Abstract
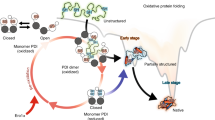
Many proteins secreted to the bacterial cell envelope contain cysteine residues that are involved in disulfide bonds. These disulfides either play a structural role, increasing protein stability, or reversibly form in the catalytic site of periplasmic oxidoreductases. Monitoring the in vivo redox state of cysteine residues, i.e., determining whether those cysteines are oxidized to a disulfide bond or not, is therefore required to fully characterize the function and structural properties of numerous periplasmic proteins. Here, we describe a reliable and rapid method based on trapping reduced cysteine residues with 4′-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS), a maleimide compound. We use theEscherichia coliDsbA protein to illustrate the method, which can be applied to all envelope proteins.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Dutton RJ, Boyd D, Berkmen M, Beckwith J (2008) Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci U S A 105:11933–8
Vertommen D, Depuydt M, Pan J, Leverrier P, Knoops L, Szikora JP, Messens J, Bardwell JC, Collet JF (2008) The disulphide isomerase DsbC cooperates with the oxidase DsbA in a DsbD-independent manner. Mol Microbiol 67:336–49
Sone M, Akiyama Y, Ito K (1997) Differentialin vivoroles played by DsbA and DsbC in the formation of protein disulfide bonds. J Biol Chem 272:10349–52
Messens J, Collet JF, Van Belle K, Brosens E, Loris R, Wyns L (2007) The oxidase DsbA folds a protein with a nonconsecutive disulfide. J Biol Chem 282:31302–7
Leverrier P, Declercq JP, Denoncin K, Vertommen D, Hiniker A, Cho SH, Collet JF (2011) Crystal structure of the outer membrane protein RcsF, a new substrate for the periplasmic protein-disulfide isomerase DsbC. J Biol Chem 286:16734–42
Denoncin K, Vertommen D, Paek E, Collet JF (2010) The protein-disulfide isomerase DsbC cooperates with SurA and DsbA in the assembly of the essential beta-barrel protein LptD. J Biol Chem 285:29425–33
Ruiz N, Chng SS, Hiniker A, Kahne D, Silhavy TJ (2010) Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc Natl Acad Sci U S A 107:12245–50
Braun M, Silhavy TJ (2002) Imp/OstA is required for cell envelope biogenesis inEscherichia coli. Mol Microbiol 45:1289–302
Bardwell JC, McGovern K, Beckwith J (1991) Identification of a protein required for disulfide bond formation in vivo. Cell 67:581–9
Martin JL, Bardwell JC, Kuriyan J (1993) Crystal structure of the DsbA protein required for disulphide bond formationin vivo. Nature 365:464–8
Collet JF, Messens J (2010) Structure, function, and mechanism of thioredoxin proteins. Antioxid Redox Signal 13:1205–16
Bardwell JC, Lee JO, Jander G, Martin N, Belin D, Beckwith J (1993) A pathway for disulfide bond formationin vivo. Proc Natl Acad Sci U S A 90:1038–42
Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC (1999) Oxidative protein folding is driven by the electron transport system. Cell 98:217–27
Grauschopf U, Winther JR, Korber P, Zander T, Dallinger P, Bardwell JC (1995) Why is DsbA such an oxidizing disulfide catalyst? Cell 83:947–55
Rietsch A, Belin D, Martin N, Beckwith J (1996) Anin vivopathway for disulfide bond isomerization inEscherichia coli. Proc Natl Acad Sci U S A 93:13048–53
McCarthy AA, Haebel PW, Torronen A, Rybin V, Baker EN, Metcalf P (2000) Crystal structure of the protein disulfide bond isomerase, DsbC, fromEscherichia coli. Nat Struct Biol 7:196–9
Joly JC, Swartz JR (1997)In vitroandin vivoredox states of theEscherichia coliperiplasmic oxidoreductases DsbA and DsbC. Biochemistry 36:10067–72
Katzen F, Beckwith J (2000) Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell 103:769–79
Rietsch A, Bessette P, Georgiou G, Beckwith J (1997) Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol 179:6602–8
Shouldice SR, Heras B, Walden PM, Totsika M, Schembri MA, Martin JL (2011) Structure and function of DsbA, a key bacterial oxidative folding catalyst. Antioxid Redox Signal 14:1729–60
Casadaban MJ (1976) Transposition and fusion of thelacgenes to selected promoters inEscherichia coliusing bacteriophage lambda and Mu. J Mol Biol 104:541–55
Miller JH (1992) A short course in bacterial genetics: laboratory manual. Cold Spring Harbor, New York
Acknowledgments
This work was supported by the European Research Council (FP7/2007–2013) ERC independent researcher starting grant 282335—Sulfenic to J.F.C. J.F.C. is a Chercheur Qualifié and PL a Chargé de Recherches of the Belgian FNRS. KD and VN are supported by a grant from the FRIA.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media New York
About this protocol
Cite this protocol
Denoncin, K., Nicolaes, V., Cho, SH., Leverrier, P., Collet, JF. (2013). Protein Disulfide Bond Formation in the Periplasm: Determination of the In Vivo Redox State of Cysteine Residues. In: Delcour, A. (eds) Bacterial Cell Surfaces. Methods in Molecular Biology, vol 966. Humana Press, Totowa, NJ. https://doi.org/10.1007/978-1-62703-245-2_20
Download citation
DOI: https://doi.org/10.1007/978-1-62703-245-2_20
Published:
Publisher Name: Humana Press, Totowa, NJ
Print ISBN: 978-1-62703-244-5
Online ISBN: 978-1-62703-245-2
eBook Packages: Springer Protocols