Abstract
The human gut microbiota consists of about 3.8 × 1013 microorganisms that play an essential role in health, metabolism, and immunomodulation. These gut microbes alter therapeutic response and toxicity to cancer therapies including cytotoxic chemotherapy, radiation therapy, kinase inhibitors, and immunotherapy agents. The gut microbiota generates short-chain fatty acids that are significant regulators of histone post-translational modifications that fundamentally regulate gene expression, linking the microbiota to cellular metabolism and transcriptional regulation. The short-chain fatty acids not only act locally but can be taken up in the blood stream to inhibit the activity of histone deacetylases, regulate gene expression in distant organs as well as the effector function of CD8+ T cells. Cancer and the treatments for it negatively impact the microbiome often resulting in dysbiosis. This can diminish a patient’s response to treatment as well as increase systemic toxicities from these therapies. In addition to the gut microbiota, microbes have been detected in tumors that can modulate chemotherapeutic drug response and can result in immune suppression. The gut microbiota and tumor-associated bacteria may be a significant contributor to the interindividual differences and heterogeneous responses to cancer therapies and drug tolerability and strategies that support and/or manipulate the microbiota to improve therapeutic outcome is an emerging area for personalized cancer treatment.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 The Gut Microbiota
Precision oncology considers the molecular characteristics of a patient’s tumor to determine an ideal approved or investigational therapy that could provide clinical benefit [1]. While prospective profiling of patient’s tumors has resulted in improved selection and response to therapies [2–4], this “tumorcentric” approach can fail to account for impact of the complex microenvironment that influences tumor growth and response to therapy. The gut microbiota is a complex ecosystem of microorganisms where the total number of bacteria in the average 70 kg person is estimated to be 3.8 × 1013 [5]. The number of bacteria in the body is of the same order of magnitude as the number of human cells and has a total mass of about 0.2 kg [6]. These microbes play fundamental roles in health and survival and have been found to play a significant role in the response to cancer therapy and susceptibility to toxic side effects of those drugs.
2 Gut Microbiota Generate Short-Chain Fatty Acids (SCFA)
The gut microbiota produces SCFA mainly through the fermentation of carbohydrates that escape digestion and absorption in the small intestine [7]. The major SCFA products produced are formate, acetate, propionate, and butyrate and these products are detectable in the circulation [7]. SCFAs are reported to directly activate G-coupled receptors, inhibit histone deacetylases (HDACs), serve as energy substrates, and promote T-cell differentiation into both effector and regulatory T cells to promote either immunity or immune tolerance [8–10]. The SCFA’s butyrate and propionate directly modulate the gene expression of CD8+ cytotoxic T lymphocytes and Tc17 cells [11]. The SCFAs appear not only optimize the function of Tregs and CD4+ T cells, but also modulate the function of CD8+ T cells to enhance anti-tumor and anti-viral activity [11, 12].
3 SCFAs as Regulators of Histone Post-translational Modifications (HPTM)
Human gut microbes regulate gene transcription using a variety of epigenetic marks (see Table 10.1, adapted from [13]). At least eleven types of HPTMs have been reported on over 60 different amino acid residues on histones, including methylation, acetylation, propionylation, butyrylation, formylation, phosphorylation, ubiquitylation, sumoylation, citrullination, proline isomerization, and ADP ribosylation [14], and the gut microbiota-generated SCFAs are involved in many of these modifications. While most consider lysine acetylation to be a predominant epigenetic event, a new histone modification, lysine crotonylation (Kcr) was found to be surprisingly abundant in the small intestine crypt and colon [15]. Crotonyl-CoA, the precursor of Kcr, is generated by Acidaminococcus fermentans [16], and depletion of gut microbiota leads to decreased histone crotonylation in the colon [15]. Class I HDAC enzymes, HDAC1, HDAC2, and HDAC3, are reported to efficiently remove the crotonyl moiety; microbiota-derived SCFAs are class I selective HDAC inhibitors and are therefore also histone decrotonylation inhibitors [15, 17]. The impact of Kcr on protein function remains to be fully elucidated; however, a variety of cancer proteins are crotonylated [18]. The gut microbiota is therefore responsible for histone post-translational modifications and alterations to the gut microbiota composition will have significant effects on transcriptional regulation and sensitivity and/or resistance to cancer therapeutics.
4 SCFAs and Response to Cancer Chemotherapy
4.1 Drug Metabolism
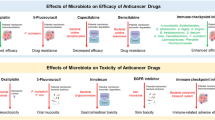
More than 40 drugs are reported to be directly metabolized by the gut microbiota including the anticancer drugs methotrexate and irinotecan [19]. The gut microbiota also directly or indirectly increases the metabolism of orally and systemically delivered drugs through SCFA modulation of cytochrome P450 (Cyp450) gene family members [20–22]. Germ-free mice demonstrate faster metabolism of many drugs suggesting the microbiota and SCFAs exert regulatory control over the rate of drug metabolism and detoxification [20]. The heterogeneity of clinical response to drug therapy and/or variable emergence of toxicities may be due in part to differences in gut microbiota composition and differential drug metabolism [23].
4.2 Response to Cancer Chemotherapy
Depletion of mouse microbiota with antibiotics results in dysbiosis that causes a drop in luminal and serum SCFAs, and increased expression of HDAC2 that has been linked to colon tumorigenesis [15, 24, 25]. Elevated HDAC2 reduces the sensitivity of non-small cell lung cancer (NSCLC) cells to cisplatin [26], melanoma cells to the alkylating drugs temozolomide, dacarbazine, and fotemustine [27], colorectal cancer cells to doxorubicin [28] and glioblastoma multiforme cells to temozolomide [29]. In breast cancer, HDAC2 overexpression is correlated with metastasis, increased Ki67, and increased multidrug resistance protein expression. HDAC2-positive breast cancer is also associated with shorter survival in patients who received chemotherapy containing anthracyclines [30]. The microbiota exerts suppressive activity on HDAC2 activity via SCFA production, suggesting the potential value of class I selective HDAC inhibitors in patients with compromised gut microbiota. Isoform-selective HDAC inhibitors could serve as SCFA-replacement therapies to support local and systemic gene regulation by acting as lysine deacetylation and decrotonylation inhibitors. The SCFA-replacement theraputics could also potentially improve clinical response to a variety of chemotherapeutic agents through the inhibition of elevated HDAC2 activity found in many cancers.
Cyclophosphamide therapeutic efficacy is due in part to the stimulation of an anti-tumor immune response. Cyclophosphamide alters the microbiota in the small intestine and causes the translocation of select Gram-positive bacteria to secondary lymphoid organs [31]. There, these bacteria stimulate the generation of pathogenic T helper 17 (pTh17) cells and memory Th1 immune responses [31]. Germ-free mice or mice treated with antibiotics showed a reduction in pTh17 cells, and tumors became resistant to cyclophosphamide [31] confirming the role of the microbiota in the anticancer mechanism for cyclophosphamide. Also, antibiotic treatment suppressed the response of subcutaneous tumors to a CpG-oligonucleotide immunotherapy and platinum chemotherapy [32]. The antibiotic treated or germ-free mice had tumor-infiltrating myeloid-derived cells that produced lower levels of cytokines after CpG-oligonucleotide treatment and produced lower amounts of reactive oxygen species (ROS) following oxaliplatin or cisplatin therapy [32]. These data demonstrate that the microbiota contributes to the modification of genotoxicity for platinum compounds independent of immunogenic cell death. Anthracyclines, alkylating agents, and camptothecins also induce ROS as part of their anticancer activity, so it is likely that the gut microbiota may influence the effectiveness of these drugs as well [32]. The role of the microbiota in modulating the response to radiation therapy needs to be characterized, but tumors in germ-free mice are less responsive to the beneficial effects of radiation when compared to normal mice with an intact microbiota; evidence in humans and experimental animals suggests that the composition of the intestinal microbiota may affect the severity of radiation-induced mucosal toxicity.
The gut microbiota also has a role in the response to tyrosine kinase inhibitors [33]. Patients with metastatic renal-cell carcinoma were treated with first-line VEGF-tyrosine kinase inhibitors and were also receiving antibiotics with either Bacteroides coverage or not. When compared to patients not receiving antibiotics, a significant improvement in PFS was observed in patients taking antibiotics that covered Bacteroides spp [33]. These data confirm a role for the gut microbiota in the clinical response to tyrosine kinase inhibitors.
5 The Gut Microbiota and the Immune System
Gut bacterial SCFAs have profound effects on the adaptive immune system, with high expression of SCFA receptors being reported on immune cells [10]. The generation of effector and regulatory T cells is influenced by the gut microbiota and is dependent on the variety of cytokines found in the microenvironment [34]. SCFAs enhance T-cell differentiation into effector T cells, such as Th1 and Th17 cells, and also anti-inflammatory IL-10þ regulatory T cells [34]. Recently, it was shown that Prevotella heparinolytica promotes the differentiation of Th17 cells colonizing the gut that migrates to the bone marrow in a transgenic mouse model of multiple myeloma [35]. In this experimental model, the commensal bacteria increase IL-17 signaling that accelerates progression of smoldering myeloma to myeloma [35].
6 Immunotherapy and the Microbiota
Approaches that modulate the patient immune system have demonstrated significant clinical activity in hematological and solid cancers. One of the first reports on the contribution of the gut microbiota on immune therapy was the reported diminished tumor response in mice receiving antibiotics, total body irradiation, and tumor-specific cytotoxic T cells [36]. In this study, the authors report that total body irradiation caused the translocation of the gut microbiota to mesenteric lymph nodes, and increased proliferation of the injected T cells in the tumor [36]. Similarly, when mice were treated with an intratumoral TLR9 agonist CpG-oligodeoxynucleotide, anticancer activity was observed; however, the anti-tumor effect in germ-free mice or mice treated with antibiotics was diminished demonstrating that an intact microbiota was required for optimal anticancer effects [32].
The role of the gut microbiota on clinical activity or resistance of immune checkpoint modulators has been reported [37–42]. In addition to the gut microbiota, there have been reports on the contribution of an intratumoral microbiome that could play a role in chemotherapy and immunotherapy resistance [43–46].
Anti-tumor immunity in patients can be reactivated by the immune checkpoint inhibitors (antibodies against cytotoxic T lymphocyte-associated antigen 4 CTLA4) and programmed cell death protein 1 (PD1) or its ligand PD1 ligand 1 (PDL1) [47]. Antibodies targeting these immune checkpoints have demonstrated significant clinical activity in patients with a variety of cancers; however, variability and duration of patient response remain an area of active investigation [48]. The gut microbiota regulates the anticancer activity of anti-CTLA4 and anti-PDL1 cancer therapies [41, 42]. Oral supplementation of either B. thetaiotaomicron or B. fragilis in microbiota-depleted mice restores the anti-tumor response to anti-CTLA4 antibodies [42]. Vancomycin enhances the efficacy of CTLA4 blockade in mice by decreasing the abundance of Gram-positive bacteria while preserving Gram-negative Bacteroidales and Burkholderiales [42]. Analysis of the fecal microbiota from patients with melanoma before and after treatment with anti‑CTLA4 showed a change in the relative proportions of three dominant enterotypes; enterotype A was dominated by Prevotella, enterotypes B and C were dominated by different Bacteroides [41, 42]. When fecal microbiota from patients with each of the three human enterotypes was transferred into tumor-bearing, germ-free mice only the enterotype C resulted in enhanced response to anti‑CTLA4 [42].
The response to anti-PDL1 was also found to be significantly associated with the gut microbiota of the Bifidobacterium genus, including Bifidobacterium breve, Bifidobacterium longum, and Bifidobacterium adolescentis [41]. Oral administration of a probiotic cocktail of Bifidobacterium including B. breve and B. longum, alone or with anti-PDL1, enhanced CD8 + T-cell-induced anti-tumor activity [41]. The effect of Bifidobacterium was abolished in CD8+ T-cell-depleted mice, indicating that Bifidobacterium action is dependent on cytotoxic T-cell activity [41]. The therapeutic effectiveness of anti-PDL1 treatment can be seen when Bifidobacterium are in higher numbers in the gut microbiota.
The anti-tumor activity of anti-PD-1 alone or when combined with anti-CTLA4 was significantly decreased when mice were treated with a broad-spectrum antibiotic combination (ampicillin + colistin + streptomycin) [38]. This experimental data were then confirmed and extended to patients with advanced NSCLC, RCC, or urothelial carcinoma (n = 42) who received PD-1/PD-L1 monoclonal antibodies. Broad-spectrum antibiotic treatment in these patients resulted in resistance to PD-1 blockade [38]. Metagenomic analysis of patient stool samples revealed correlations between clinical response to checkpoint inhibitors and the relative abundance of Akkermansia muciniphila, and in preclinical studies supplementation with A. muciniphila restored the efficacy of PD-1 blockade [38]. Other studies have reported bacterial species B. longum, Collinsella aerofaciens, and Enterococcus faecium [40] and relative abundance of the Ruminococcaceae family [49] in PD-1 blockade responding patients. Patients with a high abundance of Clostridiales, Ruminococcaceae, or Faecalibacterium in the gut had higher frequencies of effector CD4+ and CD8+ T cells in the systemic circulation and a preserved cytokine response to anti–PD-1 therapy, whereas patients with a higher abundance of Bacteroidales in the gut microbiome had higher frequencies of Tregs and myeloid-derived suppressor cells (MDSCs) in the systemic circulation, with a blunted cytokine response [49]. These findings highlight the therapeutic potential of modulating the gut microbiome in patients receiving checkpoint blockade immunotherapy, and warrant monitoring the gut microbiota in cancer clinical trials [49].
7 The Intratumoral Microbiome
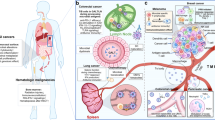
The microbiota has also emerged as a contributor to cancer development in intestinal tract malignancies, including laryngeal, esophageal, gastric, and colorectal cancers, as well as in primary liver cancer [50]. A recent report described that pancreatic cancers harbor a distinct intrapancreatic microbiome that is responsible for immune suppression and failure of immune checkpoint-targeted therapeutics [51]. When the intrapancreatic microbiome was ablated in experimental animals, immunogenic reprogramming of the tumor microenvironment occurred, including a reduction in MDSCs and an increase in M1 macrophage differentiation, promoting TH1 differentiation of CD4+ T cells and CD8+ T-cell activation [51]. There was an abundance of B. pseudolongum in gut and tumor microbiota in pancreas cancer that was associated with enhanced oncogenesis that could be reversed by ablating the microbiome [51]. The intrapancreatic microbiome has also been shown to inactivate the chemotherapeutic drug gemcitabine by Gammaproteobacteria-generated cytidine deaminase [46]. Upon examination, 113 human pancreas cancers, 86 (76%) were positive for bacteria, primarily Gammaproteobacteria, suggesting the intrapancreatic microbiome can also negatively diminish chemotherapeutic activity [46].
Recent pathological analyses have revealed a distinct microbiota that is present in breast cancer tissue that differs from normal breast tissue with a relative decreased in the genus Methylobacterium [52]. These authors also report significantly different microbiomes compared to non-cancer patients in the urinary tract characterized by increased numbers of Gram-positive bacteria [52]. The exact role of intratumoral bacteria in carcinogenesis and response to treatment in breast and urinary tract cancers is an area of active investigation.
The liver is exposed to the gut microbiota through the portal vein and recently the role of gut bacteria in anti-tumor surveillance in the liver was reported [53]. The microbiota metabolizes bile acids that recirculate back into the liver through the enterohepatic circulation [52, 53]. Antibiotic treatment of mice with vancomycin removed Gram-positive bacteria responsible for primary to-secondary bile acid metabolism causing the expression of CXCL16 and selective increase in hepatic CXCR6 positive natural killer T (NKT) cells [53]. This chemokine-dependent accumulation of hepatic NKT cells provides anti-tumor immunity in the liver, against primary and metastatic liver disease [53]. The gut microbiota increases liver anti-tumor immunosurveillance through bile acid metabolism and recruitment of immune effector cells.
In colorectal cancer, the gut microbiota translocate across compromised epithelial layers and stimulate immune cell infiltration and proinflammatory cytokine production [54]. Tumor infiltrating lymphocytes (TILs) are reported to improve survival for patients with colorectal cancer [55]. Human colorectal cancer cells from both primary tumors and established cell lines express toll-like receptors and produce significant chemokine expression when exposed to various bacterial species [55]. Antibiotic treatment of mice bearing orthotopic colorectal cancer xenografts demonstrated significantly lower levels of tumor-derived chemokines supporting the important role of the gut microbiota in tumor cell chemokine expression [55]. The extent of T-cell infiltration in primary human colorectal cancers is associated with the presence of specific bacterial families; these specific bacterial families were also associated with induction of specific immune cell attracting chemokines, suggesting the gut microbiota is directly involved in tumor cell immune cell recruitment and potentially colorectal cancer survival [55]. Some bacterial families like Fusobacteria were reported to be associated with worse clinical outcome and were found at higher levels in poorly immune cell infiltrated cancers [56]. Other bacterial families, like F. nucleatum, have been shown to inhibit natural killer and T-cell functions [57]. Taken together, these data demonstrate that the specific composition of the gut and tumor microbiota could play a key role in the attraction and/or suppression of immune effector cells in the tumor microenvironment, impacting patient outcomes.
8 Summary
It is unclear which bacterial families are required for an improved clinical response to cancer therapies, but there is no question that the variability in gut microbiota found in patients results in heterogeneous response to therapeutic interventions. Cancer patients are taking a variety of prescription and over-the-counter concomitant medications, all of which can alter the composition of the gut microbiota. For example, the COX-2 inhibitor celecoxib alters select bacterial populations in experimental animals including decreased Lactobacillaceae and Bifidobacteriaceae and increased Coriobacteriaceae [58]. Proton-pump inhibitors have been reported to significantly increase Lactobacillus spp., L. gasseri, L. fermentum, L. reuteri, and L. ruminis as well Streptococcus species [59]. Even nutraceuticals influence the gut microbiota composition, and many patients are taking a large variety of over-the-counter vitamins to supplement their prescription medications. For example, curcumin alters the gut microbiota resulting in increases in most Clostridium spp., Bacteroides spp., Citrobacter spp., Cronobacter spp., Enterobacter spp., Enterococcus spp., Klebsiella spp., Parabacteroides spp., and Pseudomonas spp. and reduced relative abundance of several Blautia spp. and most Ruminococcus spp. strains [60]. As a result, a new branch of pharmacogenomics, called pharmacomicrobiomics, has emerged to study drug–microbiome interactions [61]. One interesting question is the potential role of the regulatory authorities in requiring an assessment of new medicines effects on the microbiota during required GLP safety studies. Knowledge of the potential microbiota changes by these new medicines could have utility in identifying whether new drugs could negatively or positively impact the clinical activity of approved cancer medicines.
Studies to restore and/or enhance the gut microbiome by dietary modification, probiotics, prebiotics, post-biotics, autologous fecal microbiota transplant, and antibiotics could have therapeutic benefit for cancer patients to improve efficacy and reduce the toxicity of chemotherapy [62–65]. Dietary factors play a key role in the number and kind of bacterial taxa, and the production of a variety of epigenetic factors that regulate gene expression [66, 67], so close monitoring of the diets and supplements that cancer patients consume may be required to better understand and control for treatment outcomes.
To date, the majority of analyses of the gut and tumor microbiota have been through next-generation sequencing. However, gene/transcript presence does not necessarily indicate protein expression; therefore, directly measuring expressed proteins by metaproteomics will provide precise functional information on the microbiota [68, 69]. A thorough examination of the gut and intratumoral microbiota in cancer patients should include metaproteomic analysis which can reveal both human and microbial functional changes indicative of the host–microbiome interactions [70, 71].
Because cancer patients are already closely monitored when participating in clinical trials it will be important to add comprehensive microbiome assessments, including metaproteomic assessments to treatment protocols to fully understand baseline microbiota in cancer patients and to study the impact of therapies on specific bacterial families and their contribution to therapeutic outcomes.
References
Kurnit KC et al (2018) Precision oncology decision support: current approaches and strategies for the future. Clin Cancer Res 24(12):2719–2731
Jameson GS et al (2014) A pilot study utilizing multi-omic molecular profiling to find potential targets and select individualized treatments for patients with previously treated metastatic breast cancer. Breast Cancer Res Treat 147(3):579–588
Von Hoff DD et al (2010) Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol 28(33):4877–4883
Weiss GJ et al (2013) A pilot study using next-generation sequencing in advanced cancers: feasibility and challenges. PLoS ONE 8(10):e76438
Sender R, Fuchs S, Milo R (2016) Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14(8):e1002533
Sender R, Fuchs S, Milo R (2016) Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164(3):337–340
Morrison DJ, Preston T (2016) Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7(3):189–200
Krautkramer KA et al (2017) Metabolic programming of the epigenome: host and gut microbial metabolite interactions with host chromatin. Transl Res 189:30–50
Krautkramer KA et al (2016) Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell 64(5):982–992
Koh A et al (2016) From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165(6):1332–1345
Luu M et al (2018) Regulation of the effector function of CD8(+) T cells by gut microbiota-derived metabolite butyrate. Sci Rep 8(1):14430
Luu M, Steinhoff U, Visekruna A (2017) Functional heterogeneity of gut-resident regulatory T cells. Clin Transl Immunology 6(9):e156
Ellmeier W, Seiser C (2018) Histone deacetylase function in CD4(+) T cells. Nat Rev Immunol 18(10):617–634
Tan M et al (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146(6):1016–1028
Fellows R et al (2018) Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun 9(1):105
Cook GM et al (1994) Emendation of the description of acidaminococcus fermentans, a trans-aconitate- and citrate-oxidizing bacterium. Int J Syst Bacteriol 44(3):576–578
Wei W et al (2017) Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res 27:898–915
Huang H, Wang DL, Zhao Y (2018) Quantitative crotonylome analysis expands the roles of p300 in the regulation of lysine crotonylation pathway. Proteomics, e1700230
Haiser HJ, Turnbaugh PJ (2013) Developing a metagenomic view of xenobiotic metabolism. Pharmacol Res 69(1):21–31
Bjorkholm B et al (2009) Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS ONE 4(9):e6958
Selwyn FP et al (2016) Regulation of hepatic drug-metabolizing enzymes in germ-free mice by conventionalization and probiotics. Drug Metab Dispos 44(2):262–274
Selwyn FP, Cui JY, Klaassen CD (2015) RNA-seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab Dispos 43(10):1572–1580
Roy S, Trinchieri G (2017) Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer 17(5):271–285
Zhu P et al (2004) Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell 5(5):455–463
Ashktorab H et al (2009) Global histone H4 acetylation and HDAC2 expression in colon adenoma and carcinoma. Dig Dis Sci 54(10):2109–2117
Chen JH et al (2017) Valproic acid (VPA) enhances cisplatin sensitivity of non-small cell lung cancer cells via HDAC2 mediated down regulation of ABCA1. Biol Chem 398(7):785–792
Krumm A et al (2016) Enhanced histone deacetylase activity in malignant melanoma provokes RAD51 and FANCD2-triggered drug resistance. Cancer Res 76(10):3067–3077
Ye P et al (2016) Histone deacetylase 2 regulates doxorubicin (Dox) sensitivity of colorectal cancer cells by targeting ABCB1 transcription. Cancer Chemother Pharmacol 77(3):613–621
Zhang Z et al (2016) Silencing of histone deacetylase 2 suppresses malignancy for proliferation, migration, and invasion of glioblastoma cells and enhances temozolomide sensitivity. Cancer Chemother Pharmacol 78(6):1289–1296
Zhao H et al (2016) HDAC2 overexpression is a poor prognostic factor of breast cancer patients with increased multidrug resistance-associated protein expression who received anthracyclines therapy. Jpn J Clin Oncol 46(10):893–902
Viaud S et al (2013) The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342(6161):971–976
Iida N et al (2013) Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342(6161):967–970
Hahn AW et al (2018) Targeting bacteroides in stool microbiome and response to treatment with first-line VEGF tyrosine kinase inhibitors in metastatic renal-cell carcinoma. Clin Genitourin Cancer 16(5):365–368
Park J et al (2015) Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6 K pathway. Mucosal Immunol 8(1):80–93
Calcinotto A et al (2018) Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat Commun 9(1):4832
Paulos CM et al (2007) Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest 117(8):2197–2204
Derosa L et al (2018) The intestinal microbiota determines the clinical efficacy of immune checkpoint blockers targeting PD-1/PD-L1. Oncoimmunology 7(6):e1434468
Routy B et al (2018) Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359(6371):91–97
Zitvogel L et al (2018) The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science 359(6382):1366–1370
Matson V et al (2018) The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359(6371):104–108
Sivan A et al (2015) Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350(6264):1084–1089
Vetizou M et al (2015) Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350(6264):1079–1084
Urbaniak C et al (2014) Microbiota of human breast tissue. Appl Environ Microbiol 80(10):3007–3014
McCoy AN et al (2013) Fusobacterium is associated with colorectal adenomas. PLoS ONE 8(1):e53653
Geller LT, Straussman R (2018) Intratumoral bacteria may elicit chemoresistance by metabolizing anticancer agents. Mol Cell Oncol 5(1):e1405139
Geller LT et al (2017) Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357(6356):1156–1160
Ribas A, Wolchok JD (2018) Cancer immunotherapy using checkpoint blockade. Science 359(6382):1350–1355
Rodig SJ et al (2018) MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci Transl Med 10(450)
Gopalakrishnan V et al (2018) Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359(6371):97–103
Plottel CS, Blaser MJ (2011) Microbiome and malignancy. Cell Host Microbe 10(4):324–335
Pushalkar S et al (2018) The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov 8(4):403–416
Wang H et al (2017) Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget 8(50):88122–88138
Ma C et al (2018) Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360(6391)
Grivennikov SI et al (2012) Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491(7423):254–258
Cremonesi E et al (2018) Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 67(11):1984–1994
Mima K et al (2016) Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin Transl Gastroenterol 7(11):e200
Gur C et al (2015) Binding of the fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42(2):344–355
Montrose DC et al (2016) Celecoxib alters the intestinal microbiota and metabolome in association with reducing polyp burden. Cancer Prev Res (Phila) 9(9):721–731
Hojo M et al (2018) Gut microbiota composition before and after use of proton pump inhibitors. Dig Dis Sci 63(11):2940–2949
Peterson CT et al (2018) Effects of turmeric and curcumin dietary supplementation on human gut microbiota: a double-blind, randomized, placebo-controlled pilot study. J Evid Based Integr Med 23:2515690X18790725
ElRakaiby M et al (2014) Pharmacomicrobiomics: the impact of human microbiome variations on systems pharmacology and personalized therapeutics. OMICS 18(7):402–414
Alexander JL et al (2017) Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol 14(6):356–365
Panebianco C, Andriulli A, Pazienza V (2018) Pharmacomicrobiomics: exploiting the drug-microbiota interactions in anticancer therapies. Microbiome 6(1):92
Redman MG, Ward EJ, Phillips RS (2014) The efficacy and safety of probiotics in people with cancer: a systematic review. Ann Oncol 25(10):1919–1929
Taur Y et al (2018) Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci Transl Med 10(460)
Riscuta G et al (2018) Diet, microbiome, and epigenetics in the era of precision medicine. Methods Mol Biol 1856:141–156
Paul B et al (2015) Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin Epigenet 7:112
Erickson AR et al (2012) Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn’s disease. PLoS ONE 7(11):e49138
Verberkmoes NC et al (2009) Shotgun metaproteomics of the human distal gut microbiota. ISME J 3(2):179–189
Zhang X et al (2017) Deep metaproteomics approach for the study of human microbiomes. Anal Chem 89(17):9407–9415
Lai LA et al (2019) Metaproteomics study of the gut microbiome. Methods Mol Biol 1871:123–132
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Gately, S. (2019). Human Microbiota and Personalized Cancer Treatments: Role of Commensal Microbes in Treatment Outcomes for Cancer Patients. In: Von Hoff, D., Han, H. (eds) Precision Medicine in Cancer Therapy . Cancer Treatment and Research, vol 178. Springer, Cham. https://doi.org/10.1007/978-3-030-16391-4_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-16391-4_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-16390-7
Online ISBN: 978-3-030-16391-4
eBook Packages: MedicineMedicine (R0)