Abstract
During the evolution of Gram-negative bacteria, their outermost and major membrane components, lipopolysaccharides (LPS), the constituents of the so-called endotoxin, adapted to environmental changes. This helped pathogenic bacteria evade detection by the host immune system. The modifications were numerous and occurred in all three distinct LPS regions: lipid A, core, and O-chains.
In limited bacterial infections characterized by the presence of low LPS doses reaching bloodstream, the immune system responses are beneficial for the host and lead to the rapid clearing of pathogens. In contrast, in overwhelming infections, higher LPS doses lead to uncontrolled cytokine overproduction, and result in septic (endotoxic) shock, that is often lethal. This review describes the amazing diversity of these molecules, among genera, species and strains, and the high degree of their structural heterogeneity, together with the corresponding impact on their biological activities.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
Endotoxins are lipopolysaccharides (LPS), the major glycolipids of most Gram-negative bacterial outer membranes [1, 2] (see Fig. 3.1). LPS molecules, forming the outer layer of this membrane, represent about 50% of its weight, and cover about 75% of the bacterial surface. The general LPS architecture consists of three distinct regions: (1) The hydrophobic region, called lipid A, is a glycophospholipid that anchors LPS molecules to the outer membrane, through hydrophobic interactions with the acyl chains of phospholipids constituting the inner layer of this membrane. The outer membrane thus represents an asymmetric lipid bilayer. Its integrity is also maintained by salt bridges formed between phosphate groups of neighboring lipid A molecules, and divalent cations (Ca2+ and Mg2+). (2) The core oligosaccharide is covalently linked at its reducing end to the lipid A moiety via 3-deoxy-D-manno-oct-2-ulopyranosonic acid (Kdo). Kdo can itself be substituted by other Kdo molecules, then in most cases the sugars substituting Kdo are L-glycero-D-manno-heptopyranose (Hep) residues, some of which are substituted with phosphate (P), pyrophosphate (PP) or ethanolaminepyrophosphate (PPEA) groups. This proximal region is referred to as the “inner core”. The presence of carboxyl and phosphate groups makes this region highly negatively charged.
Salt bridges formed between neighboring acid groups via incorporation of divalent cations, as well as electrostatic interactions with positively charged amino groups, contribute to the mechanical strength and rigidity of the membrane. The distal part of the core oligosaccharide also called “outer core” is usually composed of neutral and basic hexoses. (3) An antigenic O-chain polysaccharide consisting of a variable number of repeating oligosaccharide units, of 1–10 sugar residues, is the third structurally distinct region of the LPS molecules. It is attached to the outer core and is thus the most exposed to environmental factors. The presence or absence of an O-polysaccharide chain determines whether a LPS is classified as being of a Smooth- (S) or Rough- (R) type (Fig. 3.1a, b), respectively, because of the smooth or rough appearance displayed by the bacterial colonies on agar plates.
2 Role of Lipopolysaccharides
LPSs are essential for the survival of most Gram-negative bacteria, the Rough-type (lipid A plus core) and deep rough-type LPS (lipid A plus Kdo) are described as less protective for the bacteria. However, a few examples of bacteria without LPS have been described as exceptions [3]. When present, as in the large majority of cases, the LPS monolayer contributes to the outer membrane’s integrity. In direct contact with the environment, it provides bacteria with an efficient permeability barrier to antimicrobial compounds, and protection against complement mediated lysis. Moreover, the LPS molecules are necessary for the correct anchoring and functions of the outer membrane proteins [4,5,6]. The minimal LPS structure required for bacterial growth in vitro consists of lipid A and Kdo domains [7]. For bacterial survival and virulence in natural conditions, lipid A, core and O-polysaccharide chain are required to protect the bacteria from hostile environments. Consequently, the O-antigens are present in most wild-type Gram-negative bacteria. Nevertheless, some important pathogens like Bordetella pertussis, Neisseria meningitidis, and Haemophilus influenza do not synthesize O-chains. LPS molecules of these and a few other species only display an oligosaccharide core that is often branched. They are named lipo-oligosaccharides (LOS) [8,9,10,11].
LPS molecules, in addition to being the outermost membrane layer, are highly immunogenic. While the polysaccharide moiety carries the antigenic determinants (O-chains or core side chains) [12, 13] the lipid A portion is recognized by the innate immune cell receptors (TLR4/MD2-CD14 complex of macrophages and dendritic cells). This leads to activation of signaling cascades and results in the secretion of pro- (and anti-) inflammatory cytokines, thereby initiating inflammatory and immune-defense responses [14,15,16].
Biological and physical-chemical properties of LPSs and their multimolecular assemblies are defined by their fine molecular structures [17, 18]. Being ubiquitous microorganisms living in all biotopes of the earth, bacteria have adapted to very different environmental conditions which submit them to different types of stress, both physical and chemical (temperature, ion and nutrient concentration, etc.) and biological (host-immune system). Bacterial lipopolysaccharides play important and diverse roles in this adaptation. This is likely to be the reason for the enormous structural diversity of these molecules. LPS structures can differ not only among different genera, species and strains, but also within a strain grown under different conditions. All the elements of the common LPS architecture (lipid A, core, and O-polysaccharide) contribute to this structural diversity and variability. The purpose of this chapter is to illustrate the high diversity and heterogeneity of LPS structures, and the roles of discrete LPS structural variations in bacterial adaptation and virulence.
3 Brief History of Lipopolysaccharides
The French chemist Louis Pasteur, born in 1822, elucidated the problem of fermentation and demonstrated that the theory of spontaneous generation was an erroneous doctrine. He first worked at discovering microbial infections of vines and silkworms and showed that microorganisms were at the origin of infectious diseases [19].
If the term “Endotoxin” was first introduced by Richard Pfeiffer, a German physician and bacteriologist, in 1892, to define a pyrogenic heat-stable and non-secreted toxin from Vibrio cholerea, similar substances had been previously described or cited by a few authors [20]. It was first cited as “pyrogenic material”, “putrid poison” or “toxin”. Nicolai Gamaleia, a Russian physiologist described in 1888 that heat-killed bacteria, like Burkholderia mallei, Serratia marcescens and Bacillus anthracis, could induce fever in rabbits. Sir Armand Ruffer, a German-British Paleo-pathologist and Microbiologist, demonstrated in 1889 that filtered bacterial culture supernatants could also induce fever in rabbits.
Edwin Klebs, a German and Swiss bacteriologist, attributed a possible role in military diseases and death to a pyrogenic substance from living micro-organisms that he named “Microsporum septicum”. He was followed by a Danish pathologist, Peter Ludvig Panum who reported a heat resistant, water-soluble pyrogenic toxin isolated from putrid matter. Thanks to the culture techniques developed by the German physician and microbiologist Robert Koch, in “Investigations about the etiology of wound infections” it was possible to demonstrate that some diseases were caused by different bacteria. Koch discovered many important pathogens like Bacillus anthracis, the etiologic agent of anthrax in 1876, Mycobacterium tuberculosis, the etiological agent of tuberculosis in 1882, and together with the German bacteriologist Georg Gaffky, in 1883, the agent of cholera, Vibrio cholera [21]. William B. Coley M.D., showed that a mixture of killed bacteria (Serratia marcescens and Streptococci) inducing fever were able to induce cancer remissions for soft tissue sarcoma in humans [22]. This work was at the origin of the discovery of TNF-α, decades later [23]. It was the first discovery of the association of deleterious and beneficial effects induced by pyrogenic endotoxins.
To define their chemical nature, progress in the extraction of endotoxins was crucial. Thus, André Boivin from the Pasteur institute invented the trichloro-acetic acid method and published together with his PhD student Mesrobeanu [24]. Endotoxins were then defined as being made of O-antigens glycolipid and proteins. It was later shown that the protein constituents were contaminants thanks to the phenol-water method of extraction described by Otto Lüderitz and Otto Westphal [25]. Improving the purification of LPS by solvent extraction also allowed elimination of a lipid B, at first supposed to be a LPS constituent. This non-covalently linked phospholipid is at the origin of the name given to the famous lipid A.
Anne-Marie Staub, from the Pasteur Institute established the polysaccharide nature of bacterial O-antigens [26]. Together with Otto Westphal and Otto Lüderitz, they demonstrated that oligosaccharides in the form of repeating units formed the species-specific O-antigens [27]. Following the Phenol-water extraction method, the Phenol-Chloroform-Petroleum ether (PCP) method was introduced specifically for the extraction of Rough-type LPS [28]. Other methods were introduced with more or less success, i.e., extraction with cold ethanol [29], aqueous butanol [30], methanol [31], extraction with Triton and Mg2+ [32] or extraction in water at 100 °C [33].
Extraction with hot phenol-water remains the gold standard. However, we introduced a method of extraction performed at room temperature with isobutyric acid and NH4OH (1 M) which avoids the use of toxic solvents [34].
4 LPS General Structural Architecture
Since LPS are amphipathic molecules, structural analysis of its distinct domains or testing them for their biological activities often necessitates chemical cleavage to liberate lipid A, or the core, or core plus O-chain moieties. They can be split through the acid-labile glycosidic bond of Kdo to sever the hydrophilic and hydrophobic LPS moieties.
5 O-Polysaccharides Structures
Exposed at the bacterial surface, the O-polysaccharides constitute an interface between the bacterium and its environment. They play multiple roles in the processes of colonization and virulence, including the evasion of complement and phagocytosis, adhesion to tissues, molecular mimicry, and cooperation with other virulence factors. At the same time, they are highly immunogenic and represent one of the first targets of the host immune system. They are also known as receptors for some bacteriophages as described for Pseudomonas [35]. This is why the enormous structural diversity observed in O-polysaccharides is believed to be an important factor of bacterial evolution, allowing adaptation to different niches and hosts [36]. Composed of variable numbers of repeating oligosaccharide units, from one to ten sugars, O-antigens synthesized by different species, and different serotypes of a species, differ in the type of monosaccharides and their arrangement within the repeating oligosaccharides, as well as in the presence of non-glycosidic substituents. Some examples are displayed in Fig. 3.2.
More than a hundred different monosaccharides, both common and rare, have been found in bacterial O-antigens, which form homo- and hetero-polymers with linear and branched structures of the repeating units. This makes the O-polysaccharides, not only the most variable part of the LPS molecules but also one of the most variable elements of bacterial cells. Such variability provides the basis for the serotyping schemes that identify Gram-negative bacteria. Thus far, 186 serogroups of E. coli [37], 34 serogroups of Shigella [38], 46 serogroups of Salmonella [39], and more than 200 serogroups of Vibrio cholerae [40] have been identified. Different biosynthetic pathways of O-antigen assembly determine their precise stereo regularity. A small number of O-polysaccharides occur as polymers wherein a regular repeating sequence is capped by a distinct terminal monosaccharide or capping oligosaccharide. If LPS structures are unique for a given bacterial species or strain, it is thanks to the amazing diversity of sugars and their high degree of linkage possibilities, as well as to the diversity of polymerization linkage between the oligosaccharides repeating units. The number of possibilities of linkage between sugars, versus between amino-acids, can be taken as an example.
There is only one possibility to form an amino-acid homo-dimer, but 11 possibilities with the same hexose, one possibility for an amino-acid homo-trimer, but 176 with hexoses, or 6 possibilities for an amino-acid hetero-trimer, compared to 1056 possibilities with hexoses. Fusobacterium nucleatum MJR 7757B O-chain was recently characterized [41]. It is an anaerobic bacterium already described as a dental pathogen involved in periodontis complications. It was also identified as causing colorectal cancer. The ManNAc4Lac, analogue of N-acetylmuramic acid, was found in this structure for the first time in natural sources. Xanthomonas citri is responsible for Asiatic citrus canker, the more serious citrus diseases. The LPS was involved in the pathogen virulence and shown to induce ROS accumulation [42].
5.1 Inter Genera Common or Closely-Related O-chain Structures: The Example of Brucella abortus, Yersinia enterocolitica and Vibrio cholera
If the O-chain structures are known as being unique for a given species, there are a few exceptions that prove the rule. A famous one is the structure of the O-chain of Brucella abortus, the agent of brucellosis [43]. The structure was characterized as being a linear homopolymer of 1,2-linked 4,6-dideoxy-4-formamido-α-d-mannopyranosyl residues or N-formyl-perosamine.
The serological reactivity of bovine antiserum to B. abortus 1119-3 with the lipopolysaccharides of Yersinia enterocolitica serotype 0:9 and Vibrio cholerae O1 species was shown to be due to the presence of perosamine in the O-PS repeating unit. Yersinia enterocolitica 0:9 shares an almost identical O-PS structure with that of B. abortus [44] and in V. cholerae O1 it differs by the nature of the N-acyl substituant. In V. cholerae the N-formyl group is replaced by an N-acetyl group. Of course the structures of the respective lipid A and core moieties are different, which makes the complete LPS structures different, but their serological common reactivity is due to the O-chains. This complicates the serological detection of brucellosis by bovine antiserum to B. abortus.
LPS from other genera were found to react with the B. abortus antiserum. This reactivity was due to the presence of a perosamine residue in these O-chains. It led to the characterization of many major pathogens LPS O-chain structures. N-acetyl-perosamine was first found in Perimycin an antibiotic produced by the Gram-positive Streptomyces coelicolor [45]. N-acetyl-Perosamine was found in the O-chain of the E.coli pathogen O157:H7 [46] but also in Caulobacter crescentus CB15 [47]. C. crescentus is a non-pathogenic alphaproteobacterium with an atypical lipid A structure [48] and an O-chain trapped beneath an S-layer, which complicated its structural analysis. However, it was shown to contain N-Acetyl-perosamine. This LPS contains about five repeats of the heptameric O-chain unit and its SDS-PAGE profile is particularly homogeneous with a single apparent band [49].
5.2 O-polysaccharide Structure-Function Relationships
At least 20 serogroups have been identified for Pseudomonas aeruginosa, an opportunistic pathogen and a major cause of nosocomial infections [50, 51]. This bacterium produces two types of O polysaccharides, known as the common A-band (a homopolymer), and the serospecific B-band (a heteropolymer). Several studies have demonstrated that the presence of the B-band polysaccharides confers serum resistance, and elicits a protective immune response to the bacterium [52].
Three different pathways, the Wzx/Wzy, the ABC transporter, and the synthetase pathways are described for the assembly and processing of O-antigens with the Wzx/Wzy pathway being the most common one. In this case, oligosaccharide units are synthesized on the cytoplasmic surface of the inner membrane, before translocation to its periplasmic surface where they are polymerized into the final O-antigen. The dynamic diversity of O-antigens observed within species is due to various mechanisms. New O-antigens are derived from others by mutation, lateral transfer of O-antigen coding genes, insertion or deletion of O-antigen genes mediated by insertion sequence (IS) elements, transferring O-antigen clusters, or O-antigen genes, by plasmids as well as by serotype-converting bacteriophages [36].
The role of O-antigens in bacterial virulence is multiple. The bacterial ability to evade complement thanks to the presence of long O-polysaccharide chains has been well documented [53]. O-antigen-deficient mutants are differentially attenuated in different animal models, they disseminate less easily from the initial point of infection and are more serum-sensitive than the corresponding wild-type strains. In several bacterial models, O-antigens were reported to be antiphagocytic, allowing less internalization of smooth-type strains and, in certain cases, induced better intracellular phagocyte survival [54, 55]. Another function of O-antigens contributing to bacterial virulence is molecular mimicry, i.e. structural similarity of bacterial antigenic determinants to host “self” components. The O-chains of Helicobacter pylori, the major cause of chronic gastritis and ulcers, carry Lewis antigenic epitopes (mainly LeX and LeY). From 80% to 90% of H. pylori isolates, from various geographical regions worldwide, express these antigens. Their putative biological and pathogenic roles suggested by numerous studies include escape from the control of the host-immune system, induction of autoimmune-mediated inflammation, and favoring gastric adhesion [56]. The role of LPS in bacterial adherence to host cells and mucous layers, as well as to abiotic surfaces, is now generally recognized [57]. In the case of H. pylori the O-antigen-mediated adhesion seems to be due to a specific recognition of Lewis antigens by receptors expressed on the surface of gastric epithelium [58]. However, in many cases depending on the chain length and chemical composition of the repeating oligosaccharides, O-antigens can both promote and interfere with bacterial adherence via non-specific interactions (hydrophobic, electrostatic, hydrogen bonds, etc.) as well as via masking other surface adhesins [59,60,61]. Long O-chains, necessary for bacterial protection against complement-mediated lysis, can also mask other important virulence factors.
The virulence of invasive bacteria Shigela flexnerii and Salmonella enterica critically depends on the functions of two type III secretion systems. It was shown that these bacteria possess mechanisms for adapting O-antigen length, allowing them to regulate their defensive and invasive virulence functions [62, 63].
E.coli K12, the famous laboratory strain was first thought not to produce O-antigens and consequently it had lost its capacity to survive in a natural environment where the outer core and O-chains are required. However, it was shown that the O-chains can be expressed in the wild strain [64].
5.3 Core Oligosaccharide Structures
The core oligosaccharide is built up of ten to fifteen monosaccharides arranged both in linear-and branched structures. It consists of two distinct regions: the “inner core” connected to the lipid A moiety, and the outer core to which the O-antigen can be linked [65]. The inner-core structures are generally more conserved, and in Enterobacteria they are made of Kdo and l-glycero-d-manno-heptose residues (Hep). The Kdo residue linked to the O-6’ position of lipid A through an α-ketosidic linkage is named Kdo-I, it can be glycosylated by one or two other Kdo residues (Kdo-II, and Kdo-III). Kdo-I is glycosylated at the O-4 position by Hep, designated as Hep-I, it can be decorated by P, PP or PEA, or by one or two other Hep or another sugar making the inner-core structure. The outer core in Enterobacteria consists of an oligosaccharide of up to 6 sugars with Glc, Gal, GlcN, all in pyranose form and in general having the α-anomeric configuration [2]. Figure 3.3 displays a few core examples like that of E. coli R3 core type which is present in the well-known serogroups O157, O111 and 026 (66, [67].
The major modifications of E. coli core structures involve non-stoichiometric incorporation of additional Kdo, phosphate, PEA, Rhamnose, GlcN. It was shown that when GlcN was incorporated, Hep-III was not phosphorylated. Some explanations have been given to explain the physiological impact of PEA addition, like sensitivity of the bacterium to antibiotics and detergents [68]. Another gene specific to PEA addition was involved in Ca+ hypersensitivity [69].
Proteus mirabilis, like E. coli, belongs to Enterobacteria, it is a facultative anaerobic bacterium found in soil, water, and human and animal intestinal microflora. This opportunistic bacterium is also responsible for skin wounds infections, and causes human upper and lower urinary tract infections, especially in patients with catheters [70]. A first unusual trait of the genus is to carry 4-amino-4-deoxy-ß-l-arabinose at the C-8 position of Kdo-I. Kdo is also found in the outer-core region on GlcN. These core structures also display interesting amide-linked amino-acid decorations with lysine, glycine, and alanine, but also an unusual core component like quinovosamine [71]. The LPS core structures, as well as the O-chains, can be used as markers for strain identification. Such epitopes, like terminal sugars, or non-sugar residues, are characteristic of given serogroups [70].
The Pseudomonas aeruginosa core also contains amino acids, and other non-carbohydrate substituents such as P, PPEA, acetyl and carbamoyl. Pseudomonas aeruginosa also synthesizes two types of cores, one being caped and covalently linked to the O-chains, the second uncapped and devoid of O-chain. In Pseudomonas a few modifications of the wild-type strain have been described such as a Glc residue only present in nine of the 20 serotype strains. Non-carbohydrate substitutions are the usual P, PPEA and O-acetyl groups. Another variability arises from truncation of the core structure; however such modifications are more frequently observed in laboratory strains than in clinical isolates [72].
5.4 Reducing Core Sugars
As already mentioned, in most of the described LPS structures, the reducing core sugar, linking the polysaccharide moiety to the lipid A region, is Kdo. This keto-deoxy-acid sugar is rarely present as a single element, it can be substituted by one or two other Kdo molecules, by phosphate groups [8, 40, 73] or by a GalA residue as in Xanthomonas campestris [74], thus keeping the highly negatively-charged level of this crucial region at the surface of the outer membrane. Kdo can also be substituted by Ko molecules [75], or itself being replaced by Ko [76]. All these different structures are illustrated in Fig. 3.4. The 2-deoxy-ketopyranosidic bond of Kdo being very labile in mild acidic conditions, permits the O-PS-core and lipidic moieties of LPS to be cleaved by mild hydrolytic conditions, without liberating or inducing too many modifications of other sugars or substituents. However, since the O-chain is often composed of deoxy-sugars, these can also be very labile and split at the same time, in this case the O-chain is released and this can be checked by MALDI-MS analysis.
Many different conditions have been described for Kdo anomeric bond cleavage since the beginning of the structural analyses of LPS. They varied from hydrochloric acid 1–2 M to much milder conditions including the frequently used 1% acetic acid method at pH 3.4 for one to 4 hours at 100 °C, and the mild pH 4.2 in sodium acetate buffer introduced by Rosner et al. [77]
Decades ago, LPS structures were often characterized by colorimetric tests to detect and estimate amounts of amino sugars, neutral sugars, and the thiobarbiturate test was used to detect Kdo molecules [78]. However, due to a chemical reaction, if a Kdo molecule carrying the sugar chain at position C-5 was also substituted at position C-4 by a phosphate group, a beta-elimination occurred with concomitant liberation of the phosphate and formation of a double bond between the C-3 and C-4 positions. This modification resulted in a negative response in the thiobarbiturate colorimetric test and the mistaken proposal of Kdo-less endotoxins. Among those listed at first were Vibrio LPS, Bordetella pertussis, B. parapertussis, B. bronchiseptica, Bacteroides, Aeromonas and other genera [79]. After LPS treatment with hydrofluorhydric acid to remove the Kdo phosphate groups, the colorimetric response was restored in the thiobarbiturate test, and since then the only Kdo-less endotoxins listed were those in which Kdo was replaced by Ko. Such a Kdo bearing phosphate at position C-4 was also mentioned in Haemophilus, and more recently described in Aeromonas salmonicida by detection with electrospray quadrupole orthogonal time of flight, and tandem mass spectrometry [80].
5.5 The Sugar Moiety of Bacterial Lipooligosaccharides (LOS)
As already mentioned, a number of highly virulent pathogens including Haemophilus influenzae, B. pertussis, Neisseria meningitidis, Neisseria gonorrhoeae, Campylobacter jejuni do not display an O-chain, but their complex and highly decorated core structures are responsible for serological specificity. The structure of Haemophilus influenza core is shown in Fig. 3.3. It can be decorated at the various positions, like indicated, by a glycine amino-acid substituent. It also often carries phosphoryl-choline residues [73, 81]. LOS vary with environment and culture conditions, they are often longer than usual cores, and can extend to 15 sugars. In a recent paper it was shown on non-typable H. influenzae (NTHi) that a distal Kdo or Neu5Ac could be added to the distal part of the structure on N-acetyl-lactosamine. However, Neu5Ac has to be present in the growth medium as it cannot be synthesized and has to be acquired from the environment. Studies using anti-Kdo antibodies showed that the terminal Kdo was widespread in biofilm environment and that Kdo could be used as a target of bactericidal antibodies [82]. Studies on Neisseria LOS showed that when PPEA and sialic acid are present on the core structure it induces serum resistance in vitro [83]. The Bordetella dodecasaccharide core structure will be described below in the paragraph dedicated to Bordetella LPS.
6 Core Structure-Function Relationships
The inner core structure is normally highly conserved within a genus or family. Negatively charged residues that make up this region contribute to interlink the adjacent LPS molecules, and thus to stabilize the outer membrane and prevent permeability of antimicrobial substances. Being more exposed to the external conditions, the outer-core region is more variable [65]. Such structural variability contributes to the bacterial serological specificity and may participate in bacterial adaptation to hosts and niches. Moreover, the core structure can be modified under environmental stress. Some of these modifications, e.g. substitution of the Hep-I residue with PEA and dephosphorylation of Hep-II in Salmonella, are controlled by the two component regulatory systems PmrAB. The first modification was shown to contribute to bacterial resistance against cationic antibiotics like Polymixin B, while the second has been implicated in bacterial resistance to Fe3+- mediated killing and survival in soil [84]. PEA substituting the Hep II residue in N. meningitidis LOS was identified as a site for complement C4b attachment. PEA at the 6-position of Hep II was more efficient than PEA at the C-3-position in binding C4b. Interestingly more than 70% of clinical isolates of N. meningitides express LOS molecules with PEA substituting Hep II at the C-3 position [85]. Core oligosaccharide structures are of particular importance for rough-type pathogens that do not have O-polysaccharide chains. In these bacteria, the core-oligosaccharide region is often branched and multiple lateral chains function in a similar way to O-chains in smooth-type bacteria. The phenomenon of molecular mimicry has been observed in N. meningitidis and H. influenza where their LOS structures are able to be decorated with sialic acid residues, thus mimicking the host gangliosides and reducing complement activation [11, 81, 86, 87] A. pleuropneumoniae LPS has been identified as the major molecule involved in cell adhesion, and the core oligosaccharide moiety was found to be responsible for adhesion to porcine respiratory tract cells and mucus [88,89,90,91].
7 Lipid A Structures
Lipid A is a key element and structure, for both bacterium and host, as it anchors LPS in the outer membrane of Gram-negative bacteria and carries most of the biologically deleterious and beneficial properties of LPSs. As an essential structure it is stabilized in the outer leaflet of the external membrane by two important properties, its natural charges interact to maintain molecules stability through bivalent cations bridges, and its hydrophobicity, due to the numerous fatty acids, which keep it stable in the outer membrane by interaction with other lipid molecules, mainly phospholipids.
In most Enterobacteria, and in most of the already described structures, lipid A consists in a bisphosphorylated β-1,6 glucosamine disaccharide backbone substituted via ester and amide bonds with a variable number (3–7) of fatty acids. Figures 3.5 and 3.6 display different examples of lipid A structures.
7.1 Number and Length of Fatty Acids
We demonstrated earlier that, in contrast to what was suggested by the E. coli paradigm studies, there could be considerable structural variability in lipid A structures within a genus. This was shown in the Bordetella genus, the example of lipid A heterogeneity which is presented below. The same lipid A structural variability was observed in the Yersinia genus. The best-known member of Yersiniae is Y. pestis, the agent of plague. Y. enterocolitica and Y. pseudotuberculosis cause enterogastritis [92, 93]. We have shown that different structures were present in different species grown at the same temperature although different growth temperatures could induce different structures. This result was confirmed by others and interpreted as a mechanism helping pathogenicity and host colonization as the lipid A structure grown at 37 °C is tetra-acylated and not recognized by the LPS receptor [94].
Helicobacter pylori presents relatively long FA with carbon chains of 18:0 and 16:0 [95, 96]. Some other genera present rather common FA length with one FA being of a high number of carbons as in Legionella pneumophila [97].
Halomonas pantelleriensis DSM9661Τ is a Gram-negative haloalkaliphilic bacterium isolated in the south of Italy from the sand of a volcanic lake. It is able to grow in harsh conditions in media containing 3–15% (w/v) salt and at pH as high as 9–10. LPS are thought to contribute to the restrictive membrane permeability properties of this bacterium, acting as an efficient barrier regulating the exchange with the environment ensuring survival in such harsh conditions. The lipid A structure displays a classical bis-phosphorylated disaccharide, but the FA are relatively short with four 12:0(3-OH) carrying one 12:0 and one 10:0 FA. Such structure is known for being associated to a low-level of cytokine induction [98].
7.2 Substitution of the Lipid A Phosphate Groups
The Lipid A phosphate groups can carry diverse substitutuents. ArnT (formerly PmrK) is a glycosyl transferase that has been shown to modify Salmonella and Pseudomonas lipids A with aminoarabinose [84, 99] and Francisella lipids A with GalN [100]. It was recently shown that glucosamine (GlcN) can also be a substituent on both phosphate groups in Bordetellae lipid A [101, 102].
The recently characterized lipid A of Campylobacter concisus, was shown to display a pyrophosphate group at C4’ of the GlcN backbone. This trait is different from the structure described for C. jejuni lipid A. It demonstrates that the degree of phosphorylation of the lipid A can be correlated to inflammatory potential and ability to induce immune tolerance [103]. Compared to C. jejuni, these LOS induced less TNF- α secretion in human monocytes and were less virulent. These characteristics and biological consequences strongly support the role of the LOS in pathogenicity.
7.3 Unusual Lipid A Backbone Structures
Different examples of the so-called “unusual structures” are presented in Fig. 3.6.
7.3.1 Lipid A with a 2,3-diamino-2,3-dideoxy-D-glucose (DAG) Backbones
The well-known glucosamine disaccharide backbone can be replaced by a diaminoglucose disaccharide in atypical lipid A structures such as Brucella, Legionella, Rhizobia and Ochrobactrum [104]. One of the main worldwide zoonosis also causing brucellosis Brucella abortus has a complex LPS structure [43, 105, 106]. It displays a DAG disaccharide substituted by 5–6 relatively long FA 16:0(3-OH), 18:0(3-OH), 20:0(3-OH) with PEA on one phosphate group.
Legionella pneumophila also displays the phosphorylated DAG disaccharide, but the FA, as shown in Fig. 3.5, are long and unusual [29, 97].
Bartonella henselae is a cat flea-borne Gram-negative zoonotic pathogen found in the feline reservoir host, transmitted to humans by scratch or bite. Its lipid A structure is made of a phosphorylated DAG disaccharide [107].
7.3.2 Lipid A with Mixed DAG and GlcN Backbones
Campylobacter lipid As were described to contain a mixture of three types of disaccharide backbones: the classical di-GlcN, the di-DAG backbone and a mixture of GlcN-DAG disaccharide [108].
To our knowledge, a lipid A structure with the backbone disaccharide containing GlcpN3N at the reducing and GlcpN at the non-reducing position has not been described. A lipid A composed of the mixed backbone was also identified in Acidithiobacillus thiooxidans (previously Thiobacillus thiooxidans) a proteobacterium that oxidises sulfur and produces sulfuric acid, and first isolated from the soil.
7.3.3 Lipid A with GlcN or DAG Disaccharide Backbones Without Phosphate Groups
In some lipid A structures, the glucosamine disaccharide does not carry the canonical phosphate groups, as occurs in Rhodomicrobium vannielii a phototropic bacterium [109]. The genus Rhodopseudomonas represents a major group of phototrophic purple non-sulfur (rod-shaped alpha-proteobacteria) growing in soil and aquatic environments like wastewater systems or marine costal sediments [110]. Rhodopseudomons palustris BisA53 lipid A has been recently described [111]. The DAG disaccharide is substituted by mannose (Man) and galactosaminuronic acid (GalNA) derivatives. A very long FA, C26 carries an acetyl group at C25.
The phototropic bacterium Rhodospirillum fulvum, with a classical lipid A di-GlcN backbone carries an unusual Hep residue at C-4’ and a GalNA residue at C-1 [112]. Rhizobia are Gram-negative soil bacteria capable of establishing a symbiotic relationship with legumes, the bacteria provide the plant with a source of nitrogen and is hosted by the plant in nodulas.
Rhizobia lipid A have been described as non-phosphorylated lipid As carrying long-chain FA and the GlcN 1 residue is replaced by an acylated 2-aminogluconate. Bradyrhizobium elkanii lipid A structure displays a DAG backbone with no phosphate groups, in their place the disaccharide has two mannose residues at C-4’, and one mannose at C-1 of the reducing DAG [113].
Bdellovibrio bacteriovorus are small, motile Gram-negative bacteria. They possess a predatory lifestyle and grow in the cytoplasm of other Gram-negative bacteria. They are found in the environment but also in the intestinal track of mammals, where their existence could involve a reduced count of the pathogenic Gram-negative bacteria. Their lipid A structure consist of a DAG disaccharide substituted by two Man residues [114].
Bradyrhizobium japonicum possesses lipid A molecular species with two unusual hopanoid residues [115].
Aquifex pyrophilus was isolated from hot-marine sediments, they represent a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria [116]. The structure of the lipid A of this hyperthermophilic bacterium consists in a DAG disaccharide with two 16:0(3-OH) and two 14:0(3-OH) at the amide groups. Each DAG is also substituted with one GalNA [117].
8 Lipid A Recognition Through the TLR-4 Receptor: How Structural Modifications Impact Innate Immunity and Favor Bacterial Virulence and Pathogenicity. Applications to Human Health
Pathogen-associated molecular pattern molecules (PAMPs) are characteristic molecules found on microorganisms and recognized by pattern recognition receptor (PRR), expressed by cells of the innate immune system, as well as by many epithelial cells, including dendritic cells (DC). PRR, including the Toll-like receptors (TLRs), allow the innate immune system to detect such conserved patterns and to respond, in the first line of defense, against infection by producing inflammatory cytokines. Lipid A moieties are the essential structures responsible for the activation of the TLR-4 pathway. LPS forms a complex with the myeloid differentiation factor 2 (MD-2) and the complex is recognized by the receptor inducing the biosynthesis of cytokines such as IL-1, IL-6, TNF- α, reactive oxygen species such as NO, O2 and antimicrobial peptides.
If all endotoxins are LPS, all LPS are not toxic although both names are often used as if they were interchangeable. We will comment on how some lipid A structures can be non-toxic or non-pyrogenic. The basic lipid A structures are modified during post-translational steps of LPS. Biosynthesis by the addition of different substituents on the phosphate groups with free amino compounds like EA, AraN, GalN or GlcN. Other late-stage modifications can be hydroxylation or removal of fatty acids and addition of others such as 16:0, originating from the bacterial membrane phospholipids.
The E. coli hexa-acyl lipid A structure, which is a strong cytokine inducer, can be taken as a standard. All lipid A structural modifications have a crucial impact on LPS recognition by the TLR-4/MD2 receptor complex, as illustrated by Park et al. [118] and therefore on LPS biological activities. The reactivity of the synthetic lipid IVa was tested on mouse and human TLR-4/MD2 receptors, for which solved crystal structures are known. The complexes between receptor and lipid A react differently. Lipid IVa, the synthetic tetra-acyl lipid A, reacts as an antagonist to humans, but as a weak agonist to mice [119]. When hexa-acyl lipid A and tetra-acyl lipid A complexes were compared, the tetra-acyl lipid induced a change of the di-GlcN-P backbone which was shifted upward and rotated compared to the hexa-acyl model. During this change in the rotation of the backbone the acyl chains were arranged differently in the human receptor, while it was not in the mouse receptor, which explains the difference in reactivity and confirms that animal models might not always be suitable for human medical research.
Structural modifications mostly take place under the control of the two-component systems controlling the expression of genes in response to external stimuli. A number of investigations have shown that these structural modifications contribute to bacterial adaptation and virulence, by increasing the resistance of the bacterial membrane against different stresses and antimicrobial substances, as well as by modulating the host immunity – pathogen interactions. When LPS is present in low amount during infection, this results in a low degree of inflammation suitable for activating the host human defense system in recognition of structural traits of the lipid A moiety found in most of the structures. However, when LPS are liberated in huge amounts this leads to major disorders due to a massive response and severe symptoms of sepsis are observed leading to septic shock. Therefore, LPS detection is a major and vital step when Gram-negative bacteria or LPS contamination are involved.
It has also been shown using extracted LPS that under-acylated lipid A structures are poor cytokine inducers. For example in Y. pestis grown at 37 °C this feature helps the plague agent to develop in humans, while the lipid A is hexa-acylated when grown at 27 °C in the vector flea [92]. The two Bordetella agents of whooping-cough, B. pertussis and B. parapertussis are underacylated, but also carry short-chain fatty acids (10:0(3-OH)). This also leads to poor cytokine inducing capacities due to poor stimulation of the TLR4/MD-2 complex [120]. A similar role for less-acylated lipid A structures has been described in other bacterial species, such as Francisella tularensis, Helicobacter pylori, and Porphyromonas gingivalis, the LPS of which are known as being poor cytokine inducers.
Another important structural endotoxic characteristic is the presence of the canonical phosphate groups on the lipid A GlcN disaccharide. The release, under acid treatment, of one of the phosphate groups was early demonstrated to lead to non-toxic and non-pyrogenic lipid A structures [120, 121] and also on Salmonella minnesota leading to the famous monophosphoryl lipid A (MPL) used as an adjuvant in different current vaccines [122]. In this case, the absence of the acidic group is crucial for interaction of lipid A with the receptor.
Therefore, all modifications of the canonical lipid A structure leading to hypo-acylation, addition of FA over the hexa-acyl high inducing structure, long-chain FA or short-chain ones, as well as changes at the phosphate groups help bacteria to escape the host-immune system. On the other hand, all the structure-function studies have led to an enormous amount of information which can be exploited for fighting diseases. This is the case with natural or synthetic vaccine adjuvants designed from detoxified lipid A structures. Lipid A-derived structures are also used in cancer therapy [123, 124]. LPS antigenic capacities can also be used in vaccines designed with use of core and O-chains.
In the past decades a lot of attention has been paid to the intestinal microbiome and its impact on health [125, 126]. It was shown that an increase in Gram-negative bacteria occurred in obese patients as well as in rich-diet mouse models. An increase in low-inflammatory LPS amount in the blood of patients with such metabolic diseases would explain the observed reaction leading to impact the insulin receptor [127]. We recently published structure-function relationships on Ralstonia lipid As showing that penta-acylated LPS molecules, thus low-inflammatory ones, could favor LPS diffusion through the intestinal membrane [128].
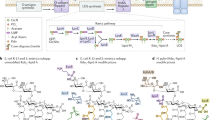
9 Brief Description of the LPS Biosynthetic Pathway
The biosynthesis pathway in E. coli and other genera has been well described in the work of the Raetz C.R.H. group, and is well-known as the “Raetz pathway” [129]. Lipid A biosynthesis is summarized in Fig. 3.7 (see also Chap. 4).
The complete LPS biosynthesis pathway starts with dimerization of GlcN followed by acylation of the amide groups, addition of phosphate and Kdo, then the lipid A-Kdo2 moiety molecule, which is first synthesized at the cytoplasmic surface of the cytoplasmic membrane. The other core sugars are added to Kdo, into the inner membrane, before the lipid A-core are flipped into the periplasmic side of the cytoplasmic membrane thanks to MsbA. The O-antigen is synthesized by cytoplasmic membrane-associated enzyme complexes using C55 Undecaprenyl-P as an acceptor for chain assembly, and is further flipped itself, to the periplasmic face of the membrane by one of the following three pathways: Wzy dependent, ABC transporter dependent and synthase dependent. When the LPS molecules have been transported across the periplasm and peptidoglycan, they are embedded into the outer membrane. The late stages of the biosynthesis, the addition of the so-called “decoration” (e.g. 16:0) happens by addition of such a residue originating from other membrane components, such as PPL or lipoproteins.
10 Origin of LPS Heterogeneity
LPS heterogeneity is well-known and some examples are described and illustrated below with sodium dodecyl sulphate electrophoresis profiles (SDS-PAGE), and mass spectrometry analysis. Heterogeneity appears at different levels of the LPS architecture and is mostly due to LPS biosynthesis in connection with culture conditions or niche adaptation.
The more important degree of heterogeneity observed in LPS, as shown by SDS-PAGE electrophoresis (Fig. 3.8) with the well-known “ladder-like” aspect of the high molecular weight species, corresponds to different lengths of the O-chain linked to the core during the biosynthetic addition step of this element. The difference between two consecutive rungs of the ladder is proportional to the number of sugars present in the oligosaccharide unit. During their biosynthesis, O-antigens are formed from polymerized chains of variable lengths added to the core on different LPS molecules. This results in the highly heterogeneous number and size of LPS molecules. In some cases, mainly for adaptive reasons related to bacterial virulence, the number of repeating units to be added to the O-chain is crucial. Examples in which a methyl group was added, as a step of biosynthesis, to the terminal sugar were described, in some other cases a different sugar can be added to the O-chain as a stop-signal [130].
Heterogeneity is well illustrated by the regular graduation of n + x O-chain oligosaccharide units as with Salmonella minnesota (2) and Actinobacillus pleuropneumoniae (3) while with E. coli O111B4 (1) the ladder rungs are missing in the middle of the profile. Such “selected” length of the O-chain is often related to virulence efficiency. The Bordetella LPS profiles [4,5,6,7,8,9] will be documented below. However, it is interesting to note here the peculiarity of B. hinzii and B.trematum LPS profile with an almost unique signal corresponding to three repeating units in addition to a linker moiety between core and O-chains for B. hinzii, and to two repeating units for B. trematum [131]. Some other examples of non ladder-like heterogeneity have been described in Rhizobia [132] and with Caulobacter crescentus while the reason of the well-define size and such unusual homogeneity of the O-chain is not yet understood.
In the last row (10) the reference Shigella flexneri mutant LPS corresponds to the shortest structure made of lipid A plus 2 Kdo molecules (see Fig. 3.4).
Other structural locations for LPS substituents, which add to the lipid A structural heterogeneity, are certainly the so-called “decorations” located on the GlcN phosphate groups like AraN and PEA with the Salmonella and other Enterobacteria [84], or constituents of a different nature, as described below in bordetellae. As these derivatives are non-stoichiometric substituents of the phosphate groups or other positions, they are another major cause of LPS heterogeneity.
Lateral gene transfer is also involved in changes in O-antigen structures, O-antigen conversion and phase variation are also involved. The modifications can be O-antigen size modification, O-acetylation of repeat-units oligosaccharides, but examples involving fucosylation or glucosylation are also described [36].
The lipid A moiety of LPS is also known for displaying peculiar molecular heterogeneity due to post-translational modifications, with the addition or removal of fatty acids by specific enzymes in late stage of the LPS biosynthesis with modifications as illustrated in Fig. 3.7. They are due to ArnT, LpxO, PagL, PagP and EptA enzymes [84, 133].
This is how the PhoP/PhoQ virulence gene regulates the expression of the lipid A PagL 3-O-deacylase, as well as the PagP palmitoyl transferase, moving 16:0 fatty acids from phospholipids, present in the outer membrane, to the lipid A moieties. The molecules being modified in this way have a decreased capacity to activate the TLR-4 signaling pathway, helping bacteria to escape the host-defense system [134]. The core structures can be heterogeneous themselves, and especially with the absence of branched sugars, and the presence or absence of “decorations” like phosphate groups, phosphoethanolamine, amino acid.
10.1 Impact of Biofilm Growth Conditions on LPS Structures
The impact of growth conditions on LPS structures has been well documented, the modifications occurring are similar to those observed in planktonic conditions with additional stress such as temperature, lack of oxygen or high-salt concentration. Therefore it is not surprising that laboratory bacterial growth conditions, aimed at favoring bacterial growth with constant addition of nutrients and oxygen, when necessary, can lead to different LPS structural profiles than those obtained for in vivo growth of pathogens.
P. aeruginosa is an opportunistic pathogen that causes different chronic infections due to its biofilm formation. Bacteria are thus protected from antimicrobial agents. The biofilms are made of a matrix, containing DNA, exopolysaccharides, lipids, and proteins. P. aeruginosa LPS structures have been compared in planktonic and biofilm growth conditions. We showed that when bacteria were grown in biofilm the isolated LPS induced slightly more inflammatory cytokines than those extracted from the planktonic counterpart. Differences in the structures were observed at the level of hydroxylation of the branched 12:0 FA [135]. Moreover, bacteria can undergo a differential expression of their O-antigens according to growth conditions or infection stage. Thus, during chronic lung infections, P. aeruginosa changes from a non-mucoid to a mucoid phenotype, that was shown to be accompanied by loss of B-band O-antigens [136]. Similar effects were observed in in vitro experiments on biofilm bacterial growth [137]. In addition to lipid A FA hydroxylation, we observed the complete loss of O-antigens by clinical isolates of P. aeruginosa grown in biofilms [135, 138]. The observed phenotype change was shown to be reversible. Once the biofilm bacteria were recultivated under batch conditions, the initial smooth-type phenotype was restored. The reversible character of the modifications suggested that the two-component regulatory systems may play an important role in regulation of O-antigen expression in response to changes of environmental conditions. It was shown, for example, that the RcsCDB Salmonella regulatory system activated by bacterial envelope stress, and exposure to CAMPs, plays a role in LPS biosynthesis and modification by regulating O-antigen production [139]. It is also known that in the Bordetella genus, O-antigen biosynthesis is controlled by the Bvg regulatory system [140].
It has been shown recently that the presence of P. aeruginosa in a biofilm favoured the growth of Fusobacterium nucleatum in the same biofilm [141]. The presence of P. aeruginosa in the ear fluid medium, reduced the level of dissolved oxygen to nearly anoxic condition within 4 hour after inoculation.
We also demonstrated that commensal and pathogenic E. coli grown in Biofilm showed increased palmitoylation in their lipid A, enhancing bacterial in vivo survival [142].
10.2 The Example of the Bordetella Genus LPS: High Structural Diversity and Heterogeneity
The Bordetella genus belongs to betaproteobacteria. A dozen species have been described currently in this genus [143, 144]. B. pertussis and B. parapertussis are the well-known agents of whooping cough [145, 146], a highly contagious disease, responsible for numerous deaths worldwide, especially in under-developed countries where vaccination is not available. The World Health Organization (WHO) has estimated that there were about 16 million cases of whooping cough and that almost 200,000 children died of the disease in 2008 [147].
Although vaccination, starting some decades ago, resulted in a dramatic decrease in disease incidence in developed countries, recent objection to adjuvants and vaccines led to new outbreaks. Serious negligence, combined with ill-informed suspicion, results in newborn infection by relatives who have not used the available boosters, and consequently transmit the disease to newborns, too young to be vaccinated.
The two major human pathogens responsible for whooping-cough evolved separately from a B. bronchiseptica-like progenitor [145, 146]. B. bronchiseptica infects many mammalian species, including humans, causing respiratory diseases [148]. B. avium, is a pathogen for birds [149]. B. hinzii [150], B. trematum B. holmesii [151], B. petrii [152] and B. ansorpi [153] were more recently described, some isolated from immunosuppressed patients [154]. An additional group of Bordetella hinzii-like species, including B. bronchialis, B. flabilis and B. sputigena [144] recently isolated from human respiratory samples, were proposed for classification according to various traits. At the same time, three new environmental Bordetella species isolated in Japan from a plaster-wall surface of 1300-year-old mural paintings were described, B. muralis, B. tumulicola, and B. tumbae [155, 156].
The SDS-PAGE LPS profiles corresponding to some of the major Bordetella species are presented in Fig. 3.8. Bordetella bronchiseptica LPS (4) is a smooth-type LPS with relatively short O-chain corresponding to a monosaccharide repeating unit, giving a smear in a well-define region, at about 6 kDa. B. hinzii Smooth-type LPSs correspond to a trisaccharide repeating unit and the O-chain displays three units of this kind. Two other units at plus and minus one repeating units give a discrete band on either side of the major unit. The reason of the selection of such a defined size of the O-chain is unknown. B. avium is also a smooth-type LPS (6) with more diffuse bands spread on a large Gaussian-type distribution of 5–13 monosaccharide repeating units. B. trematum LPS (7), like B. hinzii presents a specific O-chain unit limited to a small number of a trisaccharide repeating unit, two oligosaccharides in this case. This gives again a well-defined characteristic band with a second lighter band corresponding to lipid A plus core region in the LPS lower-mass region. B. pertussis 1414 LPS (8) gives a major band for this LOS corresponding to lipid A plus dodeca-saccharide [8]. B. pertussis A100 LPS (9) corresponding to a nona-saccharide LOS [157] migrates further in the gel.
A good example of LPS heterogeneity in the Bordetella genus is reported in a recent paper (131) where the complete structures of B. avium, B. hinzii and B. trematum are described in details thanks to new data concerning their free and O-chain-substituted cores, completing the already described lipid A [158] and O-chain structures [159,160,161,162]. It is thus described that the B. avium native LPS is particularly heterogeneous with the presence of 12 major lipid A molecular species, at least six free-core structures and four substituted core structures linked to O-chains of different variable lengths, the more diverse ones in the genus, extending from 5 to 13 repetitive units.
10.3 Lipid A
For a long time, based on the Escherichia and other Enterobacterial models, lipid A was described as the less variable structure in LPS molecules, and was described as a unique and common structure in a given genus. When the Bordetella LPS structures were explored and lipid A first characterized, it became obvious that the Bordetella genus displayed structure differences in almost each LPS species, and that this could be an exception [163].
Obviously the two human pathogens responsible for whooping cough, B. pertussis and B. parapertussis, had fatty acid patterns different from those found in B. bronchiseptica and other closely related members of the genus [158, 163,164,165,166,167].
A first trait in Bordetella lipid As is that the structure of the backbone is not symmetrical, the FA are differently distributed on the two GlcN residues.
Another trait was a reduced number of fatty acids reinforced by the presence of a shorter fatty acid carbon chain. The two human pathogens had four to five fatty acids in the various lipid A studied. In addition, they both displayed the presence of a short-chain fatty acid, the 3-hydroxydecanoic acid (10:0(3-OH)), although in different locations: on GlcN-I for B. pertussis and on GlcN-II for B. parapertussis (Fig. 3.9). These two distinctive characteristics were shown to result in a reduced degree of inflammation [120, 168] leading to the bacteria capacities to escape the host-immune system [20].
In the BP18323 strain, the lipid A structure displays two residues of 10:0(3-OH) rendering this LPS structure almost unable to induce cytokine production [169, 170] (Fig. 3.11).
The presence of longer carbon-chain fatty acids, or more FA chains, can also lead to escape host control. B. bronchiseptica lipid A is palmitoylated [166], an acylation attributed to a late biosynthetic enzymatic step mediated by PagP. This modification is required for persistent colonization of the mouse respiratory tract [166] and for resistance to antibody-mediated complement lysis during B.bronchiseptica respiratory infections. A second, more recent example, was found in human and rabbit isolates with the presence of two 16:0 fatty acid in lipid A structures related to bacterial virulence [171]. As already mentioned, the presence of 16:0 is a late biosynthetic step and uses 16:0 FA taken from membrane phospholipids after the LPS has been biosynthesized in the cytoplasm and transferred to the outer membrane. B. parapertussis lipid A can also carry two 16:0 FA [165] as shown in Fig. 3.9.
Other genera being studied in parallel by us, like the Yersinia [92, 172], the Helicobacter [95], and much later the Ralstonia [128], showed that the Bordetella genus was not an exception regarding the diversity of its lipid A structures.
The observed variations were later explained by lipid A structural modification in post-translational steps by the action of the lipid A biosynthesis enzymes.
10.4 Cores
According to B. pertussis LPS structure, the first of the genus to have been described, the core is a nonasaccharide as described in BPA100 and BP134 [157]. This core structure can be substituted by a trisaccharide turning the B-band LPS to the A-band structure as shown by SDS PAGE analysis, a transition enabled by the wlb locus. A- and B-bands were first observed and described by Peppler and [173] their size and structure were then explained and characterized by Caroff et al. [157]
B. parapertussis, the other agent of whooping-cough only displays the nonasaccharide core structure and not the distal trisaccharide [174]. B. bronchiseptica, the respiratory tract animal pathogen also found in some human infections displays the more heterogeneous LPS structure in the genus.
10.5 O-chains
The Bordetella O-chains, when present in the different species, have a peculiarity, they all contain different isomers of 2,3-diacetamido hexuronic acids (Fig. 3.10). The B. pertussis LPS, which does not produce an O-chain, contains the manno isomer of the 2,3-diacetamido uronic acid in its distal trisaccharide. We qualified this peculiarity as “Variation on a theme” in the B. avium structural characterization [159]. Effectively, LPS O-chains consisting of 1,4-linked 2,3- diacetamido- 2,3-dideoxy-l-galactopyranosyluronic acid residues were discovered in B. bronchiseptica and B. parapertussis with some heterogeneity first characterized by serology [175], then by structural analysis [176, 177]. The B. hinzii O-chain was found to be composed of trisaccharide repeating units of the gluco and galacto analogues of the same hexuronic acid [161, 162]. B. trematum O-chain is limited to two repetitive units differing in the anomeric bond of the first sugars. The structure corresponded to: ß-ManNAc3NAmA-(1-4)-ß- ManNAc3NAmA-(1-3)-α-FucNAc-(1-4)-ß-ManNAc3NAmA-(1-4)-ß-ManNAc3NAmA-(1-3)-ß-Fuc-NAc-(1-6) [160].
10.6 Bordetella LPS Biological Activities
We demonstrated earlier the significant impact of discrete structural differences on the B. pertussis lipid A on LPS recognition by TLR-4 receptor and consecutive inflammatory cytokine induction [168]. The first characterized lipid A structure in this genus was that of the vaccine strain BP1414 [163], Fig. 3.9. It was later demonstrated that this strain, in contrast to strain BP18323, has the ability to modify its lipid A phosphate groups with glucosamine, a very rare substituent in such position which seems to be a characteristic of the genus found in different other species like B. bronchiseptica [101], B. avium [158], and B. holmesii [164], This substituent on both phosphate groups greatly increased the inflammatory cytokines induction. Another modification observed in the BP18323 strain, compared to BP338, was the slight shortening of the fatty acid carbon chain at C-3’, replacing 14:0-(3OH) by the 12 or 10 carbons FA equivalents resulting in the corresponding LPS being a TLR-4 antagonist. This strain is a poor inducer in human and murine macrophages and is greatly impaired in TLR-4-mediated activation of nuclear factor–κB in transfected HEK-293 cells [169, 170].
Two inflammatory cytokine responses to the different LPS structures characterized in this work were compared (Fig. 3.11). IL-6 and TNF-α secretions were tested for the three LPS as compared to B. pertussis BP18-323 LOS, well known as a low inducer, and to Escherichia coli J5 rough-type LPS for a highly potent inducer reference. The activity of the E. coli J5 LPS was higher, as expected, than that of B. pertussis low responder LPS, then B. trematum followed by B. hinzii at almost the same inducing capacities were found at higher levels, and B. avium was the highest inducer.
LPS structure-activity relationships is a very complex topic. A non-exhaustive list of important structural parameters includes first of all, the primary lipid A structure: the number, length and distribution of fatty acids, the number of phosphate groups and presence or absence of structural elements substituting the phosphate groups (e.g. PPEA, Ara4N or GlcN). The core oligosaccharide is also important, affecting the lipid A region three-dimensional conformation and the size of the supra-molecular aggregates. The latter are considered as LPS biologically active units. Biological activities of a LOS, or of a rough-type LPS, are normally higher than those of their isolated lipid A regions [168, 178]. It is also widely accepted that activities of a smooth type LPS are generally higher than those of a rough-type LPS having the same or a similar lipid A structure. The latter is most probably due to the higher solubility of smooth-type LPS and to a smaller size of their aggregates [18]. The respiratory pathogens B. pertussis and B. bronchiseptica have a particular terminal trisaccharide in their core structures. The latter was shown to be a crucial factor for bacterial protection against bactericidal functions of pulmonary collectins [179]. A progressive loss of O-antigens was observed during a chronic infection caused by B. bronchiseptica [180]. The same was described for B. avium [181].
The data on the potencies of different LPS under study to stimulate secretion of proinflammatory cytokines could be explained by these general trends. The low inducing potency of the LOS from B. pertussis known to be due to the small number of fatty acids (from 4 to 5) and to the presence of a short-chain fatty acid 10:0(3-OH). The LPS from E. coli J5 is also a Rough-type LPS with a very potent lipid A structure. This structure contains from 4 to 6 fatty acids mostly of 14 carbon atoms, a length which is “optimal” for the TLR4 signaling. Although distributed differently over the di-glucosamine backbone, the fatty acid composition of the LPS from B. trematum, B. hinzii and B. avium is similar to that of the LPS from E. coli J5 (from 4 to 6 fatty acids containing mostly 14 carbon atoms). However, all three Bordetella LPS are of a smooth type. This feature may explain their cytokines inducing capacities being substantially higher than those of the LPS from E. coli J5. Finally, the LPS from B. avium is substantially more potent than the B. trematum and B. hinzii LPSs. This is certainly due to the high level of substitution of its lipid A phosphate groups with glucosamine. As we showed earlier in the B. pertussis model, such a modification of the lipid A structure leads to a drastic increase in NFκB transcription via TLR4 pathway and in the resulting cytokines secretion of cytokines [102].
11 Other Examples of Heterogeneity and Diversity
11.1 Example: Salmonella minnesota R595 Re LPS Heterogeneity Shown by Thin Layer Chromatography (TLC), and Matrix Assisted Laser Desorption (MALDI) Mass Spectrometry
This LPS from the Re S. minnesotta mutant strain is well-known, and became a standard for biological tests due to the fact it was supposed to be the “simplest” LPS structure, only made of lipid A plus two Kdo molecules. However, the heterogeneity of this deep-rough mutant LPS, due to different decorations is maximal and depending on batches of commercial providers, even from the same source, there is no reproducibility of the structure and this non-described heterogeneity is certainly responsible for non-reproducible activities. We highlighted this problem early when we set up the first mass spectrometry method for LPS analysis [182,183,184], as well as more recently [185]. Here, we compared the structure of three different batches from the same provider to the non-decorated Shigella flexneri Re strain, being much more homogeneous. As seen in Fig. 3.12a the numerous bands observed in the three S.minnesota Re LPS (A,B,C) illustrates a high degree of heterogeneity compared to the less heterogeneous S. flexneri sample (D).
MALDI mass spectrometry of the different samples presented on Fig. 3.12b also illustrates the high heterogeneity with information on the masses of the different molecular species. Mass differences between molecular species were observed and corresponded to fatty acids or PPEA, as well as Ara4N.
A comparison of the Limulus ameabocyte lysate LPS estimation test (Fig. 3.12c) is added together with estimation of the LPS amount by dosage of the 14:0(3-OH) content to give an idea of the influence of different “decorations” on LPS detection by varying tests. Apparently in this peculiar example the decoration on the phosphate groups impact the LAL response more than the FA estimation. In other examples, the LAL could be more accurate [38].
11.2 Example: Vibrio cholerae Lipid A Heterogeneity Shown by MALDI Mass Spectrometry
Vibrio cholera, the causative agent of cholera, has been classified in serogroups on the basis of its somatic antigens, the O-antigens. At least 206 serogroups are known [186]. In the early 1990s V. cholera O139 appeared to be the first non-O1 serogroup associated with the disease, and extensive outbreaks were recorded in Bangladesh and India. Cases caused by V. cholerae O139 have also been reported in China, Nepal, Pakistan, Thailand, Afghanistan, Kazakhstan, and Malaysia [187].
The O-antigen structure of the Smooth-type V. cholerea O1 and the Semi-Rough-type V. cholerae O139 have been described [188,189,190,191,192,193]. The lipid A structures of V. cholerae O139 and V. cholerae O1 were described as having an asymmetric distribution of FA on the bisphosphorylated- di-GlcN backbone. Two 14:0(3-OH) and two 12:0(3-OH) substitute respectively positions C-2 and C-2’, C-3 and C-3’. 12:0 and 14:0 are present in secondary acylation at C-3’ and C-2’. The 12:0 FA can be replaced by 12:0(3-OH) at C-3’ thanks to LpxN.
V. cholerae El Tor is resistant to the cationic antimicrobial polymyxin peptide while the classical biotype is sensitive to it. The difference is due to AlmG transfering Glycine 1 or 2 (Gly) to the hydroxyl group of the 12:0(3-OH) in El Tor responsible for polymyxin resistance [194,195,196].
When we compared lipid A mass spectra from the two different V. cholerae strains, we observed important heterogeneity and the presence of different molecular species illustrating the presence of one or two glycine substituents, as well as a putative phosphoglycerol substituent already described in V. fisherii as a 16:0 carrier for one or two of the FAs [197]. All these “decorations” added in a non-stoichiometric way increase LPS heterogeneity (Fig. 3.13).
12 Conclusions
LPS diversity and heterogeneity has been demonstrated and illustrated here, with some specific and previously described examples, as well as new ones. They can be interpreted in terms of evolutionary pressure and host adaptation. The structural characterization of native LPS structures is a complex task which has only been realized in the past decades with increasing efficiency thanks to improvement of methods and technologies. However, such heterogeneity will always complicate the task of LPS structural characterization, and render structure-function relationships more complicated as compared to many other molecules. Different methods have been described and their use is unavoidable for injectable drugs and endotoxin detection in patients. The use of a single method would be difficult for quantifying efficiently LPS with such diverse structures, and in such different media as required. In some peculiar conditions, like in blood, the use of a complementary method based on other LPS characteristics can be of a great help.
Abbreviations
- Ala:
-
alanine
- Lys:
-
lysine
- Ara4N:
-
4-amino-4deoxy-ß-L-arabinose
- DAG:
-
2,3-diamino-2,3-dideoxy-D-glucose
- DC:
-
dendritic cells
- EA:
-
ethanolamine
- FA:
-
fatty acid
- GalN:
-
galactosamine
- GalNA:
-
galactosaminuronic acid
- Glc:
-
glucose
- GlcN:
-
glucosamine
- GlcNAc:
-
N-acetyl-glucosamine.
- Gly:
-
glycine
- Hep:
-
L-glycero-D-manno-heptopyranose
- IL:
-
Interleukin
- Kdo:
-
3-deoxy-D-manno-oct-2-ulopyranosonic acid
- Ko:
-
D-glycero-D-talo-oct-2-ulosylonate
- LPS:
-
lipopolysaccharide
- LOS:
-
lipooligosaccharide
- Man:
-
mannose
- NO:
-
nitric oxide
- PBS:
-
phosphate buffered saline
- P:
-
phosphate
- PEA:
-
phosphoethanolamine
- PS:
-
polysaccharide
- TLR:
-
Toll-like receptor
- TLC:
-
thin-layer chromatography
- TNF:
-
tumor necrosis factor
References
Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700.
Caroff M, Karibian D. Structure of bacterial lipopolysaccharides. Carbohydr Res. 2003;338:2431–47.
Powers MJ, Trent MS. Expanding the paradigm for the outer membrane: acinetobacter baumannii in the absence of endotoxin. Mol Microbiol. 2018;107:47–56.
Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656.
Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta. 2009;1794:808–16.
Henderson JC, et al. The power of asymmetry: architecture and assembly of the Gram- negative outer membrane lipid bilayer. Annu Rev Microbiol. 2016;70:255–78.
Belunis CJ, Clementz T, Carty SM, Raetz CR. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene in Escherichia coli. J Biol Chem. 1995;270:27646–52.
Caroff M, Brisson J, Martin A, Karibian D. Structure of the Bordetella pertussis 1414 endotoxin. FEBS Lett. 2000;477:8–14.
Albitar-Nehme S, et al. Comparison of lipopolysaccharide structures of Bordetella pertussis clinical isolates from pre- and post-vaccine era. Carbohydr Res. 2013;378:56–62.
Kahler CM, Stephens DS. Genetic basis for biosynthesis, structure, and function of meningococcal lipooligosaccharide (endotoxin). Crit Rev Microbiol. 1998;24:281–334.
Phillips NJ, Apicella MA, Griffiss JM, Gibson BW. Structural studies of the lipooligosaccharides from Haemophilus influenzae type b strain A2. Biochemistry. 1993;32:2003–12.
Comstock LE, Kasper DL. Bacterial glycans: key mediators of diverse host immune responses. Cell. 2006;126:847–50.
Le Blay K, Caroff M, Blanchard F, Perry MB, Chaby R. Epitopes of Bordetella pertussis lipopolysaccharides as potential markers for typing of isolates with monoclonal antibodies. Microbiology. 1996;142(Pt 4):971–8.
Hoshino K, et al. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–52.
Ulevitch RJ, Tobias PS. Recognition of gram-negative bacteria and endotoxin by the innate immune system. Curr Opin Immunol. 1999;11:19–22.
Shimazu R, et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–82.
Caroff M, Karibian D, Cavaillon JM, Haeffner-Cavaillon N. Structural and functional analyses of bacterial lipopolysaccharides. Microbes Infect. 2002;4:915–26.
Gutsmann T, Schromm AB, Brandenburg K. The physicochemistry of endotoxins in relation to bioactivity. Int J Med Microbiol. 2007;297:341–52.
Pasteur L. Recherches sur la putrefaction. Comptes Rendus Hebd Seances Acad Sci. 1863;56:1189–94.
Cavaillon J-M. Historical links between toxinology and immunology. Pathog Dis. 2018;76(3):fty019.
Kaufmann SHE, Schaible UE. 100th anniversary of Robert Koch’s Nobel Prize for the discovery of the tubercle bacillus. Trends Microbiol. 2005;13:469–75.
Nauts HC, Swift WE, Coley BL. The treatment of malignant tumors by bacterial toxins as developed by the late William B. Coley, M.D., reviewed in the light of modern research. Cancer Res. 1946;6:205–16.
Beutler B, et al. Identity of tumour necrosis factor and the macrophage-secreted factor cachectin. Nature. 1985;316:552–4.
Boivin A, Mesrobeanu J, Mesrobeanu L. Technique pour la preparation des polysaccharides microbiens specifiques. Compt Rend Soc Biol. 1933;113:490.
Westphal O, Lüderitz O, Bister F. Über die extraktion von bakterien mit phenol/wasser. Z Für Naturforschung B. 1952;7:148–55.
Staub AM, Tinelli R. Attempted identification of O antigens of Salmonellae by means of periodic oxidation of specific polysaccharides. Comptes Rendus Hebd Seances Acad Sci. 1956;243:1460–3.
Staub AM, Tinelli R, Luderitz O, Westphal O. Immunochemical study of Salmonella. V. Role of various sugars, especially 3, 6-bis-desoxyhexoses, in the specificity of Kauffmann-White O antigens. Ann Inst Pasteur. 1959;96:303–32.
Galanos C, Lüderitz O, Westphal O. A new method for the extraction of R lipopolysaccharides. Eur J Biochem. 1969;9:245–9.
Sonesson A, Jantzen E, Bryn K, Larsson L, Eng J. Chemical composition of a lipopolysaccharide from Legionella pneumophila. Arch Microbiol. 1989;153:72–8.
Morrison DC, Leive L. Fractions of lipopolysaccharide from Escherichia coli O111:B4 prepared by two extraction procedures. J Biol Chem. 1975;250:2911–9.
Nurminen M, Vaara M. Methanol extracts LPS from deep rough bacteria. Biochem Biophys Res Commun. 1996;219:441–4.
Delahooke DM, Barclay GR, Poxton IR. A re-appraisal of the biological activity of bacteroides LPS. J Med Microbiol. 1995;42:102–12.
Eidhin DN, Mouton C. A rapid method for preparation of rough and smooth lipopolysaccharide from Bacteroides, Porphyromonas and Prevotella. FEMS Microbiol Lett. 1993;110:133–8.
Caroff M. Novel method for isolating endotoxins. WO/2004/062690. International Application N° PCT/FR2003/003617. 2004.
Latino L, Caroff M, Pourcel C. Fine structure analysis of lipopolysaccharides in bacteriophage-resistant Pseudomonas aeruginosa PAO1 mutants. Microbiol Read Engl. 2017;163:848–55.
Lerouge I, Vanderleyden J. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol Rev. 2002;26:17–47.
Stenutz R, Weintraub A, Widmalm G. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol Rev. 2006;30:382–403.
Liu B, et al. Structure and genetics of Shigella O antigens. FEMS Microbiol Rev. 2008;32:627–53.
Wang L, Andrianopoulos K, Liu D, Popoff MY, Reeves PR. Extensive variation in the O-antigen gene cluster within one Salmonella enterica serogroup reveals an unexpected complex history. J Bacteriol. 2002;184:1669–77.
Chatterjee SN, Chaudhuri K. Lipopolysaccharides of Vibrio cholerae. I Physical and chemical characterization. Biochim Biophys Acta. 2003;1639:65–79.
Vinogradov E, St Michael F, Cox AD. Structure of the LPS O-chain from Fusobacterium nucleatum strain MJR 7757 B. Carbohydr Res. 2018;463:37–9.
Di Lorenzo F, et al. Xanthomonas citri pv. citri Pathotypes: LPS structure and function as microbe-associated molecular patterns. ChemBiochem. 2017;18:772–81.
Caroff M, Bundle DR, Perry MB, Cherwonogrodzky JW, Duncan JR. Antigenic S-type lipopolysaccharide of Brucella abortus 1119-3. Infect Immun. 1984;46:384–8.
Caroff M, Bundle DR, Perry MB. Structure of the O-chain of the phenol-phase soluble cellular lipopolysaccharide of Yersinia enterocolitica serotype O:9. Eur J Biochem. 1984;139:195–200.
Lee C-H, Schaffner CP. Perimycin: chemistry of perosamine. Tetrahedron Lett. 1966;7:5837–40.
Perry MB, MacLean L, Griffith DW. Structure of the O-chain polysaccharide of the phenol-phase soluble lipopolysaccharide of Escherichia coli 0:157:H7. Biochem Cell Biol. 1986;64:21–8.
Jones MD, Vinogradov E, Nomellini JF, Smit J. The core and Opolysaccharide structure of the Caulobacter crescentus lipopolysaccharide. Carbohydr Res. 2015;402:111–7.
Smit J, et al. Structure of a novel lipid A obtained from the lipopolysaccharide of Caulobacter crescentus. Innate Immun. 2008;14:25–37.
Cabeen MT, et al. Mutations in the lipopolysaccharide biosynthesis pathway interfere with Crescentin-mediated cell curvature in Caulobacter crescentus. J Bacteriol. 2010;192:3368–78.
Lam JS, Taylor VL, Islam ST, Hao Y, Kocíncová D. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol. 2011;2:118.
Rocchetta HL, Burrows LL, Lam JS. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 1999;63:523–53.
Pier GB. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int J Med Microbiol. 2007;297:277–95.
Joiner KA. Complement evasion by bacteria and parasites. Annu Rev Microbiol. 1988;42:201–30.
Saldías MS, Ortega X, Valvano MA. Burkholderia cenocepacia O antigen lipopolysaccharide prevents phagocytosis by macrophages and adhesion to epithelial cells. J Med Microbiol. 2009;58:1542–8.
Fernandez-Prada CM, et al. Interactions between Brucella melitensis and human phagocytes: bacterial surface O-Polysaccharide inhibits phagocytosis, bacterial killing, and subsequent host cell apoptosis. Infect Immun. 2003;71:2110–9.
Moran AP. Relevance of fucosylation and Lewis antigen expression in the bacterial gastroduodenal pathogen Helicobacter pylori. Carbohydr Res. 2008;343:1952–65.
Jacques M. Role of lipo-oligosaccharides and lipopolysaccharides in bacterial adherence. Trends Microbiol. 1996;4:408–9.
Edwards NJ, et al. Lewis X structures in the O antigen side-chain promote adhesion of Helicobacter pylori to the gastric epithelium. Mol Microbiol. 2000;35:1530–9.
Clements A, et al. The major surface-associated saccharides of Klebsiella pneumoniae contribute to host cell association. PLoS One. 2008;3:e3817.
Ivanov IE, et al. Relating the physical properties of Pseudomonas aeruginosa lipopolysaccharides to virulence by atomic force microscopy. J Bacteriol. 2011;193:1259–66.
Bilge SS, Vary JC, Dowell SF, Tarr PI. Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect Immun. 1996;64:4795–801.
Hölzer SU, Schlumberger MC, Jäckel D, Hensel M. Effect of the O-antigen length of lipopolysaccharide on the functions of Type III secretion systems in Salmonella enterica. Infect Immun. 2009;77:5458–70.
West NP, et al. Optimization of virulence functions through glucosylation of Shigella LPS. Science. 2005;307:1313–7.
Liu D, Reeves PR. Escherichia coli K12 regains its O antigen. Microbiology. 1994;140(Pt 1):49–57.
Holst O. Structure of the lipopolysaccharide core region. In: Knirel YA, Valvano MA, editors. Bacterial Lipopolysaccharides. Wien: Springer; 2011. p. 21–39. https://doi.org/10.1007/978-3-7091-0733-1_2.
Amor K, et al. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect Immun. 2000;68:1116–24.
Currie CG, Poxton IR. The lipopolysaccharide core type of Escherichia coli O157:H7 and other non-O157 verotoxin-producing E. coli. FEMS Immunol Med Microbiol. 1999;24:57–62.
Yethon JA, Heinrichs DE, Monteiro MA, Perry MB, Whitfield C. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli Lipopolysaccharide and their role in the formation of a stable outer membrane. J Biol Chem. 1998;273:26310–6.
Reynolds CM, Kalb SR, Cotter RJ, Raetz CRH. A phosphoethanolamine transferase specific for the outer 3-deoxy-D-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide identification of the eptB gene and ca2+ hypersensitivity of an eptB deletion mutant. J Biol Chem. 2005;280:21202–11.
Kaca W, et al. Serotyping of Proteus mirabilis clinical strains based on lipopolysaccharide O-polysaccharide and core oligosaccharide structures. Biochem Mosc. 2011;76:851–61.
Aquilini E, Merino S, Knirel YA, Regué M, Tomás JM. Functional identification of Proteus mirabilis eptC gene encoding a core lipopolysaccharide phosphoethanolamine transferase. Int J Mol Sci. 2014;15:6689–702.
Kocincova D, Lam JS. Structural diversity of the core oligosaccharide domain of Pseudomonas aeruginosa lipopolysaccharide. Biochem Mosc. 2011;76:755–60.
Schweda EKH, Richards JC, Hood DW, Moxon ER. Expression and structural diversity of the lipopolysaccharide of Haemophilus influenzae: implication in virulence. Int J Med Microbiol IJMM. 2007;297:297–306.
Silipo A, et al. The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris. J Biol Chem. 2005;280:33660–8.
Isshiki Y, Zähringer U, Kawahara K. Structure of the core-oligosaccharide with a characteristic D-glycero-alpha-D-talo-oct-2-ulosylonate-(2-->4)-3-deoxy-D-manno-oct-2- ulosonate [alpha-Ko-(2-->4)-Kdo] disaccharide in the lipopolysaccharide from Burkholderia cepacia. Carbohydr Res. 2003;338:2659–66.
Kawahara K, Brade H, Rietschel ET, Zähringer U. Studies on the chemical structure of the core-lipid A region of the lipopolysaccharide of Acinetobacter calcoaceticus NCTC 10305. Detection of a new 2-octulosonic acid interlinking the core oligosaccharide and lipid A component. Eur J Biochem. 1987;163:489–95.
Rosner MR, Tang J, Barzilay I, Khorana HG. Structure of the lipopolysaccharide from an Escherichia coli heptose-less mutant. I. Chemical degradations and identification of products. J Biol Chem. 1979;254:5906–17.
Weissbach A, Hurwitz J. The formation of 2-keto-3-deoxyheptonic acid in extractsof Escherichia coli B. I. Identification. J Biol Chem. 1959;234:705–9.
Caroff M, Lebbar S, Szabó L. Do endotoxins devoid of 3-deoxy-D-manno-2- octulosonic acid exist? Biochem Biophys Res Commun. 1987;143:845–7.
Sioud S, Jahouh F, Nashed M, Joly N, Banoub JH. Determination of distinctive carbohydrate signatures obtained from the Aeromonas hydrophila (chemotype II) core oligosaccharide pinpointing the presence of the 4-O-phosphorylated 5-O-linked Kdo reducing end group using electrospray ionization quadrupole orthogonal time-of-flight mass spectrometry and tandem mass spectrometry. Rapid Commun Mass Spectrom. 2010;24:2475–90.
Post DMB, et al. Comparative analyses of the lipooligosaccharides from nontypeable Haemophilus influenzae and Haemophilus haemolyticus show differences in sialic acid and phosphorylcholine modifications. Infect Immun. 2016;84:765–74.
Apicella MA, et al. Nontypeable haemophilus influenzae lipooligosaccharide expresses a terminal ketodeoxyoctanoate in vivo, which can be used as a target for bactericidal antibody. MBio. 2018;9:e01401–18.
John CM, et al. Lipooligosaccharide structures of invasive and carrier isolates of Neisseria meningitidis are correlated with pathogenicity and carriage. J Biol Chem. 2015;291:3224–38. https://doi.org/10.1074/jbc.M115.666214.
Richards SM, Strandberg KL, Gunn JS. Salmonella-regulated lipopolysaccharide modifications. Subcell Biochem. 2010;53:101–22.
Ram S, et al. Neisserial lipooligosaccharide is a target for complement component C4b. Inner core phosphoethanolamine residues define C4b linkage specificity. J Biol Chem. 2003;278:50853–62.
Moran AP, Prendergast MM, Appelmelk BJ. Molecular mimicry of host structures by bacterial lipopolysaccharides and its contribution to disease. FEMS Immunol Med Microbiol. 1996;16:105–15.
Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–22.
Rioux S, Bégin C, Dubreuil JD, Jacques M. Isolation and characterization of LPS mutants of Actinobacillus pleuropneumoniae serotype 1. Curr Microbiol. 1997;35:139–44.
Rioux S, et al. Isolation and characterization of mini-Tn10 lipopolysaccharide mutants of Actinobacillus pleuropneumoniae serotype 1. Can J Microbiol. 1999;45:1017–26.
Labrie J, et al. Identification of genes involved in biosynthesis of Actinobacillus pleuropneumoniae serotype 1 O-antigen and biological properties of rough mutants. J Endotoxin Res. 2002;8:27–38.
Ramjeet M, et al. Truncation of the lipopolysaccharide outer core affects susceptibility to antimicrobial peptides and virulence of Actinobacillus pleuropneumoniae serotype 1. J Biol Chem. 2005;280:39104–14.
Aussel L, et al. Novel variation of lipid A structures in strains of different Yersinia species. FEBS Lett. 2000;465:87–92.
Therisod H, Karibian D, Perry MB, Caroff M. Structural analysis of Yersiniapseudotuberculosis ATCC 29833 lipid A. Int J Mass Spectrom. 2002;219:549–57.
Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in lipid A structure in the pathogenic yersiniae. Mol Microbiol. 2004;52:1363–73.
Thérisod H, Monteiro MA, Perry MB, Caroff M. Helicobacter mustelae lipid A structure differs from that of Helicobacter pylori. FEBS Lett. 2001;499:1–5.
Mattsby-Baltzer I, Mielniczuk Z, Larsson L, Lindgren K, Goodwin S. Lipid A in Helicobacter pylori. Infect Immun. 1992;60:4383–7.
Zähringer U, et al. The lipopolysaccharide of legionella pneumophila serogroup 1 (strain Philadelphia 1): chemical structure and biological significance. Prog Clin Biol Res. 1995;392:113–39.
Carillo S, et al. Structural characterization of the lipid A from the LPS of the haloalkaliphilic bacterium Halomonas pantelleriensis. Extremophiles. 2016;20:687–94.
Moskowitz SM, Ernst RK, Miller SI. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of Aminoarabinose to Lipid A. J Bacteriol. 2004;186:575–9.
Shaffer SA, Harvey MD, Goodlett DR, Ernst RK. Structural heterogeneity and environmentally regulated remodeling of Francisella tularensis subspecies novicida lipid A Characterized by tandem mass spectrometry. J Am Soc Mass Spectrom. 2007;18:1080–92.
Marr N, Tirsoaga A, Blanot D, Fernandez R, Caroff M. Glucosamine found as a substituent of both phosphate groups in Bordetella lipid A backbones: role of a BvgASactivated ArnT ortholog. J Bacteriol. 2008;190:4281–90.
Marr N, et al. Substitution of the Bordetella pertussis lipid A phosphate groups with glucosamine is required for robust NF-kappaB activation and release of proinflammatory cytokines in cells expressing human but not murine Toll-like receptor 4-MD-2-CD14. Infect Immun. 2010;78:2060–9.
Brunner K, et al. Novel campylobacter concisus lipooligosaccharide is a determinant of inflammatory potential and virulence. J Lipid Res. 2018;59:1893–905. https://doi.org/10.1194/jlr.M085860.
Velasco J, et al. Structural studies on the lipopolysaccharide from a rough strain of Ochrobactrum anthropi containing a 2,3-diamino-2,3-dideoxy-D-glucose disaccharide lipid A backbone. Carbohydr Res. 1998;306:283–90.
Lapaque N, et al. Characterization of Brucella abortus lipopolysaccharide macrodomains as mega rafts. Cell Microbiol. 2006;8:197–206.
Bundle DR, Cherwonogrodzky JW, Caroff M, Perry MB. The lipopolysaccharides of Brucella abortus and B. melitensis. Ann Inst Pasteur Microbiol. 1987;138:92–8.
Zähringer U, et al. Structure and biological activity of the short-chain lipopolysaccharide from Bartonella henselae ATCC 49882T. J Biol Chem. 2004;279:21046–54.
Moran AP, et al. Structural analysis of the lipid A component of Campylobacter jejuni CCUG 10936 (serotype O:2) lipopolysaccharide. Description of a lipid A containing a hybrid backbone of 2-amino-2-deoxy-D-glucose and 2,3-diamino-2,3-dideoxy-Dglucose. Eur J Biochem. 1991;198:459–69.
Holst O, Borowiak D, Weckesser J, Mayer H. Structural studies on the phosphate-free lipid A of Rhodomicrobium vannielii ATCC 17100. Eur J Biochem. 1983;137:325–32.
Okamura K, Takata K, Hiraishi A. Intrageneric relationships of members of the genus Rhodopseudomonas. J Gen Appl Microbiol. 2009;55:469–78.
Di Lorenzo F, et al. The lipid A from Rhodopseudomonas palustris strain BisA53 LPS possesses a unique structure and low Immunostimulant properties. Chem Eur J. 2017;23:3637–47.
Rau H, Seydel U, Freudenberg M, Weckesser J, Mayer H. Lipopolysaccharide of the Phototrophic BacteriumRhodospirillum fulvum. Syst Appl Microbiol. 1995;18:154–63.
Komaniecka I, Choma A, Lindner B, Holst O. The structure of a novel neutral lipid A from the lipopolysaccharide of Bradyrhizobium elkanii containing three mannose units in the backbone. Chem Weinh Bergstr Ger. 2010;16:2922–9.
Schwudke D, et al. The obligate predatory Bdellovibrio bacteriovorus possesses a neutral lipid A containing α-D-Mannoses that replace phosphate residues similarities and differences between the lipid as and the lipopolysaccharides of the wild type strain B. bacteriovorus HD100 and its host-independent derivative HI100. J Biol Chem. 2003;278:27502–12.
Komaniecka I, et al. Occurrence of an unusual hopanoid-containing lipid A among lipopolysaccharides from Bradyrhizobium species. J Biol Chem. 2014;289:35644–55.
Huber R, et al. Aquifex pyrophilus gen. nov. sp. nov., represents a novel group of marine Hyperthermophilic hydrogen-oxidizing bacteria. Syst Appl Microbiol. 1992;15:340–51.
Plötz BM, Lindner B, Stetter KO, Holst O. Characterization of a novel Lipid A Containing D-galacturonic acid that replaces phosphate residues the structure of the lipid A of the lipopolysaccharide from the hyperthermophilic bacterium Aquifex pyrophilus. J Biol Chem. 2000;275:11222–8.
Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4- MD-2 complex. Nature. 2009;458:1191–5.
Molinaro A, et al. Chemistry of lipid A: at the heart of innate immunity. Chem Weinh Bergstr Ger. 2015;21:500–19.
Caroff M, Cavaillon JM, Fitting C, Haeffner-Cavaillon N. Inability of pyrogenic, purified Bordetella pertussis lipid A to induce interleukin-1 release by human monocytes. Infect Immun. 1986;54:465–71.
Caroff M. DSc Thesis (Université Paris-Sud XI, 1982).
Masihi KN, Lange W, Brehmer W, Ribi E. Immunobiological activities of nontoxic lipid A: enhancement of nonspecific resistance in combination with trehalose dimycolate against viral infection and adjuvant effects. Int J Immunopharmacol. 1986;8:339–45.
Cluff CW. Monophosphoryl lipid A (MPL) as an adjuvant for anti-cancer vaccines: clinical results. Adv Exp Med Biol. 2010;667:111–23.
Awasthi S. Toll-like Receptor-4 modulation for cancer immunotherapy. Front Immunol. 2014;5:328.
Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–90.
Garidou L, et al. The gut microbiota regulates intestinal CD4 T cells expressing RORγt and controls metabolic disease. Cell Metab. 2015;22:100–12.
Cani PD, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–72.
Zhang-Sun W, et al. Structure function relationships in the lipids A from Ralstonia species rising in obese patients. Biochimie. 2019;159:72–80.
Raetz CR. Biochemistry of endotoxins. Annu Rev Biochem. 1990;59:129–70.
Banoub JH, Aneed AE, Cohen AM, Joly N. Structural investigation of bacterial lipopolysaccharides by mass spectrometry and tandem mass spectrometry. Mass Spectrom Rev. 2010;29:606–50.
Novikov A, Marr N, Caroff M. A comparative study of the complete lipopolysaccharide structures and biosynthesis loci of Bordetella avium, B. Hinzii, and B. Trematum. Biochimie. 2018;159:81–92. https://doi.org/10.1016/j.biochi.2018.12.011.
Tang G, Wang Y, Luo L. Transcriptional regulator LsrB of Sinorhizobium meliloti positively regulates the expression of genes involved in lipopolysaccharide biosynthesis. Appl Environ Microbiol. 2014;80:5265–73.
Trent MS, Stead CM, Tran AX, Hankins JV. Diversity of endotoxin and its impact on pathogenesis. J Endotoxin Res. 2006;12:205–23.
Kawasaki K, Ernst RK, Miller SI. Deacylation and palmitoylation of lipid A by Salmonellae outer membrane enzymes modulate host signaling through Toll-like receptor 4. J Endotoxin Res. 2004;10:439–44.
Ciornei CD, et al. Biofilm-forming Pseudomonas aeruginosa bacteria undergo lipopolysaccharide structural modifications and induce enhanced inflammatory cytokine response in human monocytes. Innate Immun. 2010;16:288–301.
Lam MY, et al. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J Clin Microbiol. 1989;27:962–7.
Beveridge TJ, Makin SA, Kadurugamuwa JL, Li Z. Interactions between biofilms and the environment. FEMS Microbiol Rev. 1997;20:291–303.
Murphy K, et al. Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J Bacteriol. 2014;196:1306–17.
Delgado MA, Mouslim C, Groisman EA. The PmrA/PmrB and RcsC/YojN/RcsB systems control expression of the Salmonella O-antigen chain length determinant. Mol Microbiol. 2006;60:39–50.
Mattoo S, Cherry JD. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev. 2005;18:326–82.
Wang JC, et al. Planktonic growth of Pseudomonas aeruginosa around a dual- species biofilm supports the growth of fusobacterium nucleatum within that biofilm. Int J Otolaryngol. 2017;2017:1–12. https://doi.org/10.1155/2017/3037191.
Chalabaev S, et al. Biofilms formed by gram-negative bacteria undergo increased lipid A palmitoylation, enhancing in vivo survival. MBio. 2014;5:e01116-14.
Le Coustumier A, Njamkepo E, Cattoir V, Guillot S, Guiso N. Bordetella petrii infection with long-lasting persistence in human. Emerg Infect Dis. 2011;17:612–8.
Vandamme PA, et al. Bordetella bronchialis sp. nov., Bordetella flabilis sp. nov. and Bordetella sputigena sp. nov., isolated from human respiratory specimens, and reclassification of Achromobacter sediminum Zhang et al. 2014 as Verticia sediminum gen. nov., comb. nov. Int J Syst Evol Microbiol. 2015;65:3674–82.
Diavatopoulos DA, et al. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 2005;1:e45.
Gueirard P, Weber C, Le Coustumier A, Guiso N. Human Bordetellabronchiseptica infection related to contact with infected animals: persistence of bacteria in host. J Clin Microbiol. 1995;33:2002–6.
World Health Organisation. https://www.who.int/immunization/monitoring_surveillance/burden/vpd/surveillance_type/passive/pertussis/en/.
Goodnow RA. Biology of Bordetella bronchiseptica. Microbiol Rev. 1980;44:722–38.
Harrington AT, Castellanos JA, Ziedalski TM, Clarridge JE, Cookson BT. Isolation of Bordetella avium and novel Bordetella strain from patients with respiratory disease. Emerg Infect Dis. 2009;15:72–4.
Cookson BT, et al. Bacteremia caused by a novel Bordetella species, ‘B. hinzii’. J Clin Microbiol. 1994;32:2569–71.
Weyant RS, et al. Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J Clin Microbiol. 1995;33:1–7.
von Wintzingerode F, et al. Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int J Syst Evol Microbiol. 2001;51:1257–65.
Ko KS, et al. New species of Bordetella, Bordetella ansorpii sp. nov., isolated from the purulent exudate of an epidermal cyst. J Clin Microbiol. 2005;43:2516–9.
Guiso N, Hegerle N. Other Bordetellas, lessons for and from pertussis vaccines. Expert Rev Vaccines. 2014;13:1125–33.
Sugiyama J, et al. Polyphasic insights into the microbiomes of the Takamatsuzuka Tumulus and Kitora Tumulus. J Gen Appl Microbiol. 2017;63:63–113.
Tazato N, et al. Novel environmental species isolated from the plaster wall surface of mural paintings in the Takamatsuzuka tumulus: Bordetella muralis sp. nov., Bordetella tumulicola sp. nov. and Bordetella tumbae sp. nov. Int J Syst Evol Microbiol. 2015;65:4830–8.
Caroff M, et al. Variations in the carbohydrate regions of Bordetella pertussis lipopolysaccharides: electrophoretic, serological, and structural features. J Bacteriol. 1990;172:1121–8.
Novikov A, et al. Complete Bordetella avium, Bordetella hinzii and Bordetella trematum lipid A structures and genomic sequence analyses of the loci involved in their modifications. Innate Immun. 2014;20:659–72.
Larocque S, Brisson J-R, Thérisod H, Perry MB, Caroff M. Structural characterization of the O-chain polysaccharide isolated from Bordetella avium ATCC 5086: variation on a theme. FEBS Lett. 2003;535(1):11–6.
Vinogradov E, Caroff M. Structure of the Bordetella trematum LPS O-chain subunit. FEBS Lett. 2005;579:18–24.
Vinogradov E. The structure of the carbohydrate backbone of the lipopolysaccharides from Bordetella hinzii and Bordetella bronchiseptica. Eur J Biochem. 2000;267:4577–82.
Vinogradov E. The structure of the core-O-chain linkage region of the lipopolysaccharide from Bordetella hinzii. Carbohydr Res. 2007;342:638–42.
Caroff M, Deprun C, Richards JC, Karibian D. Structural characterization of the lipid A of Bordetella pertussis 1414 endotoxin. J Bacteriol. 1994;176:5156–9.
Bouchez V, AlBitar-Nehmé S, Novikov A, Guiso N, Caroff M. Bordetella holmesii: lipid A structures and corresponding genomic sequences comparison in three clinical isolates and the reference strain ATCC 51541. Int J Mol Sci. 2017;18:E1080.
El Hamidi A, Novikov A, Karibian D, Perry MB, Caroff M. Structural characterization of Bordetella parapertussis lipid A. J Lipid Res. 2009;50:854–9.
Preston A, et al. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol Microbiol. 2003;48:725–36.
Basheer SM, et al. Structure activity characterization of Bordetella petrii lipid A, from environment to human isolates. Biochimie. 2016;120:87–95.
Cavaillon JM, Fitting C, Caroff M, Haeffner-Cavaillon N. Dissociation of cell associated interleukin-1 (IL-1) and IL-1 release induced by lipopolysaccharide and lipid A. Infect Immun. 1989;57:791–7.
Marr N, Novikov A, Hajjar AM, Caroff M, Fernandez RC. Variability in the lipooligosaccharide structure and endotoxicity among Bordetella pertussis strains. J Infect Dis. 2010;202:1897–906.
Shah NR, et al. Minor modifications to the phosphate groups and the C3’ acyl chain length of lipid A in two Bordetella pertussis strains, BP338 and 18-323, independently affect Toll-like receptor 4 protein activation. J Biol Chem. 2013;288:11751–60.
Basheer SM, Guiso N, Tirsoaga A, Caroff M, Novikov A. Structural modifications occurring in lipid A of Bordetella bronchiseptica clinical isolates as demonstrated by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2011;25:1075–81.
Lebbar S, Karibian D, Deprun C, Caroff M. Distribution of lipid A species between long and short chain lipopolysaccharides isolated from Salmonella, Yersinia, and Escherichia as seen by 252Cf plasma desorption mass spectrometry. J Biol Chem. 1994;269:31881–4.
Peppler MS. Two physically and serologically distinct lipopolysaccharide profiles instrains of Bordetella pertussis and their phenotype variants. Infect Immun. 1984;43:224–32.
Caroff M, et al. Structural variability and originality of the Bordetella endotoxins. J Endotoxin Res. 2001;7:63–8.
Le Blay K, Caroff M, Richards JC, Perry MB, Chaby R. Specific and crossreacting monoclonal antibodies to Bordetella parapertussis and Bordetella bronchiseptica lipopolysaccharides. Microbiology. 1994;140(Pt 9):2459–65.
Di Fabio JL, Caroff M, Karibian D, Richards JC, Perry MB. Characterization of the common antigenic lipopolysaccharide O-chains produced by Bordetella bronchiseptica and Bordetella parapertussis. FEMS Microbiol Lett. 1992;76:275–81.
Preston A, et al. Complete structures of Bordetella bronchiseptica and Bordetella parapertussis lipopolysaccharides. J Biol Chem. 2006;281:18135–44.
Lebbar S, et al. Molecular requirement for interleukin 1 induction by lipopolysaccharide-stimulated human monocytes: involvement of the heptosyl-2-keto-3-deoxyoctulosonate region. Eur J Immunol. 1986;16:87–91.
Schaeffer LM, McCormack FX, Wu H, Weiss AA. Interactions of pulmonary collectins with Bordetella bronchiseptica and Bordetella pertussis lipopolysaccharide elucidate the structural basis of their antimicrobial activities. Infect Immun. 2004;72:7124–30.
Gueirard P, Le Blay K, Le Coustumier A, Chaby R, Guiso N. Variation in Bordetella bronchiseptica lipopolysaccharide during human infection. FEMS Microbiol Lett. 1998;162:331–7.
Spears PA, Temple LM, Orndorff PE. A role for lipopolysaccharide in Turkey tracheal colonization by Bordetella avium as demonstrated in vivo and in vitro. Mol Microbiol. 2000;36:1425–35.
Caroff M, Deprun C, Karibian D, Szabó L. Analysis of unmodified endotoxin preparations by 252Cf plasma desorption mass spectrometry. Determination of molecular masses of the constituent native lipopolysaccharides. J Biol Chem. 1991;266:18543–9.
Caroff M, Deprun C, Karibian D. 252Cf plasma desorption mass spectrometry applied to the analysis of underivatized rough-type endotoxin preparations. J Biol Chem. 1993;268:12321–4.
Karibian D, Deprun C, Caroff M. Comparison of lipids A of several Salmonella and Escherichia strains by 252Cf plasma desorption mass spectrometry. J Bacteriol. 1993;175:2988–93.
Novikov A, Breton A, Caroff M. Micromethods for isolation and structural characterization of lipid A, and polysaccharide regions of bacterial lipopolysaccharides. Methods Mol Biol. 2017;1600:167–86.
Faruque SM, et al. Emergence and evolution of Vibrio cholerae O139. Proc Natl Acad Sci. 2003;100:1304–9.
Ramamurthy T, et al. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993;341:703–4.
Redmond JW. The structure of the O-antigenic side chain of the lipopolysaccharide of Vibrio cholerae 569B (Inaba). Biochim Biophys Acta. 1979;584:346–52.
Kenne L, Lindberg B, Unger P, Holme T, Holmgren J. Structural studies of the Vibrio cholerae O-antigen. Carbohydr Res. 1979;68:C14–6.
Kenne L, Lindberg B, Unger P, Gustafsson B, Holme T. Structural studies of the Vibrio cholerae O-antigen. Carbohydr Res. 1982;100:341–9.
Hisatsune K, Kondo S, Isshiki Y, Iguchi T, Haishima Y. Occurrence of 2-Omethyl-N-(3-deoxy-L-glycero-tetronyl)-D-perosamine (4-amino-4,6-dideoxy-D-mannopyranose) in lipopolysaccharide from Ogawa but not from Inaba O forms of O1 Vibrio cholerae. Biochem Biophys Res Commun. 1993;190:302–7.
Cox AD, Brisson JR, Varma V, Perry MB. Structural analysis of the lipopolysaccharide from Vibrio cholerae O139. Carbohydr Res. 1996;290:43–58.
Knirel YA, Widmalm G, Senchenkova SN, Jansson PE, Weintraub A. Structural studies on the short-chain lipopolysaccharide of Vibrio cholerae O139 Bengal. Eur J Biochem. 1997;247:402–10.
Hankins JV, et al. Elucidation of a novel Vibrio cholerae lipid A secondary hydroxyacyltransferase and its role in innate immune recognition. Mol Microbiol. 2011;81:1313–29.
Hankins JV, Madsen JA, Giles DK, Brodbelt JS, Trent MS. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in grampositive and gram-negative bacteria. Proc Natl Acad Sci U S A. 2012;109:8722–7.
Henderson JC, Herrera CM, Trent MS. AlmG, responsible for polymyxin resistance in pandemic Vibrio cholerae, is a glycyltransferase distantly related to lipid A late acyltransferases. J Biol Chem. 2017;292:21205–15.
Phillips NJ, et al. The lipid A from Vibrio fischeri lipopolysaccharide: a unique structure bearing a phosphoglycerol moiety. J Biol Chem. 2011;286:21203–19.
Acknowledgements
We are grateful to Luis Augusto from I2BC, Paris-Saclay University, for his participation to testing TNF-α and IL-6 with some of the Bordetella LPS, and to Benjamin Gensburger, from LPS-BioSciences, for performing the SDS-PAGE analysis of the Bordetella LPS. Prof. Dr. Babatosh Das from Centre for Human Microbial Ecology, Translational Health Science and Technology Institute, NCR Biotech Science Cluster, Faridabad, India, is acknowledged for providing the different V. cholerae strains studied. Prof. Dr. David R. Bundle from University of Alberta, Canada, is acknowledged for his help at editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Caroff, M., Novikov, A. (2019). LPS Structure, Function, and Heterogeneity. In: Williams, K. (eds) Endotoxin Detection and Control in Pharma, Limulus, and Mammalian Systems. Springer, Cham. https://doi.org/10.1007/978-3-030-17148-3_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-17148-3_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-17147-6
Online ISBN: 978-3-030-17148-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)