Abstract
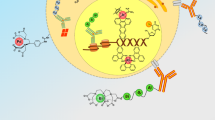
The advent of mass cytometry (CyTOF®) has permitted simultaneous detection of more than 40 antibody parameters at the single-cell level, although a limited number of metal-labeled antibodies are commercially available. Here we present optimized and scalable protocols for conjugation of lanthanide as well as bismuth ions to immunoglobulin (Ig) using a maleimide-functionalized chelating polymer and for characterization of the conjugate. The maleimide functional group is reactive with cysteine sulfhydryl groups generated through partial reduction of the Ig Fc region. Incubation of Ig with polymer pre-loaded with lanthanide ions produces metal-labeled Ig without disrupting antigen specificity. Antibody recovery rates can be determined by UV spectrophotometry and frequently exceeds 60%. Each custom-conjugated antibody is validated using positive and negative cellular control populations and is titrated for optimal staining at concentrations ranging from 0.1 to 10 μg/mL. The preparation of metal-labeled antibodies can be completed in 4.5 h, and titration requires an additional 3–5 h.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Baranov VI, Quinn Z, Bandura DR et al (2002) A sensitive and quantitative element-tagged immunoassay with ICPMS detection. Anal Chem 74:1629–1636. https://doi.org/10.1021/ac0110350
Bandura DR, Baranov VI, Ornatsky OI et al (2009) Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem 81:6813–6822. https://doi.org/10.1021/ac901049w
Bendall SC, Simonds EF, Qiu P et al (2011) Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332:687–696. https://doi.org/10.1126/science.1198704
Bodenmiller B, Zunder ER, Finck R et al (2012) Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol 30:858–867. https://doi.org/10.1038/nbt.2317
Behbehani GK, Bendall SC, Clutter MR et al (2012) Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A 81:552–566. https://doi.org/10.1002/cyto.a.22075
Fienberg HG, Simonds EF, Fantl WJ et al (2012) A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A 81:467–475. https://doi.org/10.1002/cyto.a.22067
Mingueneau M, Kreslavsky T, Gray D et al (2013) The transcriptional landscape of alphabeta T cell differentiation. Nat Immunol 14:619–632. https://doi.org/10.1038/ni.2590
Newell EW, Sigal N, Bendall SC et al (2012) Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity 36:142–152. https://doi.org/10.1016/j.immuni.2012.01.002
Lou X, Zhang G, Herrera I et al (2007) Polymer-based elemental tags for sensitive bioassays. Angew Chem Int Ed Eng 46:6111–6114. https://doi.org/10.1002/anie.200700796
Seegan GW, Smith CA, Schumaker VN (1979) Changes in quaternary structure of IgG upon reduction of the interheavy-chain disulfide bond. Immunology 76:907–911. https://doi.org/10.1073/pnas.76.2.907
Liu H, Chumsae C, Gaza-Bulseco G et al (2010) Ranking the susceptibility of disulfide bonds in human IgG1 antibodies by reduction, differential alkylation, and LC-MS analysis. Anal Chem 82:5219–5226. https://doi.org/10.1021/ac100575n
Amir ED, Davis KL, Tadmor MD et al (2013) viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 31:545–552. https://doi.org/10.1038/nbt.2594
Gibbs KD, Jager A, Crespo O et al (2012) Decoupling of tumor-initiating activity from stable immunophenotype in HoxA9-Meis1-driven AML. Cell Stem Cell 10:210–217. https://doi.org/10.1016/j.stem.2012.01.004
Spitzer MH, Carmi Y, Reticker-Flynn NE et al (2017) Systemic immunity is required for effective cancer immunotherapy. Cell 168:487–502.e15. https://doi.org/10.1016/j.cell.2016.12.022
Hartmann FJ, Bernard-Valnet R, Quériault C et al (2016) High-dimensional single-cell analysis reveals the immune signature of narcolepsy. J Exp Med 213:2621–2633. https://doi.org/10.1084/jem.20160897
Arrell DK, Terzic A (2010) Network systems biology for drug discovery. Clin Pharmacol Ther 88:120–125. https://doi.org/10.1038/clpt.2010.91
Sachs K (2005) Causal protein-signaling networks derived from multiparameter single-cell data. Science 308:523–529. https://doi.org/10.1126/science.1105809
Aghaeepour N, Jalali A, O’Neill K et al (2012) RchyOptimyx: cellular hierarchy optimization for flow cytometry. Cytometry A 81:1022–1030. https://doi.org/10.1002/cyto.a.22209
Qiu P, Simonds EF, Bendall SC et al (2011) Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 29:886–891. https://doi.org/10.1038/nbt.1991
Mair F, Hartmann FJ, Mrdjen D et al (2016) The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur J Immunol 46:34–43. https://doi.org/10.1002/eji.201545774
Ornatsky OI, Kinach R, Bandura DR et al (2008) Development of analytical methods for multiplex bio-assay with inductively coupled plasma mass spectrometry. J Anal At Spectrom 23:463. https://doi.org/10.1039/b710510j
Dulski P (1994) Interferences of oxide, hydroxide and chloride analyte species in the determination of rare earth elements in geological samples by inductively coupled plasma-mass spectrometry. Fresenius J Anal Chem 350:194–203. https://doi.org/10.1007/BF00322470
Kotecha N, Krutzik PO, Irish JM (2010) Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom Chapter 10:Unit10.17. https://doi.org/10.1002/0471142956.cy1017s53
Bigos M (2007) Separation index: an easy-to-use metric for evaluation of different configurations on the same flow cytometer. Curr Protoc Cytom Chapter 1:Unit1.21. https://doi.org/10.1002/0471142956.cy0121s40
Medvedev AE, Johnsen AC, Haux J et al (1997) Regulation of Fas and Fas-ligand expression in NK cells by cytokines and the involvement of Fas-ligand in NK/LAK cell-mediated cytotoxicity. Cytokine 9:394–404. https://doi.org/10.1006/cyto.1996.0181
Garrone P, Neidhardt EM, Garcia E et al (1995) Fas ligation induces apoptosis of CD40-activated human B lymphocytes. J Exp Med 182:1265–1273
Krutzik PO, Trejo A, Schulz KR, Nolan GP (2011) Phospho flow cytometry methods for the analysis of kinase signaling in cell lines and primary human blood samples. Methods Mol Biol 699:179–202. https://doi.org/10.1007/978-1-61737-950-5_9
Acknowledgments
We are grateful to Scott Tanner for providing useful feedback on this manuscript and to Fluidigm for supplying the chelating polymer that was used to optimize the protocol described here. F.J.H received support from the Swiss National Science Foundation (SNF Early Postdoc. Mobility), the Novartis Foundation for medical-biological research (Research Fellowship), and EMBO (Long-Term Fellowship). Also, DP5OD023056, R21AG057224 from the NIH and the Parker Institute for Cancer Immunotherapy to M.H.S. and S10OD018040 Shared Instrumentation Grant to UCSF. E.F.S. is supported by a Damon Runyon Cancer Research Foundation Fellowship (DRG-2190-14). G.P.N. is supported by the NIH grants R01CA184968, 1R01GM10983601, 1R01NS08953301, 1R01CA19665701, R01HL120724, 1R21CA183660, R33CA0183692, 1R33CA183654-01, U19AI057229, 1U19AI100627, U54-UCA149145A, N01-HV-00242, HHSN26820100034C, and 5UH2AR067676; the NIH Northrop-Grumman Corporation Subcontract 7500108142; FDA grant HHSF223201210194C; DOD grants OC110674 and W81XWH-14-1-0180; NWCRA Entertainment Industry Foundation; and Bill and Melinda Gates Foundation grant OPP1113682. S.C.B. is supported by the Damon Runyon Cancer Research Foundation Fellowship (DRG-2017-09), the NIH grants 1DP2OD022550-01, 1R01AG056287–01, 1R01AG057915-01, 1-R00-GM104148-01, 1U24CA224309-01, 5U19AI116484-02, 1U24CA224309-01, The Bill and Melinda Gates Foundation, and a Translational Research Award from the Stanford Cancer Institute.
Competing Financial Interests:
S.C.B. and E.F.S. have been paid consultants for the company Fluidigm Sciences, the manufacturers that produced some of the reagents and instrumentation described in this manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Hartmann, F.J. et al. (2019). Scalable Conjugation and Characterization of Immunoglobulins with Stable Mass Isotope Reporters for Single-Cell Mass Cytometry Analysis. In: McGuire, H., Ashhurst, T. (eds) Mass Cytometry. Methods in Molecular Biology, vol 1989. Humana, New York, NY. https://doi.org/10.1007/978-1-4939-9454-0_5
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9454-0_5
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-4939-9453-3
Online ISBN: 978-1-4939-9454-0
eBook Packages: Springer Protocols