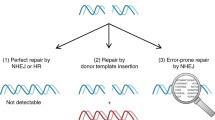
Abstract
Recent developments in gene targeting methodologies such as ZFNs, TALENs, and CRISPR/Cas9 have revolutionized approaches for gene modifications in cells, tissues, and whole animals showing great promise for translational applications. With regard to CRISPR/Cas9, a variety of repurposed systems have been developed to achieve gene knock-out, base editing, targeted knock-in, gene activation/repression, epigenetic modulation, and locus-specific labeling. A functional communality of all CRISPR/Cas9 applications is the gRNA-dependent targeting specificity of the Cas9/gRNA complex that, for gene knock-out (KO) purposes, has been shown to dictate the indel formation potential. Therefore, the objective of a CRISPR/Cas9 KO set up is to identify gRNA designs that enable maximum out-of-frame insertion and/or deletion (indel) formation and thus, gRNA design becomes a proxy for optimal functionality of CRISPR/Cas9 KO and repurposed systems. To this end, validation of gRNA functionality depends on efficient, accurate, and sensitive identification of indels induced by a given gRNA design. For in vitro indel profiling the most commonly used methods are based on amplicon size discrimination or sequencing. Indel detection by amplicon analysis (IDAA™) is an alternative sensitive, fast, and cost-efficient approach ideally suited for profiling of indels induced by Cas9/gRNA with similar sensitivity, specificity, and resolution, down to single base discrimination, as the preferred next-generation sequencing-based indel profiling methodologies. Here we provide a protocol that is based on complexed Cas9/gRNA RNPs delivered to primary peripheral blood mononuclear cells (PBMCs) isolated from healthy individuals followed by quantitative IDAA indel profiling. Importantly, the protocol described benefits from a short “sample-to-data” turnaround time of less than 5 h. Thus, this protocol describes a methodology that provides a suitable and effective solution to validate and quantify the extent of ex vivo CRISPR/Cas9 targeting in primary cells.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Chandrasegaran S, Carroll D (2016) Origins of programmable nucleases for genome engineering. J Mol Biol 428:963–989
Sfeir A, Symington LS (2015) Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway Trends Biochem Sci 40:701–714
Sehn JK (2014) Insertions and deletions (Indels). In: Clinical genomics. Elsevier, Amsterdam, pp 129–150
Deriano L, Roth DB (2013) Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet 47:433–455
van Overbeek M, Capurso D, Carter MM, Frias E, Russ C, Reece-Hoyes JS, Nye C, Vidal B, Zheng J, Hoffman GR et al (2016) DNA repair profiling reveals nonrandom outcomes at. Mol Cell 63:633–646
Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M (2016) Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533:125–129
Narimatsu Y, Joshi H, Zhang Y, Gomes C, Yen H, Lorenzetti F, Furukawa S, Schjoldager K, Hansen L, Clausen H, Bennett EP et al (2018) A validated gRNA library for CRISPR/Cas9 targeting of the human glycosyltransferase genome. Glycobiology 28(5):295–305
Lonowski LA, Narimatsu Y, Riaz A, Delay CE, Yang Z, Niola F, Duda K, Ober EA, Clausen H, Wandall HH et al (2017) Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. Nat Protoc 12:581–603
Kosicki M, Rajan SS, Lorenzetti FC, Wandall HH, Narimatsu Y, Metzakopian E, Bennett EP (2017) Dynamics of indel profiles induced by various CRISPR/Cas9 delivery methods. Prog Mol Biol Transl Sci. Elsevier Inc 152:49–67
Bennett EP, Jacobi AM, Garrett RR, Behlke MA (2018) Detection of insertion/deletion (indel) events after genome targeting: pro’s and con’s of the available methods. In: Appasani K (ed) GENOME EDITING and ENGINEERING: from Talens, ZFNs and CRISPRs to molecular surgery. Cambridge University Press, Cambridge, pp 181–194
Yang Z, Steentoft C, Hauge C, Hansen L, Thomsen AL, Niola F, Vester-Christensen MB, Frodin M, Clausen H, Wandall HH et al (2015) Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Res 43:e59. 1–8
Sandmann S, De Graaf AO, Karimi M, Van Der Reijden BA, Hellström-Lindberg E, Jansen JH, Dugas M (2017) Evaluating variant calling tools for non-matched next-generation sequencing data. Sci Rep 7:1–12
Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ (2012) Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30:434–439
Brinkman EK, Chen T, Amendola M, van Steensel B (2014) Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42:e168–e168
Hsiau T, Maures T, Waite K, Yang J, Kelso R, Holden K, Stoner R (2018) Inference of CRISPR edits from sanger trace data. bioRxiv 1101:10. https://doi.org/10.1101/251082
Jamuar S, Lam A-TN, Kircher M, D’Gama A, Wang J, Barry BJ, Zhang X, Hill RS, Partlow JN, Topcu M et al (2014) Somatic mutations in cerebral cortical malformations. N Engl J Med 371:733–743
Mohamed S, Penaranda G, Gonzalez D, Camus C, Khiri H, Boulmé R, Sayada C, Philibert P, Olive D, Halfon P (2014) Comparison of ultra-deep versus Sanger sequencing detection of minority mutations on the HIV-1 drug resistance interpretations after virological failure. AIDS 28:1315–1324
Yang Z, Wang S, Halim A, Schulz MA, Frodin M, Rahman SH, Vester-Christensen MB, Behrens C, Kristensen C, Vakhrushev SY et al (2015) Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat Biotechnol 33:2014–2017
Cornu TI, Mussolino C, Cathomen T (2017) Refining strategies to translate genome editing to the clinic. Nat Med 23:415–423
Clark JM (1988) Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res 16:9677–9686
Hu G (1993) DNA polymerase-catalyzed addition of nontemplated extra nucleotides to the 3′ end of a DNA fragment. DNA Cell Biol 12:763–770
Smith JR, Carpten JD, Brownstein MJ, Ghosh S, Magnuson VL, Gilbert DA, Trent JM, Collins FS (1995) Approach to genotyping errors caused by nontemplated nucleotide addition by Taq DNA polymerase. Genome Res 5:312–317
Sentmanat MF, Peters ST, Florian CP, Connelly JP, Pruett-Miller SM (2018) A survey of validation strategies for CRISPR-Cas9 editing. Sci Rep 8(1):888
Magnuson VL, Ally DS, Nylund SJ, Karanjawala ZE, Rayman JB, Knapp JI, Lowe AL, Ghosh S, Collins FS (1996) Substrate nucleotide-determined non-templated addition of adenine by Taq DNA polymerase: implications for PCR-based genotyping and cloning. Biotechniques 21(4):700–9
Acknowledgments
We thank Vasili Korol and Ilia A. Solov’yov from the University of Southern Denmark, Department of Physics, Chemistry and Pharmacy, for development of ProfileIt™ and Camilla Andersen from Copenhagen Center for Glycomics, Department of Odontology, University of Copenhagen, for excellent technical assistance. This work was supported by the Witten/Herdecke University internal research promotion Grant No. IFF2017-12, the German Duchenne Foundation “Aktion Benni & Co.” starting grant to E.E.-S., the Danish National Research Foundation [DNRF107], the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 765269, and the German Federal Ministry of Education and Research (BMBF). Z.Y. received support from the Lundbeck Foundation and H.H.W. received support from ERC-2017-COG Type of action: ERC-COG; 772735; GlycoSkin.
Conflict of Interest Statement: E.P.B. declares that a patent application covering the IDAA™ method is pending, and acts as scientific advisor for Cobo Technologies Aps.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
König, S., Yang, Z., Wandall, H.H., Mussolino, C., Bennett, E.P. (2019). Fast and Quantitative Identification of Ex Vivo Precise Genome Targeting-Induced Indel Events by IDAA. In: Luo, Y. (eds) CRISPR Gene Editing. Methods in Molecular Biology, vol 1961. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-9170-9_4
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9170-9_4
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-9169-3
Online ISBN: 978-1-4939-9170-9
eBook Packages: Springer Protocols