is and what is not [2], since KG have emerged as a unifying technology in several areas of artificial intelligence, including Natural Language Processing and Semantic Web, and the scope of what constitutes a KG has continued to broaden [3]. Most KGs seek to aggregate knowledge from third party sources, either from external databases, from data aggregated through crawling the Web, or through the application of entity and relationship extraction methods [4]. KGs are not simply aggregations of Resource Description Frameworks (RDFs)Footnote 1 or Linked Data (LD) [5]. Instead they provide critical time-invariant information about entities of general interest. Their structures tend to be focused on a limited set of relations adhering to a coherent knowledge model, setting them apart from the LD cloud in general, which usually has relied on the open framework of the SW to accommodate a completely free-form use of vocabularies and ontologies.
The ocean and life sciences, in general, yielded an amount of data that is not only huge in volume, but also highly heterogeneous both in types and formats, and scattered across distributed data repositories [6]. For individual researchers, this situation presents a difficult challenge regarding discovery, access and integration of the data required to conduct scientific inquiries. This also introduces difficult knowledge management issues that must be overcome by the whole research community [7]. We can mention a couple related works which partially address these issues through KGs. The first is a proof-of-concept of a KG for the Australian fauna, combining taxonomic classifications and scientific publications. The latter is a datasetFootnote 2 including information from oceanographic cruises, physical samples, and technical reports from the geoscience metadata repositories in the United States. In both cases, data has been published according to best practices for linked data and are publicly available via a SPARQL endpoint. None of them integrate biodiversity and biogeography data as proposed here [8] and GeoLink [9].
In this short contribution we present the initial efforts to develop OceanGraph KG. OceanGraph has leveraged linked data principles to create a KG that allows users to seamlessly query and reason over some of the largest oceanographic data repositories such as the National Marine Data System (NMDS)Footnote 3, Global Biodiversity Information Facility (GBIF)Footnote 4 and Ocean Biogeographic Information System (OBIS)Footnote 5. As an illustration, we present a use case that shows how OceanGraph allows to relate species occurrences from GBIF to environmental variables from OBIS, a fundamental requirement of macroecological analyses [10], particularly those considering environmental drivers of species distributions, and how distributions are expected to shift as the climate changes [11].
2 OceanGraph Data Providers
The datasets that make up OceanGraph originate from areas of ocean science, Biodiversity/Biogeography, scientific publications, locations, and environmental data. Most of the datasets have information funded by the Argentine government, although there are others that belong to third parties. The datasets that currently comprise OceanGraph are the following:
-
Marine Biodiversity/Biogeography. As mentioned earlier, part of the information comes from GBIF and OBIS, two of the most important international databases. Both databases use Darwin Core standard (DwC) [12] to represent species information. For additional information see [13] where is described how this information was converted and published as Linked Open Data.
-
Oceanographic campaigns. National Marine Data System is a web platform that allows publishing datasets of oceanographic campaigns that was sampled in Argentine sea. These datasets are composed of (i) metadata of the oceanographic campaigns (name of the campaign, vessel, dates, people and institutions involved, geographical coverage among others), and (ii) data recorded by the vessel in its trajectory, which contains the information of the measured variables (pressure, salinity, temperature, depth, positions where the variable was sampled among others). In [14] the complete process of conversion and publication of these data sets is described.
-
Publications. Springer Nature SciGraph [15] a Linked Open Data platform for the scholarly domain which aggregates data sources from Springer Nature and key partners from the scholarly domain. The Linked Open Data platform collates information from across the research landscape, for example funders, research projects, conferences, affiliations and publications. Data in Springer Nature SciGraph is projected to contain 1.5 to 2 billion triples (as of January 2019).
-
Environmental variables. Data generated by fixed stations belonging to Comodoro conocimiento agencyFootnote 6 created by the City Government of Comodoro Rivadavia (Argentina) has maritime buoys for environmental monitoring. The aim is to monitor the mariculture zones in the San Jorge Gulf and validate data from oceanographic campaigns in the area. These buoys measure radiation, wind speed, temperature, humidity, oxygen, conductivity, salinity and fluorescence, among others.
-
Locations. GeoNamesFootnote 7 is a free and open source geographical database. Primarily for developers wanting to integrate the project into web services and applications, it combines world-wide geographical data including names of places in various languages, elevation, population, and all latitude/longitude coordinates. Data is accessible through a number of web services and a daily database export.
3 OceanGraph Development
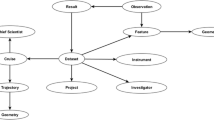
The general structure of OceanGraph is based on the relationships established among datasets of Fig. 1. The core entities are campaigns, species, publications, people, environmental variables and locations. For instance, if the knowledge graph is queried for a particular scientist, results might include the oceanographic campaigns they participated in (from NMDS), datasets they collected (from OBIS/GBIF), and papers they has written (from SciGraph). Similarly, if a particular species is queried for, the user can determine who collected it, when, where, and under which oceanographic campaign.
OceanGraph can be accessed through GraphDBFootnote 8, which is a highly efficient and robust graph database. It allows users to explore the hierarchy of RDF classes (Class hierarchy), where each class can be browsed to explore its instances. Similarly, relationships among these classes also can be explored giving an overview about how many links exist between instances of the two classes (Class relationship). The user can visually explore the dataset, accessing the URL http://web.cenpat-conicet.gob.ar:7200/login using the credentials (user: oceangraph password: ocean.user). After successful authentication, select the repository OceanGraph.
3.1 Underlying Vocabularies and Ontologies
The description and management of information resources have to obey well-known standards to ensure that they will be made available for various communities of users. In this section, we will describe the main resources related to geospatial data, Biodiversity/Biogeography, oceanography and environmental data. In addition to information resources management, we selected existing standards to manage information about agents and domain entities. Several data providers use their own ontologies and existing vocabularies such as FOAFFootnote 9, Dublin CoreFootnote 10 and Prov-OFootnote 11.
-
NERC Vocabulary Server [16] Natural Environment Research Council (NERC) Vocabulary Server provides access to lists of standardized terms that cover a broad spectrum of disciplines of relevance to the oceanographic and wider community. Using standardized sets of terms in metadata and to label data solves the problem of ambiguities associated with data markup, for example, sometimes data-level errors may occur, which are caused by differences that occur in data domains due to multiple possible representations, similar data interpretations, or even spelling errors e.g.Oxygen, O2, Oxgen.
-
GeoSPARQL [17] is a standard for representing and querying of geospatial linked data for the Semantic Web from the Open Geospatial Consortium (OGCFootnote 12). The definition of a small ontology based on well-understood OGC standards is intended to provide a standardized exchange basis for geospatial RDF data which can support both qualitative and quantitative spatial reasoning and querying with the SPARQLFootnote 13 database query language.
-
Darwin Core Standard [12] includes a glossary of terms intended for sharing information about biological diversity by providing reference definitions, examples, and commentaries. In OceanGraph, we use it to describe properties, elements, fields, columns, attributes and concepts.
-
Geolink [18] describes an ontological design pattern (ODP) for oceanographic cruises using Web Ontology Language (OWL). This pattern was specified as a combination and reuse of the existing patterns: trajectory, event and information object. We consider that this ODP is sufficiently generic and adapts well to our requirements, and for this reason will be adopted to define the relationships and classes that we will designate in our data set.
-
SSN and SOSA ontologies [19] to describe sensors and their observations we use the Semantic Sensor Network (SSN), specially the self-contained ontology SOSA (Sensor, Observation, Sample and Actuator) that describes elementary classes and properties. Both ontologies can be used for a wide range of applications and use cases for example, satellite imagery, large-scale scientific monitoring and the Web of Things among others. We use SOSA to describe the process of gathering information from fixed stations.
3.2 Cross-Linking
Cross-linking the OceanGraph datasets in a semi-automated way is crucial aiming at facilitating data integration by linking overlapping contents existing in many of the OceanGraph repositories. For example, people involved in an oceanographic campaign can also be authors of scientific publications or, for example, marine species observed during a campaign are published in OBIS or GBIF. Linking the same people or species in different repositories is the key feature that enables integrated querying and makes OceanGraph so useful. To do this, we use SILK frameworkFootnote 14 to express heuristics for deciding whether a semantic relationship exists between two entities. For instance, to relate people involved in an oceanographic campaign with their contributions in OBIS or GBIF, the Levenshtein distance is used to disambiguate two inputs through computing the similarity between them. This operator receives inputs such as dwc:recordedByFootnote 15 (property used in OBIS/GBIF) and foaf:name and returns the links between them by using the owl:sameAs axioms. Figure 2 shows the relationships used to integrate OceanGraph datasets.
4 Use Case
As mentioned earlier, relating species occurrences with environmental variables is a very common requirement of macroecological analyses [10]. This use case describes how this problem can be addressed in a simplified way. To do this we define a SPARQL query that associates the occurrences of a marine species (a fish for example) with water body temperature in a specific marine region. Firstly, we define the marine region called San Matias Gulf, type (geo:Polygon), secondly we retrieve the observations of the defined species as points using (geo:point). Since GeoSPARQL allows performing spatial operations, we can query if a point is contained within a polygon using the function (geof:sfWithin). Finally we retrieve the measured environmental variable (also georeferenced by a point). NERC provides URIs for each of the variables, so we only need to retrieve the URI for the variable temperature of the water body http://vocab.nerc.ac.uk/collection/P02/current/TEMP/. After authenticating, the query can be executed in GraphDB using the following linkFootnote 16. Figure 3 shows the results after executing the query.
Although this is a simple example, it is important to highlight that KGs used as tools to integrate information, allow us to answer questions that require an integrated management of heterogeneous information sources. As OceanGraph begins to be disseminated in the oceanographic research community, we hope that the use of data by third parties will continue to grow and generate new answers.
5 Conclusions and Future Work
Currently the publication of KGs grew substantially in diverse areas, however, there is still much work to be done in the domain of ocean science. In this paper, we presented an overview of our initial effort to create an oceanographic KG called OceanGraph, reusing specific vocabularies and ontologies of this domain. This initiative will allow to model a public and freely available source of ocean science data composed of largest data repositories in this domain, and thus building applications on data reconciliation, data augmentation, and meta-analyses in these fields. Particularly, as future work we need to work on a user-friendly interface together with searching engines and visualizations that allows non-expert users to explore the data. In this same direction, we plan to link our dataset to other ones from diverse domains, f.e., fisheries [20].
Notes
- 1.
- 2.
- 3.
- 4.
- 5.
- 6.
- 7.
- 8.
- 9.
- 10.
- 11.
- 12.
- 13.
- 14.
- 15.
- 16.
References
Berners-Lee, T., Hendler, J., Lassila, O., et al.: The Semantic Web. Scientific American (2001)
Paulheim, H.: Knowledge graph refinement: a survey of approaches and evaluation methods. Semant. Web J. 8, 489–508 (2016)
Sequeda, J.F., Kejriwal, M., Lopez, V.: Construction, management and querying. Semant. Web J. (2018). Special Issue on Knowledge Graphs
Wöß, W., Ehrlinger, L.: Towards a definition of knowledge graphs. In: 12th International Conference on Semantic Systems - SEMANTiCS 2016 (2016)
Bizer, C., Heath, T., Berners-Lee, T.: Linked data-the story so far. In: Semantic Services, Interoperability and Web Applications: Emerging Concepts (2009)
Malik, T., Foster, I.: Addressing data access needs of the long-tail distribution of geoscientists. In: 2012 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), pp. 5348–5351. IEEE (2012)
Campbell, P.: Data’s shameful neglect. Nature 461(7261), 145 (2009)
Page, R.D.M.: Ozymandias: a biodiversity knowledge graph. bioRxiv (2018)
Cheatham, M., et al.: The GeoLink knowledge graph. Big Earth Data (2018)
Muller-Karger, F.E., et al.: Advancing marine biological observations and data requirements of the complementary essential ocean variables (EOVS) and essential biodiversity variables (EBVS) frameworks. Front. Mar. Sci. 5, 211 (2018)
Pearson, R.G., Dawson, T.P.: Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Glob. Ecol. Biogeogr. 12(5), 361–371 (2003)
Wieczorek, J., et al.: Darwin core: an evolving community-developed biodiversity data standard. PLoS ONE 7, 1–8 (2012)
Zárate, M., Braun, G., Fillottrani, P.: Adding biodiversity datasets from argentinian patagonia to the web of data (2017)
Zárate, M., Rosales, P., Fillottrani, P., Delrieux, C., Lewis, M.: Oceanographic data management: towards the publishing of Pampa Azul oceanographic campaigns as linked data (2018)
Springer Nature SciGraph. http://www.springernature.com/gp/researchers/scigraph. Accessed 24 Jan 2019
Leadbetter, A., Lowry, R., Clements, D.O.: The NERC vocabulary server: version 2.0. In: Geophysical Research Abstracts, vol. 14 (2012)
Battle, R., Kolas, D.: Enabling the geospatial semantic web with parliament and GeoSPARQL. Semant. Web 3(4), 355–370 (2012)
Krisnadhi, A., et al.: An ontology pattern for oceanographic cruises: towards an oceanographer’s dream of integrated knowledge discovery (2014)
W3C. Semantic Sensor Network Ontology (SSN) W3C recommendation (2017)
Froese, R., Pauly, D., et al.: Fishbase (2010)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Zárate, M., Rosales, P., Braun, G., Lewis, M., Fillottrani, P.R., Delrieux, C. (2019). OceanGraph: Some Initial Steps Toward a Oceanographic Knowledge Graph. In: Villazón-Terrazas, B., Hidalgo-Delgado, Y. (eds) Knowledge Graphs and Semantic Web. KGSWC 2019. Communications in Computer and Information Science, vol 1029. Springer, Cham. https://doi.org/10.1007/978-3-030-21395-4_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-21395-4_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-21394-7
Online ISBN: 978-3-030-21395-4
eBook Packages: Computer ScienceComputer Science (R0)