Abstract
The Lpp lipoprotein of Escherichia coli is the first identified protein with a covalently linked lipid. It is chemically bound by its C-terminus to murein (peptidoglycan) and inserts by the lipid at the N-terminus into the outer membrane. As the most abundant protein in E. coli (106 molecules per cell) it plays an important role for the integrity of the cell envelope. Lpp represents the type protein of a large variety of lipoproteins found in Gram-negative and Gram-positive bacteria and in archaea that have in common the lipid structure for anchoring the proteins to membranes but otherwise strongly vary in sequence, structure, and function. Predicted lipoproteins in known prokaryotic genomes comprise 2.7% of all proteins. Lipoproteins are modified by a unique phospholipid pathway and transferred from the cytoplasmic membrane into the outer membrane by a special system. They are involved in protein incorporation into the outer membrane, protein secretion across the cytoplasmic membrane, periplasm and outer membrane, signal transduction, conjugation, cell wall metabolism, antibiotic resistance, biofilm formation, and adhesion to host tissues. They are only found in bacteria and function as signal molecules for the innate immune system of vertebrates, where they cause inflammation and elicit innate and adaptive immune response through Toll-like receptors. This review discusses various aspects of Lpp and other lipoproteins of Gram-negative and Gram-positive bacteria and archaea.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
The first identified and fully characterized protein with a covalently linked lipid was the murein lipoprotein Lpp of Escherichia coli, also called the major lipoprotein or Braun’s lipoprotein. It is the type protein of a large variety of proteins in Gram-negative and Gram-positive bacteria and in archaea. These proteins are abundant in the cells and all have in common a lipid structure through which they anchor the proteins to a membrane outside of the cytoplasm. However, their sequence, structure, and function strongly vary. They are involved in membrane protein incorporation, protein secretion, signal transduction, conjugation, cell wall metabolism, antibiotic resistance, biofilm formation, adhesion to host tissues, and signaling for the innate immune system of vertebrates. This review will discuss various aspects of E. coli Lpp and highlight features of other lipoproteins from other bacterial and archaeal species. The early work on Lpp has been summarized in (Braun 1975; Braun and Hantke 1974; Braun and Wu 1994).
E. coli Lpp
Discovery and Components of Lpp
The Lpp lipoprotein covalently bound to murein (peptidoglycan) of Escherichia coli was discovered in a search for a bond that upon cleavage with trypsin decreased the absorbance of the isolated cell envelope much more rapidly than when any other protease was used (Braun and Rehn 1969). Cell envelopes treated with trypsin separated into two membranes, namely the outer membrane and cytoplasmic membrane, as observed by electron microscopy of ultra-thin sections. Samples not treated with trypsin were closely attached to each other. Since trypsin released only small amounts of protein into the medium, the trypsin cleavage site was sought in the insoluble part.
It had been shown earlier that protein was associated with isolated murein, but the protein material was not investigated further because the focus was on the chemical composition of murein (Weidel and Pelzer 1964). Treatment of cell envelopes with a hot sodium dodecyl sulfate solution left murein as the sole insoluble component. It became morphologically thinner when treated with proteases, but kept the size and rod form of the exponentially growing cells from which it was isolated. Acid hydrolysis of murein of exponentially growing cells yielded the constituents of murein and a mixture of all but six common amino acids (Cys, His, Pro, Gly, Phe, and Trp; by contrast, in stationary phase cells, the proteins associated with murein consist of all amino acids). The specific lack of these common amino acids suggested that a defined protein was associated with murein. After trypsin treatment, only lysine remained associated with murein, which indicated that it linked the protein and murein. Further analysis showed that the lysine is bound through the ε-amino group to the optical l-center of diaminopimelic acid of the murein peptide side chain, where it replaces d-alanine (Braun and Bosch 1972). The lysine residue is at the C-terminal end of Lpp (Fig. 3.1a).
a Amino acid sequence of the E. coli murein lipoprotein (Lpp) with bound murein (peptidoglycan) subunits. The heptad repeats (3 + 4) are in boldface. The N-terminal lipid attachment site and the C-terminal murein attachment site (in italics) are outside the heptad register. b High-resolution crystal structure of the Lpp trimer (residues 2–56) comprising a parallel three-stranded coiled coil and two helix capping motifs (Shu et al. 2000). c Chemical structure of glycerylcysteine with three bound fatty acids (mostly palmitic acid)
The mature Lpp protein consists of 57 residues and an additional N-terminal modified cysteine. The amino acid sequence reveals 14 repeats composed of seven residues, of which every third or fourth residue is hydrophobic (Fig. 3.1a). Such a periodicity is typical for coiled-coils of α-helices in which seven residues (heptad repeats) form two turns of a helix and that the side chains of parallel helices interlock systematically. Lpp was the first coiled-coil protein to be sequenced (Lupas et al. 2017). Its high α-helical content was deduced from circular dichroism spectra (Braun et al. 1976b). Recombinant Lpp devoid of the lipid forms a long parallel α-helical trimer. The X-ray crystal structure reveals a three-stranded coiled-coil domain from residue 5 to 53, with the heptad repeat in register throughout the entire region (Fig. 3.1b) (Shu et al. 2000). The exceedingly high trypsin cleavage rate is explained by the accumulation of three trypsin cleavage sites at the very C-terminus and their exposure outside the triple helix. The sequences Tyr-Arg-Lys and Tyr-Lys-Lys are most frequently found in lipoproteins linked to murein.
Lipid Covalently Bound to the Protein
When structural work on Lpp began, lipoproteins were not defined molecules but rather mixtures of proteins with non-covalently linked mixtures of lipids. But in Lpp, lipid was associated with a protein that could not be removed with detergents or organic solvents and was therefore considered to be covalently bound. The chemical structure of the protein lipid attachment site was determined by chemically degrading isolated Lpp, incorporation of radioactively labeled suspected precursors, and finally by chemical synthesis of S-glycerylcysteine thioether (Hantke and Braun 1973). The structure was shown to be a glyceryl group with a fatty acid composition similar to that of phospholipids and bound as a thioether to the sulfhydryl group of cysteine, and was named glycerylcysteine (Fig. 3.1c). The amino group of the N-terminal cysteine is bound to a fatty acid, mainly palmitic acid. This unique lipid structure was later shown to be part of all bacterial lipoproteins with a covalently bound lipid. The first three-dimensional structure of triacylated cysteine in a lipid bilayer was recently found in the electron cryomicroscopy structure of a cytochrome oxidase (Sun et al. 2018). In this structure, the lipid anchors at the ActB and ActE proteins are tilted with respect to the plane of the lipid bilayer, thereby restricting the ability of other lipids to pack around them. The lipids are localized close to the entry point of menaquinol in the complex.
Linkage of Lpp to Murein (Peptidoglycan)
In E. coli, one-third of Lpp is covalently bound to murein (Braun and Rehn 1969; Inouye et al. 1972). The linkage is formed between the carboxyl group of the optical l-center of meso-diaminopimelate (Dpm) of the murein tetrapeptide and the ε-amino group of the carboxyl-terminal lysine (Lys) of lipoprotein (Fig. 3.1a) (Braun and Bosch 1972). The reaction is catalyzed by three very similar l-d-transpeptidases, namely ErfK, YcfS, and YbiS (Magnet et al. 2007), whereby the d-Ala linked to Dpm is replaced by Lys and the peptide energy of the Dpm-Ala bond is preserved to form the Dpm-Lys bond. The genes encoding all three enzymes must be deleted to obtain murein free of Lpp, but YbiS plays the major transpeptidation because the deletion of its gene largely prevents binding of Lpp to murein. The presence of several transpeptidases is not surprising because redundant enzymes in murein synthesis and murein modification have been frequently observed (Pazos et al. 2017). Murein is essential for the structure of cells that they cannot afford a defective synthesis. In addition, the structure of murein changes under different growth condition which requires different enzyme activities (Vollmer et al. 2008).
The C-terminal Lys in the Tyr-Arg-Lys triad plays a critical role in linkage of Lpp to murein (Zhang and Wu 1992; Zhang et al. 1992). By contrast, alterations at the N-terminus, such as lack of lipid modification, addition of a signal peptide, and fusion of the outer membrane protein OmpF, reduce but do not abolish covalent attachment of Lpp to murein (Zhang et al. 1992). The amount of Lpp bound to murein can vary; for example, in stationary phase cells, Lpp binding increases by 70% (Glauner et al. 1988).
Free Lpp
Free Lpp not attached to murein was detected when proteins of cell envelopes labeled with radioactive arginine or histidine were separated by SDS-polyacrylamide gel electrophoresis (Inouye et al. 1972). A fast migrating band, i.e., a small protein, contained labeled arginine but not histidine. The only protein known to be devoid of histidine in E. coli was the small protein Lpp. Its identity was confirmed by double labeling with other amino acids present and absent in Lpp. The electrophoretic mobility of free Lpp was faster than that of Lpp released from murein by lysozyme; the latter preparation also contains muropeptides. The amount of free Lpp was twice as high as that of murein-bound Lpp.
Distinct Arrangement of Bound and Free Lpp in the Outer Membrane
The early proposal (Braun and Rehn 1969) that Lpp extends from murein across the periplasm into the outer membrane was supported by the immunological determination of Lpp in separated outer and cytoplasmic membranes (Bosch and Braun 1973). Lpp is bound by the C-terminal lysine to diaminopimelic acid of murein and is fixed to the outer membrane by insertion of the lipid part into the inner lipid leaflet of the outer membrane. It was assumed that free lipoprotein occupies the same location as the bound form in the cell envelope. This assumption was supported by cross-linking the free form with the bound form and non-covalent binding of the free form by the C-terminal lysine to murein (Choi et al. 1987). However, distinct cellular locations of bound and free Lpp were discovered over two decades later (Cowles et al. 2011). Free Lpp spans the outer membrane and its C-terminus is surface-exposed, whereas bound Lpp resides in the periplasm. This conclusion was drawn from differential labeling with membrane-impermeable biotin reagents and proteolytic removal of the labels at the cell surface. The biotin label was attached to the C-terminal Lys55 and Lys58 residues. In addition, the C-terminal FLAG epitope (DYKDDDDK) of a constructed Lpp derivative could be proteolytically removed by treating cells with trypsin, which showed that the C-terminus of free Lpp is exposed at the cell surface.
The question arises how the free form inserts into and is partially translocated across the outer membrane. The parallel three-stranded coiled-coil structure of Lpp is too hydrophilic to insert spontaneously into a lipid bilayer. Since the free form is first synthesized and then converted to the bound form (Inouye et al. 1972), the two forms might transiently associate with each other, which would explain their cross-linkage. The free form is either translocated by the Bam complex, or by a specific translocase. Specific translocases were identified for various proteins, e.g., NalP or RcsF with the Bam complex, pullulanase with the type II secretion system, and in Neisseria Slam1 with TbPB, LbpB, fHb1, and Slam2 with HpuA (see section “Lipoproteins in Other Gram-Negative Bacteria”). The translocation of lipoproteins to the cell surface has recently been reviewed (Hooda and Moraes 2018).
Biosynthesis of Lpp
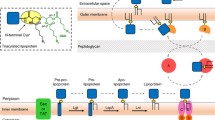
E. coli Lpp is synthesized in the cytoplasm, modified with lipids at the outer leaflet of the cytoplasmic membrane, and translocated to and inserted into the outer membrane. All these steps are mediated by dedicated proteins.
Unmodified lipoprotein can be synthesized in vitro with an mRNA of about 250 nucleotides (Hirashima et al. 1974). The mRNA has an unusually long average half-life of 11.5 min, in contrast to the half-life of 2.1 min of cytoplasmic protein mRNAs (Hirashima and Inouye 1973). The long half-life is suitable for allowing synthesis of this most abundant protein in E. coli (about 106 molecules per cell).
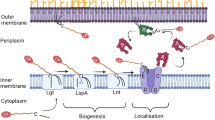
Lpp is synthesized in the cytoplasm with a signal sequence that serves to transfer the protein across the cytoplasmic membrane by the Sec translocon. The cysteine residue located at the border between the signal sequence and the mature protein is strictly conserved in all lipoproteins. It is surrounded by a partially conserved sequence in lipoproteins—Leu(Ala/Val)-Leu-Ala(Ser)-Gly(Ala)-Cys. This so-called lipobox is recognized by lipid-modifying enzymes. Lipid modification takes place at the periplasmic side of the cytoplasmic membrane. First, diacylglycerol is added to the sulfhydryl group of the cysteine by phosphatidylglycerol/prolipoprotein diacylglyceryl transferase (Lgt) (Fig. 3.2). Then, a special lipoprotein signal peptidase (Lsp or signal peptidase II) cleaves the signal peptide in front of the Cys residue (Innis et al. 1984). Signal peptidase II preferentially recognizes didecanoyl glycerol as the optimal lipid length and exclusively the enantio (R) form of the diacylglycerol (Kitamura and Wolan 2018). The protein remains attached to the cytoplasmic membrane via the diacylglycerol. Finally, a fatty acid is transferred from phospholipids to the amino group of the Cys residue by membrane-bound phospholipid/apolipoprotein transacylase (Lnt) (Tokunaga et al. 1982; Noland et al. 2017).
Genes encoding the three enzymes Lgt, Lsp, and Lnt are essential for E. coli and most other Gram-negative bacteria. The crystal structure of the diacylglyceryl transferase Lgt shows a central cavity about 20 Å deep, with a major opening to the periplasmic side and two clefts exposed to the membrane lipid (Mao et al. 2016). Phosphatidylglycerol is positioned with the two acyl chains in the lower hydrophobic part of the cleft and the hydrophilic head protruding into the interface between the outer leaflet of the cytoplasmic membrane and the periplasm. It is predicted that the two clefts (6 and 10 Å wide) are entrances for phosphatidylglycerol and the lipobox of the lipoprotein. Substrates and product enter and leave laterally relative to the lipid bilayer.
The crystal structure of the Lsp signal peptidase II from Pseudomonas aeruginosa complexed with the inhibitor globomycin has been determined (Vogeley et al. 2016). The enzyme consists of a periplasmic domain fixed to the cytoplasmic membrane by a domain of four transmembrane helices. Globomycin acts as a noncleavable peptide that sterically blocks the active site. It mimics diglyceride Lpp in that it is composed of a 19-member cyclic depsipeptide that includes an α-methyl-β-hydroxy fatty acid. It is first partitioned in the cytoplasmic membrane and then diffuses laterally into the active site of the enzyme. The lipopeptide helix of Lpp fits into the space between membrane helices 12 and 14, which positions the Cys residue of the lipobox into the active site while leaving the protein in the periplasm. It is predicted that Lsp is an aspartyl endopeptidase with two Asp residues suitably positioned to form the catalytic diad. After release of the signal peptide, lipoprotein moves out of the active site but remains bound to the cytoplasmic membrane by the diacylglyceride moiety.
The crystal structures of the N-acylase Lnt of E. coli (Noland et al. 2017) and P. aeruginosa (Wiktor et al. 2017) have been determined. Both structures are composed of a large periplasmic domain that is fixed to the cytoplasmic membrane by eight transmembrane helices. The active site with the catalytic Cys387 is positioned at the base of a long channel above the outer leaflet of the cytoplasmic membrane. It contains an opening towards the membrane for substrates and products; this opening serves as the portal through which phospholipid and glycerylcysteine of lipoprotein enter the active site. The Lnt enzyme transfers a fatty acid from a phospholipid, preferentially phosphatidylethanolamine, to form S-acyl cysteine. The acyl chain, usually palmitate, is transferred to the α-amino group of S-diacyl-glyceryl lipoprotein. Triacylated lipoprotein leaves the active site and enters the membrane, and the active site is ready for the next round of lipoprotein acylation.
Transfer of Lpp from the Cytoplasmic Membrane to the Outer Membrane
Sorting of the lipoprotein from the cytoplasmic membrane to the outer membrane was studied mainly with E. coli Lpp and P. aeruginosa Pal (Okuda and Tokuda 2011; Narita and Tokuda 2017). The final destination of Lpp and Pal is the inner leaflet of the outer membrane; the protein moiety remains in the periplasm, but the N-terminal lipid anchors the lipoprotein to the outer membrane. Five proteins, designated LolA, LolB, LolC, LolD, and LolE, mediate lipoprotein sorting from the cytoplasmic membrane into the outer membrane (Fig. 3.3). LolCDE belong to the ABC transporter superfamily; LolC and LolE are the transmembrane subunits and LolD is the ATPase. One LolD subunit is bound to LolC, and one is bound to LolE. The LolCDE complex releases lipoprotein from the cytoplasmic membrane by ATP hydrolysis. Lipoprotein attaches to the soluble periplasmic protein LolA, and is then handed over to lipoprotein LolB, which is attached to the outer membrane by its lipid moiety. The affinity of LolB for lipoprotein is higher than that of LolA. The triacyl group of LolB possibly distorts the packing of the lipid bilayer, as has been found for cytochrome oxidase (Sun et al. 2018), and by this means facilitates entry of the triacyl group of lipoprotein into the inner leaflet of the outer membrane. LolE of E. coli binds lipoprotein and interacts with LolA, whereas LolC does neither (Okuda and Tokuda 2009).
Crystal structures of LolA and LolB have been determined and are similar despite their different amino acid sequences (Takeda et al. 2003). Both structures contain hydrophobic cavities that serve as binding sites for the acyl chains of lipoproteins. The sequence similarity between the periplasmic regions of LolC and LolE and between LolA and LolB suggest the presence of cavities in LolC and LolE similar to those in LolA and LolB. It is likely that lipoprotein is transferred from LolCDE to LolA to LolB through their hydrophobic cavities (Okuda and Tokuda 2009). Only triacylated lipoprotein is transferred across the periplasm by the Lol proteins. Deletion of any of the genes encoding the Lol proteins are not tolerated by the cells, presumably because aberrant mislocated lipoproteins accumulate.
Modified Lipoproteins that Remain in the Cytoplasmic Membrane
When Ser2 of mature Lpp is replaced with Asp, Lpp is retained in the cytoplasmic membrane; other amino acids direct sorting into the outer membrane (Yamaguchi et al. 1988). The Asp-dependent retention of Lpp in the cytoplasmic membrane is influenced by the residue at position 3, and certain combinations of residues at positions 2 and 3 act as cytoplasmic membrane signals. Asp2 interferes with recognition of N-acyl S-diacylglyceryl Cys by LolCDE and therefore prevents Lpp release from the cytoplasmic membrane. Lpp with Asp at position2 also does not form a complex with LolA. In P. aeruginosa, residues at positions 3 and 4 but also Asp2 determine membrane localization (Narita and Tokuda 2007; Lewenza et al. 2008). Structural requirements around the Cys residue for the release of Lpp by LolA from the cytoplasmic membrane have been studied in detail with point mutants (summarized in Tokuda et al. 2007). The lipoprotein signal peptidase Lsp specifically recognizes the enantio (R) form of the diacylglycerol (Kitamura and Wolan 2018).
Alternative Lol Pathway
The essentiality of the Lol pathway in transferring lipoproteins from the cytoplasmic membrane into the outer membrane was recently questioned (Grabowicz and Silhavy 2017b). A chromosomal lolB deletion mutant carrying an arabinose-inducible lolB gene on a plasmid does not grow when lolB transcription is not induced. However, growth of the lolB deletion strain is partially restored by deleting lpp. Growth is further enhanced by deleting in addition rcsF, which encodes an outer membrane lipoprotein that controls the Rcs stress response. lolB deletion leads to a toxic activation of the Rcs stress response caused by accumulation of mislocalized Lpp. The lpp rcsF double deletion mutant is markedly more viable during LolB depletion because the Rcs regulon is not activated. Furthermore, suppressor mutations that allow growth of the lolB mutant are in cpxA. CpxA is part of the Cpx stress regulon. An lpp rcsB double deletion mutant that carries an activating cpxA mutation is still viable when lolB is deleted. CpxA alleviates stress caused by Lpp trafficking defects. In addition, the Bam complex, which contains the lipoproteins BamB, BamC, BamD, and BamE attached from the periplasm to the BamA outer membrane barrel protein is fully functional in the lpp cpxA rcsB triple mutant. In this triple mutant, LolB is not required for the delivery of lipoproteins from the cytoplasmic membrane to the outer membrane. Under conditions in which LolB is not essential, LolA is also not required. LolCDE are still essential and cannot be bypassed. These data suggest the presence of alternative LolAB-independent lipoprotein pathways with LolCDE as a constituent. Such pathways might exist in α- and ε-proteobacterial classes of the major Gram-negative phyla in which no LolB homolog is apparent.
Control of Lpp Biosynthesis
Since Lpp is by far the most abundant protein in E. coli (approximately 106 copies per cell), its synthesis must be controlled to avoid upon overexpression depletion of substrates and enzymes of RNA and protein synthesis required for other cell components and overloading of the processing and assembly machineries. Synthesis of the essential lipoproteins BamD of the Bam outer membrane protein assembly complex and LptE of LPS assembly directly competes with the lipid modification and Lol-dependent transfer across the periplasm of Lpp. Sigma E is the major regulator of outer membrane biogenesis. It controls transcription of 100 genes, including all of the machinery involved in transport and assembly of outer membrane proteins (Guo et al. 2014). Upon induction of sigma E synthesis, translation of the mRNA encoding Lpp is repressed specifically through the small sRNA MicL attached to the Hfq protein (Guo et al. 2014). Transient overproduction of Lpp decreases insertion of the BamD and LptE lipoproteins into the outer membrane and assembly of their respective machineries. This disrupts insertion of LPS and proteins into the outer membrane, and the accumulated LPS and outer membrane proteins trigger sigma E activation. Down-regulation of lpp mRNA translation increases synthesis of these and other outer membrane components to restore outer membrane homeostasis. Upon stress and during transition to stationary growth phase, sigma E upregulates MicL, which down-regulates lpp mRNA translation. Inhibition of lpp mRNA translation is more effective than inhibition of lpp transcription because the amount of lpp mRNA is high (10% of all cellular mRNA) (Guo et al. 2014) and the mRNA has an unusually long half-life (Hirashima and Inouye 1973; Nilsson et al. 1984).
E. coli controls extracytoplasmic stress using several systems that respond to aberrant structural and functional alterations in the cell envelope (Grabowicz and Silhavy 2017a, b). The RcsF lipoprotein functions as a sensor that recognizes outer membrane defects and transmits the information into the periplasm to activate the stress response. The N-terminal lipid of RcsF in the outer membrane is exposed to the cell surface, and the carboxyl group is in the periplasm, where it interacts with the major outer membrane proteins OmpA, OmpC, and OmpF (Konovalova et al. 2014, 2016). Inhibition of the Lol pathway activates the Rcs phosphorelay system, which results in enhanced transcription of the lolA gene (Tao et al. 2012). Mislocalized lipoproteins in the cytoplasmic membrane caused by Lol defects or mutations in RcsF activate the RcsF stress response.
The Cpx regulon consists of a two-component system that responds to signaling that originates, among others, from the NlpE outer membrane lipoprotein. When lipoprotein trafficking is impaired, CpxA induces a protective response through NlpE that improves cell viability. CpxA is a histidine kinase that spans the cytoplasmic membrane and activates CpxR, which up-regulates a number of genes for periplasmic chaperons and proteases. Cpx is necessary to alleviate stress caused by lipoprotein trafficking defects.
Function of Lpp in E. coli
An lpp deletion mutant was fortuitously isolated during construction of an F′ plasmid (Hirota et al. 1977). The mutant grows and divides normally and remains susceptible to phages. It is hypersensitive to EDTA, cationic dyes, and detergents. Enzymes leak out of the periplasm, and outer membrane vesicles (blebs) are formed and released into the medium, which suggests that the integrity of the outer membrane is disturbed. This phenomenon was originally observed when Lpp was released from murein by treating cell envelopes with trypsin, whereby the outer membrane detaches from the cytoplasmic membrane (Braun and Rehn 1969). A phenotype similar to that of the lpp deletion mutant is observed for mutants lacking the C-terminal lysine or that are devoid of the three l,d-transpeptidases that link Lpp to murein (Asmar et al. 2017).
Lpp plays an important structural role in that it fixes the outer membrane to murein, which also affects interaction of the cytoplasmic membrane with murein and the outer membrane. Recently, it was found that murein-bound Lpp determines the distance between the outer membrane and the cytoplasmic membrane (Asmar et al. 2017). Insertion of 14 or 21 amino acids into Lpp increases the distance between the outer membrane and cytoplasmic membrane by 3 nm and 4 nm, respectively, as revealed by electron cryomicroscopy. Response to stress elicited by β-lactams depends on the correct distance between the outer membrane and the cytoplasmic membrane, which is determined by the correct (wild-type) length of Lpp. The correct distance is required for the stress-responsive RcsF protein in the outer membrane to contact the stress-transferring IgaA protein in the cytoplasmic membrane and to confer antibiotic resistance. If signaling is interrupted, the antibiotics A22 and mecillinam cause cell lysis.
The length of the flagellum rod (25 nm) is determined by the width of the space between the outer membrane and the cytoplasmic membrane, as shown in studies of Salmonellla typhimurium. The length of the flagellum rod of non-motile flagellin flgG mutants is higher (up to 60 nm). Selection for motile suppressor mutants of S. typhimurium unexpectedly identified deletions in lppA. In five flgG mutants tested, deletion of lppA resulted in a significant increase in swim diameter on soft agar plates. An flgG wild-type lppA deletion mutant is impaired in swimming through liquid (individual cell behavior) and in swarming motility (moving of a swarm of flagellated bacteria across a hydrated surface). Further investigations revealed that the length of LppA determines the width of the periplasm; the width decreased in LppA mutants lacking 21 amino acids and increased in LppA mutants having an additional 21 amino acids (Cohen et al. 2017). Purified flagellum rods of the LppA mutant lacking 21 amino acids were shorter than those of the LppA wild-type. All LppA length mutants (+21, +42, +63, and −21) could not swarm, but three of the mutants (+21, +42, and +63) could swim. The growing flagellum senses when it hits the outer membrane and stops growing. Without LppA, the space between murein and the outer membrane is less constricted and allows FlgG distal rods to be positioned perpendicular to the outer membrane as they grow longer than 25 nm, which permits formation of the periplasmic flagellar ring-lipopolysaccharide complex.
An lpp deletion mutant of uropathogenic E. coli CFT073 is impaired in its ability to produce capsular polysaccharides. In this lpp mutant, the amount of KpsD—the putative outer membrane translocon for type 2 capsular polysaccharide export—is reduced. Shortage of KpsD decreases polysaccharide capsule formation and as a consequence serum resistance in vitro and complement-mediated clearance in vivo (Diao et al. 2017). Binding of Lpp to murein is important as indicated by the similar phenotypes of both an lpp mutant lacking the C-terminal lysine and the lpp deletion mutant. Detailed studies of these specific systems revealed the important function of Lpp, which was less obvious in earlier studies of lpp deletion mutants that multiplied normally but exhibited outer membrane defects. The findings also indicate the importance of a correct cell envelope architecture, specifically a precise distance between the outer membrane and the cytoplasmic membrane. This distance is determined by the abundant Lpp which acts as a spacer.
Rojas et al. (2018) recently showed that the outer membrane of Gram-negative bacteria is an essential load-bearing element. It, along with murein, contributes to the mechanical properties of the cell envelope. An lpp deletion mutant strongly contracts upon plasmolysis and lysis; the authors concluded that the coupling between murein and the outer membrane was diminished.
Lipoprotein Prediction Algorithms
Most algorithms for predicting Lpp in prokaryotes were published before 2010. T2003hey were based mainly on the increasing number of data sets obtained with experimentally proven E. coli lipoproteins (Juncker et al. ; Gonnet et al. 2004; Babu et al. 2006). An extended list with 90 experimentally proven lipoproteins of E. coli was published (Tokuda et al. 2007). Unfortunately, the proven negative results were not recorded. The website DOLOP (Babu et al. 2006) is dedicated to lipoproteins; its prediction program attempts to integrate the advances of its predecessors. However, the site has not been modified since 2005. A recent unpublished analysis (September 7, 2018) based on the prediction algorithm of Juncker et al. (2003) examined 126,697,114 gene sequences, of which 3,472,106 genes (2.7%) encode proteins with a lipobox (Jens Baßler, Max Planck Institute for Developmental Biology).
To date, most predicted lipoproteins have not been experimentally proven and a certain number of false positives is obtained. Determination of a lipid anchor is not routine in proteome analyses of cell envelopes, and special analyses are required to identify the lipid anchor and the fatty acid composition (Haake and Zückert 2017; Asanuma et al. 2011). An old but still efficient method is to use radioactively labeled precursors, e.g., glycerol or palmitate. The lipoproteins are often enriched by Triton X-114 partitioning before protease digestion and mass spectroscopy analysis (Asanuma et al. 2011). Even if some internal peptides of a predicted lipoprotein are found in a proteome analysis, this only indicates synthesis of the protein and not its lipidation, which must still be shown. Therefore, even for the well-studied E. coli, proof for the lipid modification of every predicted lipoprotein has not been obtained. In addition, the expression of 96 true and predicted lipoproteins was tested under three different growth conditions, aerobic and anaerobic ± nitrate. The mRNA of 21 lipoproteins was not expressed under any condition (Brokx et al. 2004). This indicates that many of these unknown predicted lipoproteins may have no discernible function for the strain.
In Gram-positive bacteria, many substrate-binding proteins are anchored by a lipid to the outer leaflet of the cytoplasmic membrane. This type of modification has also been predicted and found for the binding protein MetQ of a methionine uptake system in E. coli (Tokuda et al. 2007). However, when metQ was recently cloned and expressed in E. coli, the crystallized protein isolated for X-ray structure determination did not contain the lipid anchor; the signal peptide was split off after Gly at position 16 (the Cys at position 23 was not lipidated) (Nguyen et al. 2015b). Unfortunately, the authors did not discuss this discrepancy to the lipidation observed by Tokuda et al. (2007). It is possible that only part of the protein is lipidated under certain conditions. Such a case has been described by Hayashi et al. (1988) for FtsI, the penicillin-binding protein 3 (PBP3). They found that less than 15% of plasmid-encoded overproduced FtsI was lipidated. Lipidation of FtsI in wild-type E. coli K-12 was below the detection limit (Hayashi et al. 1988) and further studies excluded that the observed lipidation is important in vivo. In addition, no N-terminal processing was observed.
PRED-LIPO is a special Lpp prediction program for Gram-positive bacteria (Bagos et al. 2008) that was trained with a set of lipoproteins from Gram-positive bacteria. Interestingly, Oenococcus oeni is listed there as a bacterium without a lipoprotein. A blast search for Lgt and LspA homologues in the family Leuconostocaceae (which includes species of Leuconostoc, Oenococcus, and Weissella) revealed that these enzymes seem to be missing in this group of bacteria. This supports the prediction that these bacteria do not have lipoproteins in contrast to other lactic acid bacteria. How this evolved is an interesting but unsolved question. Another interesting observance is that mitochondria and plastids have lost this type of protein lipidation during evolution, which makes this structure a specific bacterial recognition signal.
Functions of E. coli Lipoproteins Other Than Murein Lipoprotein Lpp
The activity of lipoproteins is as multifaceted as the multitude of tasks the cell envelope has to fulfill for the cell. In nearly every task of the envelope of E. coli, a lipoprotein is involved (Table 3.1). Of the 116 lipoproteins listed, 38 have an unknown function or evidence of their function is inconclusive. In the following, we will discuss some of the lipoproteins with interesting functions.
In Gram-negative bacteria, lipoproteins are located either in the outer leaflet of the cytoplasmic membrane or in the inner leaflet of the outer membrane and exposed to the cell surface. The sorting processes are catalyzed by specific chaperones, e.g., LolA and LolB. LolB is an essential protein. Being itself a lipoprotein, it helps to insert other lipoproteins into the outer membrane (Szewczyk and Collet 2016).
The Bam proteins are another essential protein complex necessary for the assembly of the outer membrane. The β-barrel protein BamA and the associated proteins BamB, BamC, BamD, and BamE integrate β-barrel proteins into the outer membrane. All Bam proteins except BamA are anchored by a lipid on the periplasmic site of the outer membrane (Konovalova et al. 2017; Botos et al. 2017).
The 14 lipoproteins AmiC, Lpp, MepS, MltABCDE, NlpCDI,YgeR, YraP, LppA participate in the handling of murein. Remarkable is NlpD, which is located in the outer membrane at the divisome and activates the amidase AmiC in the process of cell division.
LPS is exported to the outer leaflet of the outer membrane, while phospholipids are mainly found in the inner leaflet of the outer membrane. This asymmetry of lipid distribution in the outer membrane is critical for resistance to detergents in the presence of EDTA (Malinverni and Silhavy 2009). LPS is integrated into the outer membrane by LptD and the lipoprotein LptE. The asymmetry of the phospholipid distribution is accomplished by the lipoprotein MlaA in collaboration with outer membrane β-barrel proteins OmpC or OmpF, MlaC in the periplasm, and the ABC transporter MlaFEDB in the inner membrane. Additional factors, such as the Tol-Pal system (Pal is an outer membrane lipoprotein) (Shrivastava et al. 2017) and the phospholipase PldA (May and Silhavy 2018) also play a role. The molecular mechanisms are mostly not understood.
RcsF is one of the lipoproteins whose lipid is inserted in the outer leaflet of the outer membrane; the transmembrane part is threaded through the pore of outer membrane β-barrel proteins with the C-terminus in the periplasm. It serves as a sensor for envelope perturbations and interacts with the periplasmic domain of RcsC, a histidine kinase, and the Rcs system regulator YrfF in the inner membrane. Capsular polysaccharide synthesis, biofilm formation, and other cell surface properties depend on the Rcs regulatory system (Sato et al. 2017).
YcaL is a predicted lipoprotein with protease activity against misfolded outer membrane proteins. It possibly “cleans” the Bam complex of unsuitable substrates at a certain stage, thereby helping to avoid clogging of the Bam pathway (Soltes et al. 2017).
The Iss lipoproteins residing in the outer membrane confer serum resistance to E. coli and are encoded by ColV/B plasmids. Related proteins, called Bor, are encoded by lambdoid phages, e.g., on the defective DLP12 prophage in E. coli K-12 (Johnson et al. 2008).
The TraT lipoproteins encoded on the F plasmid and related plasmids are surface-exposed proteins responsible for surface exclusion, which prevents futile matings between bacteria with similar or identical plasmids. In addition, they might also contribute to serum resistance (Arutyunov and Frost 2013).
The group A colicins are released by concomitantly synthesized lysis lipoproteins. Some mature slowly, but all seem to be exported to the outer membrane by the Lol system. It has been shown that the release from the cell of a group B colicin lacking a specific lysis protein is stimulated by the concomitantly induced lysis protein of a lambdoid phage (Nedialkova et al. 2016). Possibly this type of colicin is dependent on the complementation by a phage lysis protein or by another colicin lysis protein.
Lipoproteins in Other Gram-Negative Bacteria
Of the great variety of lipoproteins (2.7% of all predicted proteins), only some examples can be presented in this review.
Borrelia Spp.
Lipoproteins of pathogenic bacteria have attracted special interest because they stimulate the immune system of vertebrates. For example, various species of Borrelia, the causative agents of Lyme disease, alter their surface proteins when they switch hosts from ticks to vertebrates. In the mammalian host, the lipoproteins OspA and OspB are repressed by BosR, a copper-dependent Fur family regulator. In this connection, it is interesting to note that ticks have copper-chelating antimicrobial peptides to control their bacterial inhabitants, which might generate surroundings with low copper availability for Borrelia spp. By contrast, the synthesis of the Borrelia spp. lipoproteins OspC and VlsE in the mammalian host are enhanced by pH, temperature, and short-chain fatty acids. In Borrelia burgdorferi, the lipoprotein VlsE (variable major protein-like sequence) becomes a dominating surface protein (Bankhead 2016) that is an important immune evasion factor of the spirochete. Through antigenic variation, the bacteria escape the immune defense of their host, which may lead to persistence and chronic infection. Multiple variants of VlsE are constantly produced, and it is assumed that VlsE shields dominant epitopes of other outer membrane proteins so that they cannot be recognized by protective antibodies (Batool et al. 2018).
In addition to the lipoproteins that have been studied intensely as vaccine candidates (see later), B. burgdorferi has a lipoproteome of at least 125 proteins; 86 are surface exposed, and only 8 seem to reside in the inner leaflet of the outer membrane and 31 in the cytoplasmic membrane (Dowdell et al. 2017).
B. burgdorferi contains several episomal prophages of the cp32 family. These plasmids contain different replication/segregation loci that allow stable maintenance of several episomes in one cell. The erp locus on these plasmids encodes surface-exposed lipoproteins with a conserved N-terminal peptide and very variable protein sequences attached (Brisson et al. 2013). The different erp loci also contribute to the variability in the appearance of B. burgdorferi.
Species of the Phylum Bacteroidetes
Bacteroidetes have many surface-exposed lipoproteins that are mainly engaged in the utilization of polysaccharides. One of the first systems studied in detail was the starch utilization system (Sus). Lipoproteins on the cell surface bind the polysaccharide, and a lipoprotein glycosidase sets oligosaccharides free. The oligosaccharides are transported by a TonB-dependent protein into the periplasm, where glycoside hydrolases split the oligosaccharides to disaccharides and monosaccharides. These are taken up by dedicated transport systems in the cytoplasmic membrane (Foley et al. 2016).
Genome sequences revealed that certain Bacteroides species have about 100 Sus-like polysaccharide utilization systems. An X-ray structure of a SusD-like substrate-binding lipoprotein in complex with the corresponding SusC-like TonB-dependent transporter has been solved. SusC is capped by the lipoprotein SusD, and the substrate is bound in the interface between the two proteins (Glenwright et al. 2017).
Results of a bioinformatic analysis led Lauber et al. (2016) to postulate a new lipoprotein export system that determines the cell-surface localization of lipoproteins in species of the phylum Bacteroidetes. The N-terminal consensus sequence CXK(DD or EE) was found in surface-exposed Sus-like lipoproteins of Capnocytophaga canimorsus. These amino acids allowed export of the periplasmic lipoprotein sialidase to the cell surface. Although the transporter-associated Sus-like proteins have a certain resemblance to neisserial outer membrane lipoproteins, such as TbpB and HupA, the lipoproteins seem to be exported by different means. In species of Neisseria , Slam proteins support this process (see below). Species of the phylum Bacteroidetes lack Slam-like proteins, and the system that recognizes the above-mentioned consensus sequence remains to be identified.
Neisseria Spp.
The first better-known examples of surface-exposed lipoproteins were found in Neisseria spp. in complex with TonB-dependent receptors, where they help in binding host iron-containing molecules (TbpB, transferring binding protein; LbpB, lactoferrin binding protein) and facilitate the uptake of iron. In a screening for Neisseria meningitidis mutants that do not expose the lipoprotein TbpB on the cell surface, a mutation was identified that also prevented the surface exposure of LbpB and fHbp (factor H binding protein). The encoded protein was named Slam (surface lipoprotein assembly modulator (Hooda et al. 2016).
Slam homologues were found in pathogenic species of, e.g., Moraxella, Acinetobacter, Haemophilus, Vibrio, and other proteobacteria but not in E. coli. Heterologous expression of the gene encoding Slam in E. coli results in exposure of TbpB, LbpB, and fHbp on the cell surface. Interestingly, Slam proteins have substrate specificity. For example, another Slam protein in N. meningitidis causes exposure of only HpuA (hemoglobin-haptoglobin binding protein) on the cell surface, but not of TbpB, LbpB, or fHbp (Hooda et al. 2016). The sequence identity between the two Slam proteins of N. meningitidis (Slam1 and Slam2) is only 26% (Hooda et al. 2017).
The function of Slam was challenged by (Fantappie et al. 2017), who found fHbp exposed on the surface of the outer membrane of E. coli independent of Slam. However, with Slam present, the amount of surface-exposed fHbp clearly increased, which indicates that Slam is at least a facilitator of surface exposure in E. coli. The authors proposed that certain uncharacterized structure determinants allow the Slam-independent surface exposure of these proteins.
Gram-Positive Bacteria and Archaea
Low-GC Gram-Positive Bacteria
Homologues of the apolipoprotein N-acyltransferase Lnt have not been found in low-GC Gram-positive bacteria. Therefore, it was generally assumed that these bacteria have lipoproteins with only two ester-bound fatty acids on the S-glycerolcysteine. However, Toll-like-receptor (TLR)-dependent stimulation of macrophages provided the first hint that Staphylococcus aureus might indeed have triacylated lipoproteins. Analysis of MntC (SitC), a binding protein of a manganese ABC transporter, revealed N-acylation of MntC in S. aureus. This was also shown for other S. aureus lipoproteins (Kurokawa et al. 2009). However, lipoproteins of S. aureus cells in the stationary phase grown under certain stress conditions contain only two ester-bound fatty acids and no N-acylation (Kurokawa et al. 2012b). Another important difference in lipidation is found between commensal S. aureus and non-commensal Staphylococcus carnosus. The lipoproteins of S. carnosus are N-acetylated and lead to a tenfold stronger immune response (Nguyen et al. 2017). It is assumed that the low immune response of the S. aureus lipoprotein is an evolutionary adaptation to the commensal lifestyle.
These results stimulated Kurokawa et al. (2012b) to analyze the lipid structure in other low-GC Gram-positive bacteria. As expected, they found lipoprotein with only two ester-bound fatty acids in Listeria monocytogenes, but to their surprise, three other lipoprotein modifications were detected in other bacteria. In Enterococcus faecalis and several other species (e.g., Bacillus cereus, Lactobacillus bulgaricus, Streptococcus sanguinis), a lysoform, i.e., an N-acylated lipoprotein with one fatty acid esterified to the glycerol residue, was detected. However, in several species of Bacillus (B. subtilis, B. licheniformis, B. halodurans), two fatty acids were bound to glycerol and the amino group was acetylated. The structure of some lipoproteins of Mycoplasma fermentans is unusual in that the signal peptidase cleaves at two residues before the S-diacyl-glyceryl-cysteine. However, in two other lipoproteins of M. fermentans, the S-diacyl-glycerylcysteine is the N-terminal amino acid (Kurokawa et al. 2012b). The various structures found are critically reviewed in detail in Nakayama et al. (2012).
The genesis of the unusual lyso-lipoprotein structures was explained 5 years later. In an attempt to clone the gene responsible for the unknown Lnt activity that leads to N-acylation in E. faecalis, Armbruster and Meredith (2018) devised an lnt complementation screening method in E. coli. The enzyme that they identified intramolecularly transacylated an ester-bound fatty acid to the α-amino group of the cysteine. The identified protein was called lipoprotein intramolecular transacylase (Lit) (Armbruster and Meredith 2017). Interestingly, homologues of this protein are found in species of Bacillus and Lactobacillus, but not in staphylococci. Therefore, the enzyme that acylates the amino group in S. aureus still remains to be identified.
High-GC Gram-Positive Bacteria
In contrast to many low-GC Gram-positive bacteria, species of the high-GC Gram-positive genera Mycobacteria and Streptomyces encode Lnt homologues and have lipoproteins with three fatty acids (Tschumi et al. 2009; Brulle et al. 2013). Streptomyces coelicolor encodes about 200 predicted lipoproteins; the majority are solute-binding proteins of transport systems, as generally found in Gram-positive bacteria. About a quarter of the 200 lipoproteins are not exported by the Sec system, but instead via the twin-arginine translocation (Tat) system. Another peculiarity is that two Lgt homologues are encoded, both of which are active. In addition, all species of Streptomyces studied contain two Lnt homologues. Both homologues in the potato pathogen Streptomyces scabies can be deleted without yielding a growth, differentiation, or virulence phenotype (Thompson et al. 2010).
Mycobacteria
Several lipoproteins of Mycobacterium tuberculosis have been studied as potential virulence factors and vaccine candidates (extensively reviewed and discussed in (Becker and Sander 2016). As in other pathogens, not all lipoproteins stimulate the immune response. Some even have immune-suppressing properties and help M. tuberculosis survive in the host. These latter lipoproteins are not suitable for a vaccine because they exacerbate the disease, as shown in challenged, previously vaccinated mice.
Of the 100 predicted lipoproteins, we will mention only a few well-studied cases. The LppX protein assists in localization of a group of complex lipids—the phthiocerol dimycocerosates—in the outer lipid-rich membrane of M. tuberculosis (Sulzenbacher et al. 2006). The crystal structure shows structural similarities to that of LolA and LolB, which also transport a lipidic substrate. Without LppX, phthiocerol dimycocerosates are not released from the cell surface into the Triton-X100-containing medium, which indicates that the lipid has not been properly inserted into the outer membrane. The correct positioning of phthiocerol dimycocerosates seems to be important, because this lipid helps in hiding pathogen-associated molecular patterns from the innate immune system of the host (Cambier et al. 2014).
The lipoprotein LprG similarly helps in positioning lipoarabinomannan on the cell surface, which inhibits phagosome-lysosome fusion, an important component in the survival strategy of M. tuberculosis (Shukla et al. 2014). LprG also has immune-suppressing activity.
The lipoprotein RpfB is necessary for resuscitation, which is an important pathogenicity factor of M. tuberculosis. The enzyme has a lysozyme fold and lytic transglycosylase activity. In a joint action of RpfB with an endopeptidase anhydro-muramic acid containing muropeptides were set free from cell walls. The muropeptides stimulated resuscitation of dormant mycobacteria (Nikitushkin et al. 2015).
LpqH and LprN are lipoproteins with immune-suppressing functions. They contribute to the immune escape and late immune response of the host, which allows propagation of the infecting bacteria to high numbers (Becker and Sander 2016). Mass spectrometry of LpqH revealed that it contains, in addition to the N-terminal N-acyl-S-diacylglycerolcysteine, up to nine hexoses on the N-terminal peptide (Parra et al. 2017). Of the 41 proteins enriched from the growth medium with ConA-lectin, 34 were lipoproteins (Gonzalez-Zamorano et al. 2009). The glycosylation of lipoproteins influences envelope permeability and consequently antibiotic resistance in addition to the immunogenic properties (van Els et al. 2014).
Archaea
The first hint that archaea synthesize lipoproteins was obtained in 1994 by the analysis of halocyanin from Natronobacterium pharaonis of the phylum Euryarchaeota. The protein had an appropriate molecular mass and contained a lipobox and a blocked N-terminal end, which are good indicators of a lipid modification (Mattar et al. 1994).
Of the archaea, only members of the phylum Euryarchaeota seem to make lipoproteins (Storf et al. 2010). Additional experimental evidence comes mainly from the analysis of lipoproteins of Haloferax volcanii that are exported by the Tat system. The presence of lipoproteins was unexpected because Lgt, Lsp, and Lnt homologues have not been detected in archaeal genomes. However, an appropriate lipobox with cysteine was predicted for about 50% of the predicted Tat substrates and for a few Sec substrates in members of the class Haloarchaea (Storf et al. 2010). A cysteine-to-serine mutation in a maltose-binding protein or an iron-binding protein resulted in Tat-dependent secretion into the medium, which is good evidence for cysteine-dependent lipidation. In addition, globomycin inhibits both growth of the cells and maturation of the maltose-binding protein (Gimenez et al. 2007). However, an in-depth structural analysis of the lipid part is still lacking.
Immunological Properties of E. coli Lpp
Lpp as an Antigen
E. coli Lpp spontaneously coats erythrocytes, which are then lysed upon addition of complement (Bosch and Braun 1973). This method was used to determine the distribution of Lpp between the outer membrane and the cytoplasmic membrane. More than 10- to 16-fold Lpp was found in the outer membrane than in the cytoplasmic membrane. When entire cells are used as antigens, anti-Lpp titers are higher when the cells are of rough mutants lacking O-antigen and parts of the LPS core region, possibly because Lpp might be better exposed when portions of LPS are lacking. Antibodies raised against Lpp released from murein by lysozyme recognize the C-terminus with the attached murein subunits. Lpp released from murein with trypsin [trypsin-Lpp; trypsin cleaves the Lys(55)-Tyr(56) and Arg(57)-Lys(58) bonds] does not react with antiserum raised against Lpp released by lysozyme. Anti-Lpp raised against trypsin-Lpp only reacts with trypsin-Lpp. The antiserum specifically recognizes the C-terminal Lys. Removal of Lys with carboxypeptidase B inactivates the antigenicity. It is remarkable that antisera raised against lysozyme-Lpp and antisera raised against trypsin-Lpp contain no antibodies against the polypeptide chain. By contrast, antisera raised against cells react with both lysozyme-Lpp and trypsin-Lpp. In this case, antibody formation is elicited by the polypeptide integrated into the membrane. Release of the ester-linked fatty acids on glycerylcysteine does not affect antigenicity of isolated Lpp. LPS as a control does not interfere with the Lpp immunogenicity and antigenicity determinations (Braun et al. 1976a).
Lpp: A B-Lymphocyte Mitogen
Mitogens serve as tools to understand processes during proliferation and differentiation of B-cells into plasma cells. Mitogenic stimulation can be monitored by an increase in thymidine uptake, the appearance of plaque-forming cells assayed with densely coupled trinitrophenylated sheep red blood cells, and increase in synthesis and secretion of radiolabeled IgM compared to synthesis and secretion of other cellular proteins. These methods show that splenic lymphocytes of mice are activated by lipoprotein released from murein by lysozyme or trypsin. Removal of the ester-linked fatty acids by alkaline hydrolysis abolishes the mitogenic activity. Anti-immunoglobulin antibodies inhibit the mitogenic stimulation of B-cells by lipoprotein. Activation is T-cell independent and does not need serum factors or adherent cells. Small resting B-cells display the highest lipoprotein sensitivity (Melchers et al. 1975).
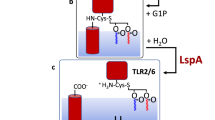
Lpp and Lpp-Derived Lipopeptides Act as Strong Adjuvants in the Innate Immune Response
The mitogenic activity of Lpp prompted further studies on the immunogenic activities of Lpp and the immunogenic domains of Lpp. Various lipopeptides were synthesized and tested for their B lymphocyte mitogenicity in an in vitro cell culture system (Bessler et al. 1985). The lipopeptides consisted of S-[2,3-bis-(palmitoleyloxy)propyl)-N-palmitoleylcysteine (Pam3C) with the following C-terminal extensions: cysteine methylester, cysteine-serine, cysteine-serine-serine, cysteine-serine-serine-asparagine, and cysteine-serine-serine-asparagine-alanine (see structure in Fig. 3.1a). To be fully active, the compounds had to carry at least one serine. The lipopeptide-induced stimulation of B lymphocytes is as strong as stimulation by the complete lipoprotein.
Using synthetic lipid, it has been shown that of the two possible diastereometric configurations (R and S) at position C2 of the propyl moiety, the R configuration is the most likely one in the native molecule (Wiesmüller et al. 1983). A later systematic study showed that lipopeptides with the R configuration activate B-cells more than the S forms. Induction of specific CD8(+) T-cells is significantly higher in mice injected with the R forms than with the S forms (Khan et al. 2009).
Tests of a range of peptides revealed that for full mitogenic activity and polyclonal stimulation, the hydrophilic dipeptide structure is necessary. Further investigations revealed that synthetic lipopeptides constitute potent macrophage activators. Co-administration of lipopeptides with haptens or low-molecular-weight antigens that are not immunogenic per se, or covalent coupling, result in highly immunogenic conjugates. Such conjugates might serve as synthetic vaccines and provide protection by enhancing the antibody-mediated immune response in vivo (Bessler et al. 1997). Many studies have shown that lipopeptides derived from Lpp promote immunological defense, inflammation, or sepsis. Without the use of adjuvant (self-adjuvanting activity) lipopeptides generate potent immune responses (summarized in Asmar and Collet 2018; Becker and Sander 2016; Moyle and Toth 2008; Nguyen and Götz 2016; Steinhagen et al. 2011; Zaman and Toth 2013).
Lipopeptides constitute potent immunoadjuvants in various species in parental immunizations. Lipopeptide adjuvants are comparable or even superior to Freund’s adjuvant without showing side effects. Lipopeptide Pam3CSK4 also enhances serum antibody responses to various proteins when administered in combination with antigens via the nasal route in mice (Baier et al. 2000). Thus, they constitute strong adjuvants for mucosal immunizations.
Lpp and Lpp-Derived Lipopeptides Act as Ligands of Toll-like Receptors TLR2-TLR1 and TLR2-TLR6
Lpp and lipopeptides derived from Lpp are recognized by the innate immune system, which represents the first line of defense against invasive infectious pathogens. They bind to Toll-like receptor TLR2 together with TLR1 and TLR6 (Aliprantis et al. 1999). The degree of acylation at the lipid moiety discriminates the interaction of TLR2 with TLR1 and TLR-6; tri-acylated lipopeptide binds to the heterodimer TLR2/TLR1, and di-acylated lipopeptide interacts with TLR2/TLR6 (Kang et al. 2009). Signals are transduced from the lipopeptide-loaded TLR receptors to nuclear factor NF-kappaB by a cascade of phosphorylations, resulting in transcription and synthesis of proinflammatory cytokines and chemokines. Lipopeptides of various structures are important ligands for activation of the TLR receptors, which are essential for induction of the innate and adaptive immune responses.
TLRs are composed of three domains: an extracellular ligand-binding domain, a single transmembrane helix, and an intracellular signaling domain. The crystal structure of the extracellular domain of human TLR1-TLR2 heterodimer with bound tri-acylated lipopeptide Pam3CSK4 has been determined (Jin et al. 2007). The three lipid chains mediate formation of an M-shaped heterodimer. The two ester-linked lipid chains are inserted in a pocket of TLR2, while the amide-bound lipid chain is inserted in a hydrophobic channel in TLR1. Heterodimer formation of TLR6 with TLR2 is triggered by binding of diacyl-Pam2CSK4. The resulting structure is similar to that of the extracellular domain of TLR1-TLR2-Pam3CSK4 (Kang et al. 2009). The binding site of the amide-linked acyl chain is blocked by two phenylalanines, which explains the preferred binding of diacyl lipids versus triacyl lipids. The structures of Toll-like receptors with various ligands are discussed in Kang and Lee (2011).
Although low-GC Gram-positive bacteria usually contain diacylated lipoprotein, the abundant lipoprotein SidC of the Gram-positive S. aureus carries a triacylated lipid (Kurokawa et al. 2009, 2012a). Lipoprotein lipid structures of commensal and non-commensal staphylococci induce different immune responses (Nguyen et al. 2017). TheTLR2 response induced by commensal S. aureus and Staphylococcus epidermidis is almost ten times lower than that induced by non-commensal S. carnosus. The N-terminus of S. aureus lipoprotein and of S. epidermidis lipoprotein carries a long acyl chain (C17), whereas S. carnosus lipoprotein contains a short acetyl group. The long-chain N-acylated lipoprotein recognized by TLR-2/TLR-1 receptors decreases the innate and adaptive immune responses, which results in immune evasion, whereas the short chain N-acetylated lipoproteins recognized by TLR2/TLR6 receptor boosts IL8 production.
Low-GC Gram-positive bacteria, e.g., Bacillus cereus, Enterococcus faecalis, Streptococcuc sanguinis, and Lactobacillus bulgaricus, have a modified lipoprotein that carries the structure N-acyl-S-monoacyl-glycerylcysteine (lyso-form) (Kurokawa et al. 2012b). The purified lipoprotein induces proinflammatory cytokine production from mice peritoneal macrophages in a TLR2-dependent and TLR1-independent manner. Lyso-lipoprotein from B. cereus induces TLR2-dependent and TLR1- and TLR6-independent tumor necrosis factor (TNF-α) and interleukin-6. By contrast, E. faecalis lyso-lipoprotein induces TLR2/TLR6-dependent and TLR1-independent cytokine secretion, as does N-acetyl-S-diacyl-glyceryl-cysteine lipoprotein from B. subtilis.
S. aureus lipoproteins activate Toll-like receptors when lipoproteins are released from the cytoplasmic membrane by phenol-soluble modulin (PSM) peptides (Hanzelmann et al. 2016). The different capacities of S. aureus clinical isolates in activating TLR2 depends on high-level production of PSM peptides. PSM peptides have a strong impact on the severity of sepsis, and TLR2 plays an important role in systemic S. aureus infections only for strains that produce PSM peptides.
TLR-Independent Lipopeptide Immunogenicity
Although the action of Lpp and other lipopeptides is usually mediated through interaction with TLR2 associated with either TLR6 or TLR1, lipopeptides can be immunogenic without TLR1 and TLR6 interaction. For example, diacylated lipopeptides, e.g., Pam2CSK4, induce TLR6-independent B lymphocyte proliferation and TNF-α secretion in macrophages, as determined with cells from TLR6-deficient mice (Buwitt-Beckmann et al. 2005). Diacylated Pam2CSK4 and triacylated Pam3 CGNNEDESNISFKEK stimulate murine spleen cells isolated from TLR1- and TLR6-deficient mice. Thus, distinct lipopeptides are recognized by TLR2 in a TLR1- and TLR6-independent manner (Buwitt-Beckmann et al. 2006). Even a TLR-independent enhancement of infection by lipopeptides has been observed with various virus infection models. The respiratory syncytial virus (RSV) elicits respiratory tract infections and bronchiolitis, which are associated with bacterial co-infections (Nguyen et al. 2010). Therefore, the authors examined whether bacterial Toll-like receptor agonists influence RSV infections in human primary cells or cell lines. Pam3-CSK4 enhanced RSV infection in primary epithelial, myeloid, and lymphoid cells. The lipopeptide not only enhanced RSV but also HIV-1, measles virus, and human metapneumovirus infection independent of TLR signaling.
Cytotoxic T lymphocytes (CTLs) play an important role in the immune response to viral infections. CTLs recognize peptides derived from viral proteins together with the major histocompatibility complex class I molecules at the surface of infected cells. They usually require in vivo priming with infectious virus. Synthetic viral peptides covalently linked to tripalmitoyl-S-glycerylcysteinyl-Ser-Ser (Pam3CSS) efficiently prime influenza-virus-specific cytotoxic T lymphocytes in vivo to a level as high as infectious virus. Lipopeptide priming is MHC class I-restricted (Deres et al. 1989).
Suppression of Immune Response by Lipopeptides
Lipopeptides not only enhance the immune response but can also suppress immune reactions. For example, skin exposed to synthetic diacylated lipopeptides suppresses the immune response by interleukin-6-dependent induction of granulocytic and monocytic myeloid-derived suppressor cells. Triacylated lipopeptides are inactive in this assay (Skabytska et al. 2014). As discussed before, the long-chain N-acylated lipoprotein recognized by TLR2/TLR1 receptors of S. aureus decreases innate and adaptive immune responses, resulting in immune evasion. Of the many surface-exposed lipoproteins of Mycobacterium tuberculosis (Becker and Sander 2016), LpqH promotes immune evasion by inhibiting MHC class II expression and interference with cytokine production in macrophages. LprG is immunosuppressive in that it inhibits TLR2-mediated MHC II antigen presentation.
Virulence Enhancement by Lpp
Salmonella enterica serovar Typhimurium encodes in tandem two lipoproteins of the murein lipoprotein type (Lpp). All mice orally infected with an lppA lppB double mutant survive. The mutant triggers robust innate and adaptive immune responses (Erova et al. 2016). Mice immunized with the mutant are completely protected against a lethal oral challenge dose of wild-type S. typhimurium. Since the mutant is among the most attenuated and immunogenic strains, it is considered an excellent candidate for a vaccine against S. typhimurium infection. Also single lpp mutants of S. enterica are highly attenuated in in vitro and in vivo models of pathogenesis (Fadl et al. 2005). The virulence potential of a double lpp knockout mutant was examined in cell cultures of intestinal epithelial cells, a macrophage cell line, and mice infected intraperitoneally (Sha et al. 2004). The mutant is defective in invading and inducing cytotoxic effects. Induction of proinflammatory cytokines tumor necrosis factor-α and interleukin 8 is significantly reduced. Intracellular survival and replication of Salmonella sp. is not affected. Similar results have been obtained with Yersinia pestis. Deletion of lpp reduces the virulence of Y. pestis lacking plasmid pPCP1 in a mouse model of pneumonic plague. pPCP1 encodes virulence-associated traits, such as the plasminogen-activating protease. Mice infected with an lpp deletion mutant survive longer than mice infected with an lpp wild-type strain (Agar et al. 2009). In addition, the levels of most cytokines and chemokines strongly decrease. Transfer of sera and splenocytes from mice immunized with the lpp mutant to naive animals provides protection against a lethal dose of the Y. pestis parent strain (Liu et al. 2010). Deletion of lpp provides a strong attenuation in a mouse model of pneumonic plague.
Species of Staphylococcus encode between 55 and 70 lipoproteins arranged in tandem. Deletion of the entire cluster results in a decreased TLR2-dependent stimulation of proinflammatory cytokines in human monocytes, macrophages, and keratinocytes. The lipoprotein cluster contributes to invasion of S. aureus into human keratinocytes and mouse skin; transfer of the gene cluster into S. carnosus, which is devoid of the cluster, renders S. carnosus invasive (Nguyen et al. 2015a).
All lipoproteins can be mutated at once by inactivation of the lgt gene, which attaches the diacylglyceryl group to the invariant cysteine (Stoll et al. 2005). The mutants show a decreased induction of the proinflammatory cytokines IL6 and IL8 and monocytic chemoattractant protein in human monocytic, epithelial, and endothelium cells compared to that of the wild-type. They are also affected in induction of early cytokines via TLR2 in murine peritoneal macrophages and are severely affected in pathogenicity. However, one has to bear in mind that these deletion mutants are sick even when the lgt mutants contain the lipid-free proteins still attached to the cytoplasmic membrane. Additional changes in the composition of the cell envelope can be predicted, and these might contribute to the immunogenic properties of the mutants. The same conclusion applies to lspA mutants of Mycobacterium tuberculosis, which do not release the signal peptides from the predicted 99 prolipoproteins. The mutants show a decreased intracellular multiplication in mouse macrophages and a decreased growth in lungs and spleens (Sander et al. 2004).
Mycoplasma synthesize a number of lipoproteins. The lipopeptide MALP-2 of Mycoplasma fermentans displays an extremely high immunologic activity. It stimulates macrophages at concentrations as low as 0.2 ng/ml, whereas lipoproteins of other bacterial sources require concentrations of 1 µg/ml for a half-maximal response. It consists of an N-terminal diacylglycerol cysteine linked to the amino acid sequence GNNDESNISFKEK (Mühlradt et al. 1997). Synthetic lipopeptide displays the same activity as lipopeptide isolated from M. fermentans. It stimulates murine macrophages to release TNF, IL-1, IL6, prostaglandins, and nitric oxide from IFN-primed murine macrophages. In addition, MALP-2 down-regulates class II MHC expression on macrophages, which results in impaired antigen presentation to T-cells.
The examples given above demonstrate that natural and synthetic lipopeptides are potent macrophage and B cell activators. Synthetic conjugates consisting of lipopeptides with T helper cell and CTL epitopes from viral and bacterial proteins are efficient low-molecular-weight vaccines with strong adjuvant activity.
Lpp-Derived Peptides as Adjuvants in Vaccines
A vaccine that induces a long-lasting high protection against foot-and-mouse disease and serotype-specific virus-neutralizing antibodies in guinea pigs after a single administration contains the lipopeptide Pam3CSS N-terminally bound to VPI and a synthetic lipopeptide of 20 residues derived from the FMDV protein, which serves as amphiphilic α-helical T-cell epitope (Wiesmüller et al. 1989). A vaccine containing Pam3C linked to the outer surface protein A (OspA) of Borrelia burgdorferi, the causative agent of Lyme disease, has been clinically tested in over 20,000 volunteers (Steinhagen et al. 2011). The induction of protective immunity correlated with the development of antibodies against an epitope on the C-terminus of OspA. Three doses of vaccine induced protective immunity in over 70% of subjects. It was licensed by the FDA in 1998, but withdrawn three years later by the manufacturer because the media reported possible autoimmune side effects. However, neither the FDA nor the CDC found a connection between the vaccine and the development of autoimmunity (Nigrovic and Thompson 2007).
Pam3C has been used as adjuvant in a vaccine against malaria. The vaccine contains multiple B cell epitopes and a universal T cell epitope derived from the Plasmodium falciparum circumsporozoite protein CSP. After three immunizations, all ten volunteers developed peptide specific IgG1, IgG3, and IgG4 antibodies, whereas a vaccine lacking P3C induced only IgG1 and IgG3 isotypes (Nardin et al. 2001).
Mutants of Mycobacterium tuberculosis deficient in the surface lipoprotein LpqS are attenuated and are highly protective against a M. tuberculosis challenge in guinea pigs (Sakthi et al. 2016).
A totally synthetic vaccine activates TLR2 of dendritic cells (Jackson et al. 2004). It consists of a single helper T-cell epitope, a target epitope that is either recognized by B cells or CD8+ T cells, and Pam2Cys inserted between the two epitopes to form a branched configuration. The CD8+ epitopes are from influenza virus and Listeria monocytogenes. Each of the vaccines induces either CD8+ cell or antibody-mediated immune responses in mice. The lipidated vaccines but not the non-lipidated vaccines mediate protection against viral or bacterial infections and confer prophylactic and therapeutic anticancer activity. Additional studies have shown that a conserved minimal peptide from the M protein (J14) of group A streptococci linked to Pam2C and a universal T cell epitope administered intranasally to mice protects them from lethal respiratory challenge with group A streptococci and induces J14-specific mucosal immunoglobulins (Batzloff et al. 2006). These results demonstrate anti-disease and transmission-inhibiting activities, which could form the basis for development of an anti-streptococcal vaccine.
Lipopeptides as adjuvants were investigated in vaccines against influenza virus, group A streptococci, hepatitis C virus, leishmaniasis, Listeria monocytogenes, and other disease-causing microorganisms (summarized in Moyle and Toth 2008; Zaman and Toth 2013). Although strong lipopeptide-dependent immune reactions were elicited, no vaccine is in use. The induction of a lasting CD8+ cytotoxic lymphocyte response was presumably too low to elicit a satisfactory immunity.
Weak Adaptive Immune Response to Lipoproteins
The adaptive immune response to lipoproteins, in contrast to the native immune response, has been studied in healthy S. aureus human carriers and non-carriers (Vu et al. 2016). Specific B-cell and T-cell responses were determined. In contrast to expectations derived from the strong innate immune response described above, the titers of antibodies (IgG) binding to lipoproteins were low. Proliferation assays and cytokine profiling data revealed only subtle responses of T-cells. It remains to be determined why in this study the adaptive immune system reacts only weakly to lipoproteins (Kretschmer et al. 2016). Access of the lipoproteins in cells might not be the cause of low antigenicity, as most isolated recombinant S. aureus lipoproteins are also weak antigens. Impaired processing of lipoproteins by antigen-presenting cells might be a reason and opens new avenues of immune research. The results question the use of lipoprotein and its derived peptides as antigens for production of vaccines against S. aureus.
References
Agar SL, Sha J, Baze WB, Erova TE, Foltz SM, Suarez G, Wang S, Chopra AK (2009) Deletion of Braun lipoprotein gene (lpp) and curing of plasmid pPCP1 dramatically alter the virulence of Yersinia pestis CO92 in a mouse model of pneumonic plague. Microbiology 155(Pt 10):3247–3259. https://doi.org/10.1099/mic.0.029124-0
Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, Klimpel GR, Godowski P, Zychlinsky A (1999) Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285(5428):736–739
Armbruster KM, Meredith TC (2017) Identification of the lyso-form N-acyl intramolecular transferase in low-GC firmicutes. J Bacteriol 199(11). https://doi.org/10.1128/jb.00099-17
Armbruster KM, Meredith TC (2018) Enrichment of bacterial lipoproteins and preparation of N-terminal lipopeptides for structural determination by mass spectrometry. J Vis Exp (135). https://doi.org/10.3791/56842
Arutyunov D, Frost LS (2013) F conjugation: back to the beginning. Plasmid 70(1):18–32. https://doi.org/10.1016/j.plasmid.2013.03.010
Asanuma M, Kurokawa K, Ichikawa R, Ryu KH, Chae JH, Dohmae N, Lee BL, Nakayama H (2011) Structural evidence of alpha-aminoacylated lipoproteins of Staphylococcus aureus. FEBS J 278(5):716–728. https://doi.org/10.1111/j.1742-4658.2010.07990.x
Asmar AT, Collet JF (2018) Lpp, the Braun lipoprotein, turns 50-major achievements and remaining issues. FEMS Microbiol Lett 365(18). https://doi.org/10.1093/femsle/fny199
Asmar AT, Ferreira JL, Cohen EJ, Cho SH, Beeby M, Hughes KT, Collet JF (2017) Communication across the bacterial cell envelope depends on the size of the periplasm. PLoS Biol 15(12):e2004303. https://doi.org/10.1371/journal.pbio.2004303
Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, Sankaran K (2006) A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J Bacteriol 188(8):2761–2773. https://doi.org/10.1128/JB.188.8.2761-2773.2006
Bagos PG, Tsirigos KD, Liakopoulos TD, Hamodrakas SJ (2008) Prediction of lipoprotein signal peptides in gram-positive bacteria with a Hidden Markov model. J Proteome Res 7(12):5082–5093. https://doi.org/10.1021/pr800162c
Baier W, Masihi N, Huber M, Hoffmann P, Bessler WG (2000) Lipopeptides as immunoadjuvants and immunostimulants in mucosal immunization. Immunobiol 201(3-4):391–405
Bankhead T (2016) Role of the VlsE lipoprotein in immune avoidance by the Lyme disease spirochete Borrelia burgdorferi. For Immunopathol Dis Therap 7(3–4):191–204. https://doi.org/10.1615/ForumImmunDisTher.2017019625
Batool M, Caoili SEC, Dangott LJ, Gerasimov E, Ionov Y, Piontkivska H, Zelikovsky A, Waghela SD, Rogovskyy AS (2018) Identification of surface epitopes associated with protection against highly immune-evasive VlsE-expressing Lyme disease spirochetes. Infect Immun 86(8). https://doi.org/10.1128/iai.00182-18
Batzloff MR, Hartas J, Zeng W, Jackson DC, Good MF (2006) Intranasal vaccination with a lipopeptide containing a conformationally constrained conserved minimal peptide, a universal T cell epitope, and a self-adjuvanting lipid protects mice from group A streptococcus challenge and reduces throat colonization. J Infect Dis 194(3):325–330. https://doi.org/10.1086/505146
Becker K, Sander P (2016) Mycobacterium tuberculosis lipoproteins in virulence and immunity—fighting with a double-edged sword. FEBS Lett 590(21):3800–3819. https://doi.org/10.1002/1873-3468.12273
Bessler WG, Cox M, Lex A, Suhr B, Wiesmüller KH, Jung G (1985) Synthetic lipopeptide analogs of bacterial lipoprotein are potent polyclonal activators for murine B lymphocytes. J Immunol 135(3):1900–1905
Bessler WG, Heinevetter L, Wiesmüller KH, Jung G, Baier W, Huber M, Lorenz AR, Esche UV, Mittenbühler K, Hoffmann P (1997) Bacterial cell wall components as immunomodulators–I. Lipopeptides as adjuvants for parenteral and oral immunization. Int J Immunopharmacol 19(9–10):547–550
Bosch V, Braun V (1973) Distribution of murein-lipoprotein between the cytoplasmic and outer membrane of Escherichia coli. FEBS Lett 34(2):307–310
Botos I, Noinaj N, Buchanan SK (2017) Insertion of proteins and lipopolysaccharide into the bacterial outer membrane. Philos Trans R Soc Lond B Biol Sci 372(1726). https://doi.org/10.1098/rstb.2016.0224
Braun V (1975) Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim Biophys Acta 415(3):335–377
Braun V, Bosch V (1972) Repetitive sequences in the murein-lipoprotein of the cell wall of Escherichia coli. Proc Natl Acad Sci USA 69(4):970–974
Braun V, Hantke K (1974) Biochemistry of bacterial cell envelopes. Annu Rev Biochem 43:89–121. https://doi.org/10.1146/annurev.bi.43.070174.000513
Braun V, Rehn K (1969) Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem 10(3):426–438
Braun V, Wu HC (1994) Lipoproteins, structure, function, biosynthesis and model for protein export. In: Ghuysen J-M, Hagenbeck R (eds) Bacterial cell wall. Elsevier, Amsterdam, pp 319–341
Braun V, Bosch V, Klumpp ER, Neff I, Mayer H, Schlecht S (1976a) Antigenic determinants of murein lipoprotein and its exposure at the surface of Enterobacteriaceae. Eur J Biochem 62(3):555–566
Braun V, Rotering H, Ohms JP, Hagenmaier H (1976b) Conformational studies on murein-lipoprotein from the outer membrane of Escherichia coli. Eur J Biochem 70(2):601–610
Brisson D, Zhou W, Jutras BL, Casjens S, Stevenson B (2013) Distribution of cp32 prophages among Lyme disease-causing spirochetes and natural diversity of their lipoprotein-encoding erp loci. Appl Environ Microbiol 79(13):4115–4128. https://doi.org/10.1128/AEM.00817-13
Brokx SJ, Ellison M, Locke T, Bottorff D, Frost L, Weiner JH (2004) Genome-wide analysis of lipoprotein expression in Escherichia coli MG1655. J Bacteriol 186(10):3254–3258
Brulle JK, Tschumi A, Sander P (2013) Lipoproteins of slow-growing Mycobacteria carry three fatty acids and are N-acylated by apolipoprotein N-acyltransferase BCG_2070c. BMC Microbiol 13:223. https://doi.org/10.1186/1471-2180-13-223
Buwitt-Beckmann U, Heine H, Wiesmüller KH, Jung G, Brock R, Akira S, Ulmer AJ (2005) Toll-like receptor 6-independent signaling by diacylated lipopeptides. Eur J Immunol 35(1):282–289. https://doi.org/10.1002/eji.200424955
Buwitt-Beckmann U, Heine H, Wiesmüller KH, Jung G, Brock R, Akira S, Ulmer AJ (2006) TLR1- and TLR6-independent recognition of bacterial lipopeptides. J Biol Chem 281(14):9049–9057. https://doi.org/10.1074/jbc.M512525200
Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, Cosma CL, Ramakrishnan L (2014) Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505(7482):218–222. https://doi.org/10.1038/nature12799
Choi DS, Yamada H, Mizuno T, Mizushima S (1987) Molecular assembly of the lipoprotein trimer on the peptidoglycan layer of Escherichia coli. J Biochem 102(5):975–983
Cohen EJ, Ferreira JL, Ladinsky MS, Beeby M, Hughes KT (2017) Nanoscale-length control of the flagellar driveshaft requires hitting the tethered outer membrane. Science 356(6334):197–200. https://doi.org/10.1126/science.aam6512
Cowles CE, Li Y, Semmelhack MF, Cristea IM, Silhavy TJ (2011) The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol Microbiol 79(5):1168–1181. https://doi.org/10.1111/j.1365-2958.2011.07539.x
Deres K, Schild H, Wiesmüller KH, Jung G, Rammensee HG (1989) In vivo priming of virus-specific cytotoxic T lymphocytes with synthetic lipopeptide vaccine. Nature 342(6249):561–564. https://doi.org/10.1038/342561a0
Diao J, Bouwman C, Yan D, Kang J, Katakam AK, Liu P, Pantua H, Abbas AR, Nickerson NN, Austin C, Reichelt M, Sandoval W, Xu M, Whitfield C, Kapadia SB (2017) Peptidoglycan association of murein lipoprotein is required for KpsD-dependent group 2 capsular polysaccharide expression and serum resistance in a Uropathogenic Escherichia coli isolate. MBio 8(3). https://doi.org/10.1128/mbio.00603-17
Dowdell AS, Murphy MD, Azodi C, Swanson SK, Florens L, Chen S, Zückert WR (2017) Comprehensive spatial analysis of the Borrelia burgdorferi lipoproteome reveals a compartmentalization bias toward the bacterial surface. J Bacteriol 199(6). https://doi.org/10.1128/jb.00658-16
Erova TE, Kirtley ML, Fitts EC, Ponnusamy D, Baze WB, Andersson JA, Cong Y, Tiner BL, Sha J, Chopra AK (2016) Protective immunity elicited by oral immunization of Mice with Salmonella enterica Serovar Typhimurium Braun Lipoprotein (Lpp) and Acetyltransferase (MsbB) mutants. Front Cell Infect Microbiol 6:148. https://doi.org/10.3389/fcimb.2016.00148
Fadl AA, Sha J, Klimpel GR, Olano JP, Niesel DW, Chopra AK (2005) Murein lipoprotein is a critical outer membrane component involved in Salmonella enterica serovar typhimurium systemic infection. Infect Immun 73(2):1081–1096. https://doi.org/10.1128/IAI.73.2.1081-1096.2005
Fantappie L, Irene C, De Santis M, Armini A, Gagliardi A, Tomasi M, Parri M, Cafardi V, Bonomi S, Ganfini L, Zerbini F, Zanella I, Carnemolla C, Bini L, Grandi A, Grandi G (2017) Some gram-negative lipoproteins keep their surface topology when transplanted from one species to another and deliver foreign polypeptides to the bacterial surface. Mol Cell Proteomics 16(7):1348–1364. https://doi.org/10.1074/mcp.M116.065094
Foley MH, Cockburn DW, Koropatkin NM (2016) The Sus operon: a model system for starch uptake by the human gut Bacteroidetes. Cell Mol Life Sci 73(14):2603–2617. https://doi.org/10.1007/s00018-016-2242-x
Gimenez MI, Dilks K, Pohlschroder M (2007) Haloferax volcanii twin-arginine translocation substates include secreted soluble, C-terminally anchored and lipoproteins. Mol Microbiol 66(6):1597–1606. https://doi.org/10.1111/j.1365-2958.2007.06034.x
Glauner B, Holtje JV, Schwarz U (1988) The composition of the murein of Escherichia coli. J Biol Chem 263(21):10088–10095
Glenwright AJ, Pothula KR, Bhamidimarri SP, Chorev DS, Basle A, Firbank SJ, Zheng H, Robinson CV, Winterhalter M, Kleinekathofer U, Bolam DN, van den Berg B (2017) Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 541(7637):407–411. https://doi.org/10.1038/nature20828
Gonnet P, Rudd KE, Lisacek F (2004) Fine-tuning the prediction of sequences cleaved by signal peptidase II: a curated set of proven and predicted lipoproteins of Escherichia coli K-12. Proteomics 4(6):1597–1613. https://doi.org/10.1002/pmic.200300749
Gonzalez-Zamorano M, Mendoza-Hernandez G, Xolalpa W, Parada C, Vallecillo AJ, Bigi F, Espitia C (2009) Mycobacterium tuberculosis glycoproteomics based on ConA-lectin affinity capture of mannosylated proteins. J Proteome Res 8(2):721–733. https://doi.org/10.1021/pr800756a
Grabowicz M, Silhavy TJ (2017a) Envelope stress responses: an interconnected safety net. Trends Biochem Sci 42(3):232–242. https://doi.org/10.1016/j.tibs.2016.10.002
Grabowicz M, Silhavy TJ (2017b) Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc Natl Acad Sci USA 114(18):4769–4774. https://doi.org/10.1073/pnas.1702248114
Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G (2014) MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev 28(14):1620–1634. https://doi.org/10.1101/gad.243485.114
Haake DA, Zückert WR (2017) Spirochetal lipoproteins in pathogenesis and immunity. Curr Top Microbiol Immunol. https://doi.org/10.1007/82_2017_78
Hantke K, Braun V (1973) Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. Eur J Biochem 34(2):284–296
Hanzelmann D, Joo HS, Franz-Wachtel M, Hertlein T, Stevanovic S, Macek B, Wolz C, Gotz F, Otto M, Kretschmer D, Peschel A (2016) Toll-like receptor 2 activation depends on lipopeptide shedding by bacterial surfactants. Nat Commun 7:12304. https://doi.org/10.1038/ncomms12304
Hayashi S, Hara H, Suzuki H, Hirota Y (1988) Lipid modification of Escherichia coli penicillin-binding protein 3. J Bacteriol 170(11):5392–5395
Hirashima A, Inouye M (1973) Specific biosynthesis of an envelope protein of Escherichia coli. Nature 242(5397):405–407
Hirashima A, Wang S, Inouye M (1974) Cell-free synthesis of a specific lipoprotein of the Escherichia coli outer membrane directed by purified messenger RNA. Proc Natl Acad Sci USA 71(10):4149–4153
Hirota Y, Suzuki H, Nishimura Y, Yasuda S (1977) On the process of cellular division in Escherichia coli: a mutant of E. coli lacking a murein-lipoprotein. Proc Natl Acad Sci USA 74(4):1417–1420
Hooda Y, Moraes TF (2018) Translocation of lipoproteins to the surface of gram negative bacteria. Curr Opin Struct Biol 51:73–79. https://doi.org/10.1016/j.sbi.2018.03.006
Hooda Y, Lai CC, Judd A, Buckwalter CM, Shin HE, Gray-Owen SD, Moraes TF (2016) Slam is an outer membrane protein that is required for the surface display of lipidated virulence factors in Neisseria. Nat Microbiol 1:16009. https://doi.org/10.1038/nmicrobiol.2016.9
Hooda Y, Shin HE, Bateman TJ, Moraes TF (2017) Neisserial surface lipoproteins: structure, function and biogenesis. Pathog Dis 75(2). https://doi.org/10.1093/femspd/ftx010
Innis MA, Tokunaga M, Williams ME, Loranger JM, Chang SY, Chang S, Wu HC (1984) Nucleotide sequence of the Escherichia coli prolipoprotein signal peptidase (lsp) gene. Proc Natl Acad Sci USA 81(12):3708–3712
Inouye M, Shaw J, Shen C (1972) The assembly of a structural lipoprotein in the envelope of Escherichia coli. J Biol Chem 247(24):8154–8159
Jackson DC, Lau YF, Le T, Suhrbier A, Deliyannis G, Cheers C, Smith C, Zeng W, Brown LE (2004) A totally synthetic vaccine of generic structure that targets Toll-like receptor 2 on dendritic cells and promotes antibody or cytotoxic T cell responses. Proc Natl Acad Sci USA 101(43):15440–15445. https://doi.org/10.1073/pnas.0406740101
Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO (2007) Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130(6):1071–1082. https://doi.org/10.1016/j.cell.2007.09.008
Johnson TJ, Wannemuehler YM, Nolan LK (2008) Evolution of the iss gene in Escherichia coli. Appl Environ Microbiol 74(8):2360–2369. https://doi.org/10.1128/AEM.02634-07
Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A (2003) Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci 12(8):1652–1662. https://doi.org/10.1110/ps.0303703
Kang JY, Lee JO (2011) Structural biology of the Toll-like receptor family. Annu Rev Biochem 80:917–941. https://doi.org/10.1146/annurev-biochem-052909-141507
Kang JY, Nan X, Jin MS, Youn SJ, Ryu YH, Mah S, Han SH, Lee H, Paik SG, Lee JO (2009) Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31(6):873–884. https://doi.org/10.1016/j.immuni.2009.09.018
Keseler IM, Mackie A, Santos-Zavaleta A, Billington R, Bonavides-Martinez C, Caspi R, Fulcher C, Gama-Castro S, Kothari A, Krummenacker M, Latendresse M, Muniz-Rascado L, Ong Q, Paley S, Peralta-Gil M, Subhraveti P, Velazquez-Ramirez DA, Weaver D, Collado-Vides J, Paulsen I, Karp PD (2017) The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res 45(D1):D543–D550. https://doi.org/10.1093/nar/gkw1003
Khan S, Weterings JJ, Britten CM, de Jong AR, Graafland D, Melief CJ, van der Burg SH, van der Marel G, Overkleeft HS, Filippov DV, Ossendorp F (2009) Chirality of TLR-2 ligand Pam3CysSK4 in fully synthetic peptide conjugates critically influences the induction of specific CD8+ T-cells. Mol Immunol 46(6):1084–1091. https://doi.org/10.1016/j.molimm.2008.10.006
Kitamura S, Wolan DW (2018) Probing substrate recognition of bacterial lipoprotein signal peptidase using FRET reporters. FEBS Lett 592(13):2289–2296. https://doi.org/10.1002/1873-3468.13155
Konovalova A, Perlman DH, Cowles CE, Silhavy TJ (2014) Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of beta-barrel proteins. Proc Natl Acad Sci USA 111(41):E4350–4358. https://doi.org/10.1073/pnas.1417138111
Konovalova A, Mitchell AM, Silhavy TJ (2016) A lipoprotein/beta-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. Elife 5. https://doi.org/10.7554/elife.15276
Konovalova A, Kahne DE, Silhavy TJ (2017) Outer membrane biogenesis. Annu Rev Microbiol 71:539–556. https://doi.org/10.1146/annurev-micro-090816-093754
Kretschmer D, Hanzelmann D, Peschel A (2016) Lipoprotein immunoproteomics question the potential of Staphylococcus aureus TLR2 agonists as vaccine antigens. Proteomics 16(20):2603–2604. https://doi.org/10.1002/pmic.201600351
Kurokawa K, Lee H, Roh KB, Asanuma M, Kim YS, Nakayama H, Shiratsuchi A, Choi Y, Takeuchi O, Kang HJ, Dohmae N, Nakanishi Y, Akira S, Sekimizu K, Lee BL (2009) The triacylated atp binding cluster transporter substrate-binding lipoprotein of Staphylococcus aureus functions as a native ligand for toll-like receptor 2. J Biol Chem 284(13):8406–8411. https://doi.org/10.1074/jbc.M809618200
Kurokawa K, Kim MS, Ichikawa R, Ryu KH, Dohmae N, Nakayama H, Lee BL (2012a) Environment-mediated accumulation of diacyl lipoproteins over their triacyl counterparts in Staphylococcus aureus. J Bacteriol 194(13):3299–3306. https://doi.org/10.1128/JB.00314-12
Kurokawa K, Ryu KH, Ichikawa R, Masuda A, Kim MS, Lee H, Chae JH, Shimizu T, Saitoh T, Kuwano K, Akira S, Dohmae N, Nakayama H, Lee BL (2012b) Novel bacterial lipoprotein structures conserved in low-GC content gram-positive bacteria are recognized by Toll-like receptor 2. J Biol Chem 287(16):13170–13181. https://doi.org/10.1074/jbc.M111.292235
Lauber F, Cornelis GR, Renzi F (2016) Identification of a new lipoprotein export signal in gram-negative bacteria. MBio 7(5). https://doi.org/10.1128/mbio.01232-16
Lewenza S, Mhlanga MM, Pugsley AP (2008) Novel inner membrane retention signals in Pseudomonas aeruginosa lipoproteins. J Bacteriol 190(18):6119–6125. https://doi.org/10.1128/JB.00603-08
Liu T, Agar SL, Sha J, Chopra AK (2010) Deletion of Braun lipoprotein gene (lpp) attenuates Yersinia pestis KIM/D27 strain: role of Lpp in modulating host immune response, NF-kappaB activation and cell death. Microb Pathog 48(1):42–52. https://doi.org/10.1016/j.micpath.2009.09.002
Lupas AN, Bassler J, Dunin-Horkawicz S (2017) The structure and topology of alpha-helical coiled coils. Subcell Biochem 82:95–129. https://doi.org/10.1007/978-3-319-49674-0_4
Magnet S, Bellais S, Dubost L, Fourgeaud M, Mainardi JL, Petit-Frere S, Marie A, Mengin-Lecreulx D, Arthur M, Gutmann L (2007) Identification of the L,D-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J Bacteriol 189(10):3927–3931. https://doi.org/10.1128/JB.00084-07
Malinverni JC, Silhavy TJ (2009) An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci USA 106(19):8009–8014. https://doi.org/10.1073/pnas.0903229106
Mao G, Zhao Y, Kang X, Li Z, Zhang Y, Wang X, Sun F, Sankaran K, Zhang XC (2016) Crystal structure of E. coli lipoprotein diacylglyceryl transferase. Nat Commun 7:10198. https://doi.org/10.1038/ncomms10198
Mattar S, Scharf B, Kent SB, Rodewald K, Oesterhelt D, Engelhard M (1994) The primary structure of halocyanin, an archaeal blue copper protein, predicts a lipid anchor for membrane fixation. J Biol Chem 269(21):14939–14945
May KL, Silhavy TJ (2018) The Escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. MBio 9(2). https://doi.org/10.1128/mbio.00379-18
Melchers F, Braun V, Galanos C (1975) The lipoprotein of the outer membrane of Escherichia coli: a B-lymphocyte mitogen. J Exp Med 142(2):473–482
Moyle PM, Toth I (2008) Self-adjuvanting lipopeptide vaccines. Curr Med Chem 15(5):506–516
Mühlradt PF, Kiess M, Meyer H, Süssmuth R, Jung G (1997) Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J Exp Med 185(11):1951–1958
Nakayama H, Kurokawa K, Lee BL (2012) Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J 279(23):4247–4268. https://doi.org/10.1111/febs.12041
Nardin EH, Calvo-Calle JM, Oliveira GA, Nussenzweig RS, Schneider M, Tiercy JM, Loutan L, Hochstrasser D, Rose K (2001) A totally synthetic polyoxime malaria vaccine containing Plasmodium falciparum B cell and universal T cell epitopes elicits immune responses in volunteers of diverse HLA types. J Immunol 166(1):481–489
Narita S, Tokuda H (2007) Amino acids at positions 3 and 4 determine the membrane specificity of Pseudomonas aeruginosa lipoproteins. J Biol Chem 282(18):13372–13378. https://doi.org/10.1074/jbc.M611839200
Narita SI, Tokuda H (2017) Bacterial lipoproteins; biogenesis, sorting and quality control. Biochim Biophys Acta Mol Cell Biol Lipids 1862(11):1414–1423. https://doi.org/10.1016/j.bbalip.2016.11.009
Nedialkova LP, Sidstedt M, Koeppel MB, Spriewald S, Ring D, Gerlach RG, Bossi L, Stecher B (2016) Temperate phages promote colicin-dependent fitness of Salmonella enterica serovar Typhimurium. Environ Microbiol 18(5):1591–1603. https://doi.org/10.1111/1462-2920.13077
Nguyen MT, Götz F (2016) Lipoproteins of gram-positive bacteria: key players in the immune response and virulence. Microbiol Mol Biol Rev 80(3):891–903. https://doi.org/10.1128/MMBR.00028-16
Nguyen DT, de Witte L, Ludlow M, Yuksel S, Wiesmüller KH, Geijtenbeek TB, Osterhaus AD, de Swart RL (2010) The synthetic bacterial lipopeptide Pam3CSK4 modulates respiratory syncytial virus infection independent of TLR activation. PLoS Pathog 6(8):e1001049. https://doi.org/10.1371/journal.ppat.1001049
Nguyen MT, Kraft B, Yu W, Demircioglu DD, Hertlein T, Burian M, Schmaler M, Boller K, Bekeredjian-Ding I, Ohlsen K, Schittek B, Götz F (2015a) The nuSa alpha specific lipoprotein like cluster (lpl) of S. aureus USA300 contributes to immune stimulation and invasion in human cells. PLoS Pathog 11(6):e1004984. https://doi.org/10.1371/journal.ppat.1004984
Nguyen PT, Li QW, Kadaba NS, Lai JY, Yang JG, Rees DC (2015b) The contribution of methionine to the stability of the Escherichia coli MetNIQ ABC transporter-substrate binding protein complex. Biol Chem 396(9–10):1127–1134. https://doi.org/10.1515/hsz-2015-0131
Nguyen MT, Uebele J, Kumari N, Nakayama H, Peter L, Ticha O, Woischnig AK, Schmaler M, Khanna N, Dohmae N, Lee BL, Bekeredjian-Ding I, Götz F (2017) Lipid moieties on lipoproteins of commensal and non-commensal staphylococci induce differential immune responses. Nat Commun 8(1):2246. https://doi.org/10.1038/s41467-017-02234-4
Nigrovic LE, Thompson KM (2007) The Lyme vaccine: a cautionary tale. Epidemiol Infect 135(1):1–8. https://doi.org/10.1017/S0950268806007096
Nikitushkin VD, Demina GR, Shleeva MO, Guryanova SV, Ruggiero A, Berisio R, Kaprelyants AS (2015) A product of RpfB and RipA joint enzymatic action promotes the resuscitation of dormant mycobacteria. FEBS J 282(13):2500–2511. https://doi.org/10.1111/febs.13292
Nilsson G, Belasco JG, Cohen SN, von Gabain A (1984) Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature 312(5989):75–77
Noland CL, Kattke MD, Diao J, Gloor SL, Pantua H, Reichelt M, Katakam AK, Yan D, Kang J, Zilberleyb I, Xu M, Kapadia SB, Murray JM (2017) Structural insights into lipoprotein N-acylation by Escherichia coli apolipoprotein N-acyltransferase. Proc Natl Acad Sci USA 114(30):E6044–E6053. https://doi.org/10.1073/pnas.1707813114
Okuda S, Tokuda H (2009) Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc Natl Acad Sci USA 106(14):5877–5882. https://doi.org/10.1073/pnas.0900896106
Okuda S, Tokuda H (2011) Lipoprotein sorting in bacteria. Annu Rev Microbiol 65:239–259. https://doi.org/10.1146/annurev-micro-090110-102859
Parra J, Marcoux J, Poncin I, Canaan S, Herrmann JL, Nigou J, Burlet-Schiltz O, Riviere M (2017) Scrutiny of Mycobacterium tuberculosis 19 kDa antigen proteoforms provides new insights in the lipoglycoprotein biogenesis paradigm. Sci Rep 7:43682. https://doi.org/10.1038/srep43682
Pazos M, Peters K, Vollmer W (2017) Robust peptidoglycan growth by dynamic and variable multi-protein complexes. Curr Opin Microbiol 36:55–61. https://doi.org/10.1016/j.mib.2017.01.006
Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, Chang F, Weibel DB, Theriot JA, Huang KC (2018) The outer membrane is an essential load-bearing element in gram-negative bacteria. Nature 559(7715):617–621. https://doi.org/10.1038/s41586-018-0344-3
Sakthi S, Palaniyandi K, Gupta UD, Gupta P, Narayanan S (2016) Lipoprotein LpqS deficient M. tuberculosis mutant is attenuated for virulence in vivo and shows protective efficacy better than BCG in guinea pigs. Vaccine 34(6):735–743. https://doi.org/10.1016/j.vaccine.2015.12.059
Sander P, Rezwan M, Walker B, Rampini SK, Kroppenstedt RM, Ehlers S, Keller C, Keeble JR, Hagemeier M, Colston MJ, Springer B, Bottger EC (2004) Lipoprotein processing is required for virulence of Mycobacterium tuberculosis. Mol Microbiol 52(6):1543–1552. https://doi.org/10.1111/j.1365-2958.2004.04041.x
Sato T, Takano A, Hori N, Izawa T, Eda T, Sato K, Umekawa M, Miyagawa H, Matsumoto K, Muramatsu-Fujishiro A, Matsumoto K, Matsuoka S, Hara H (2017) Role of the inner-membrane histidine kinase RcsC and outer-membrane lipoprotein RcsF in the activation of the Rcs phosphorelay signal transduction system in Escherichia coli. Microbiology 163(7):1071–1080. https://doi.org/10.1099/mic.0.000483
Sha J, Fadl AA, Klimpel GR, Niesel DW, Popov VL, Chopra AK (2004) The two murein lipoproteins of Salmonella enterica serovar Typhimurium contribute to the virulence of the organism. Infect Immun 72(7):3987–4003. https://doi.org/10.1128/IAI.72.7.3987-4003.2004
Shrivastava R, Jiang X, Chng SS (2017) Outer membrane lipid homeostasis via retrograde phospholipid transport in Escherichia coli. Mol Microbiol 106(3):395–408. https://doi.org/10.1111/mmi.13772
Shu W, Liu J, Ji H, Lu M (2000) Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 A resolution. J Mol Biol 299(4):1101–1112. https://doi.org/10.1006/jmbi.2000.3776
Shukla S, Richardson ET, Athman JJ, Shi L, Wearsch PA, McDonald D, Banaei N, Boom WH, Jackson M, Harding CV (2014) Mycobacterium tuberculosis lipoprotein LprG binds lipoarabinomannan and determines its cell envelope localization to control phagolysosomal fusion. PLoS Pathog 10(10):e1004471. https://doi.org/10.1371/journal.ppat.1004471
Skabytska Y, Wölbing F, Günther C, Köberle M, Kaesler S, Chen KM, Guenova E, Demircioglu D, Kempf WE, Volz T, Rammensee HG, Schaller M, Röcken M, Götz F, Biedermann T (2014) Cutaneous innate immune sensing of Toll-like receptor 2-6 ligands suppresses T cell immunity by inducing myeloid-derived suppressor cells. Immunity 41(5):762–775. https://doi.org/10.1016/j.immuni.2014.10.009
Soltes GR, Martin NR, Park E, Sutterlin HA, Silhavy TJ (2017) Distinctive roles for periplasmic proteases in the maintenance of essential outer membrane protein assembly. J Bacteriol 199(20). https://doi.org/10.1128/jb.00418-17
Steinhagen F, Kinjo T, Bode C, Klinman DM (2011) TLR-based immune adjuvants. Vaccine 29(17):3341–3355. https://doi.org/10.1016/j.vaccine.2010.08.002
Stoll H, Dengjel J, Nerz C, Götz F (2005) Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect Immun 73(4):2411–2423. https://doi.org/10.1128/IAI.73.4.2411-2423.2005
Storf S, Pfeiffer F, Dilks K, Chen ZQ, Imam S, Pohlschröder M (2010) Mutational and bioinformatic analysis of haloarchaeal lipobox-containing proteins. Archaea 2010. https://doi.org/10.1155/2010/410975
Sulzenbacher G, Canaan S, Bordat Y, Neyrolles O, Stadthagen G, Roig-Zamboni V, Rauzier J, Maurin D, Laval F, Daffe M, Cambillau C, Gicquel B, Bourne Y, Jackson M (2006) LppX is a lipoprotein required for the translocation of phthiocerol dimycocerosates to the surface of Mycobacterium tuberculosis. EMBO J 25(7):1436–1444. https://doi.org/10.1038/sj.emboj.7601048
Sun C, Benlekbir S, Venkatakrishnan P, Wang Y, Hong S, Hosler J, Tajkhorshid E, Rubinstein JL, Gennis RB (2018) Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 557(7703):123–126. https://doi.org/10.1038/s41586-018-0061-y
Szewczyk J, Collet JF (2016) The journey of lipoproteins through the cell: one birthplace, multiple destinations. Adv Microb Physiol 69:1–50. https://doi.org/10.1016/bs.ampbs.2016.07.003
Takeda K, Miyatake H, Yokota N, Matsuyama S, Tokuda H, Miki K (2003) Crystal structures of bacterial lipoprotein localization factors. LolA and LolB. EMBO J 22(13):3199–3209. https://doi.org/10.1093/emboj/cdg324
Tao K, Narita S, Tokuda H (2012) Defective lipoprotein sorting induces lolA expression through the Rcs stress response phosphorelay system. J Bacteriol 194(14):3643–3650. https://doi.org/10.1128/JB.00553-12
Thompson BJ, Widdick DA, Hicks MG, Chandra G, Sutcliffe IC, Palmer T, Hutchings MI (2010) Investigating lipoprotein biogenesis and function in the model gram-positive bacterium Streptomyces coelicolor. Mol Microbiol 77(4):943–957. https://doi.org/10.1111/j.1365-2958.2010.07261.x
Tokuda H, Matsuyama S, Tanaka-Masuda K (2007) Structure, function, and transport of lipoproteins in Escherichia coli. In: Ehrmann M (ed) The periplasm. ASM, Washington D.C., pp 67–80
Tokunaga M, Tokunaga H, Wu HC (1982) Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc Natl Acad Sci USA 79(7):2255–2259
Tschumi A, Nai C, Auchli Y, Hunziker P, Gehrig P, Keller P, Grau T, Sander P (2009) Identification of apolipoprotein N-acyltransferase (Lnt) in mycobacteria. J Biol Chem 284(40):27146–27156. https://doi.org/10.1074/jbc.M109.022715
van Els CA, Corbiere V, Smits K, van Gaans-van den Brink JA, Poelen MC, Mascart F, Meiring HD, Locht C (2014) Toward understanding the essence of post-translational modifications for the mycobacterium tuberculosis immunoproteome. Front Immunol 5:361. https://doi.org/10.3389/fimmu.2014.00361
Vogeley L, El Arnaout T, Bailey J, Stansfeld PJ, Boland C, Caffrey M (2016) Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin. Science 351(6275):876–880. https://doi.org/10.1126/science.aad3747
Vollmer W, Blanot D, de Pedro MA (2008) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32(2):149–167. https://doi.org/10.1111/j.1574-6976.2007.00094.x
Vu CH, Kolata J, Stentzel S, Beyer A, Gesell Salazar M, Steil L, Pane-Farre J, Ruhmling V, Engelmann S, Gotz F, van Dijl JM, Hecker M, Mader U, Schmidt F, Volker U, Broker BM (2016) Adaptive immune response to lipoproteins of Staphylococcus aureus in healthy subjects. Proteomics 16(20):2667–2677. https://doi.org/10.1002/pmic.201600151
Weidel W, Pelzer H (1964) Bagshaped macromolecules—a new outlook on bacterial cell walls. Adv Enzymol Relat Subj Biochem 26:193–232
Wiesmüller KH, Bessler W, Jung G (1983) Synthesis of the mitogenic S-[2,3-bis(palmitoyloxy)propyl]-N-palmitoylpentapeptide from Escherichia coli lipoprotein. Hoppe Seylers Z Physiol Chem 364(5):593–606
Wiesmuller KH, Jung G, Hess G (1989) Novel low-molecular-weight synthetic vaccine against foot-and-mouth disease containing a potent B-cell and macrophage activator. Vaccine 7(1):29–33
Wiktor M, Weichert D, Howe N, Huang CY, Olieric V, Boland C, Bailey J, Vogeley L, Stansfeld PJ, Buddelmeijer N, Wang M, Caffrey M (2017) Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis. Nat Commun 8:15952. https://doi.org/10.1038/ncomms15952
Yamaguchi K, Yu F, Inouye M (1988) A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53(3):423–432
Zaman M, Toth I (2013) Immunostimulation by synthetic lipopeptide-based vaccine candidates: structure-activity relationships. Front Immunol 4:318. https://doi.org/10.3389/fimmu.2013.00318
Zhang WY, Wu HC (1992) Alterations of the carboxyl-terminal amino acid residues of Escherichia coli lipoprotein affect the formation of murein-bound lipoprotein. J Biol Chem 267(27):19560–19564
Zhang WY, Inouye M, Wu HC (1992) Neither lipid modification nor processing of prolipoprotein is essential for the formation of murein-bound lipoprotein in Escherichia coli. J Biol Chem 267(27):19631–19635
Acknowledgements
The authors work was supported by institutional funds of the Max Planck Society (V.B.) and the German Science Foundation (V.B., K.H.). We thank Jens Baßler for his lipoprotein sequence analysis. V.B. thanks Andrei Lupas for his generous hospitality in his Department of Protein Evolution in the Max Planck Institute for Developmental Biology. K.H. thanks Karl Forchhammer and Joachim E. Schultz for generous hospitality.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Braun, V., Hantke, K. (2019). Lipoproteins: Structure, Function, Biosynthesis. In: Kuhn, A. (eds) Bacterial Cell Walls and Membranes . Subcellular Biochemistry, vol 92. Springer, Cham. https://doi.org/10.1007/978-3-030-18768-2_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-18768-2_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-18767-5
Online ISBN: 978-3-030-18768-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)