Abstract
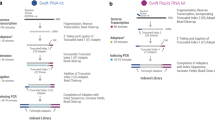
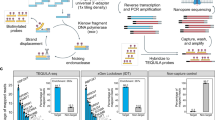
Gene expression profiling of samples from biobanks requires a method that can be used with intact as well as partially degraded RNA. High throughput applications can benefit from reducing the number of processing steps including eliminating the poly(A) selection and ribosomal depletion steps. When performing targeted capture, we have found that we can eliminate the upfront poly(A) selection/ribosomal depletion steps that cause bias in standard mRNA-Seq workflows. This target enrichment solution allows for whole transcriptome or customized content to characterize differential gene expression patterns (especially for mid/low level transcripts). Protocol modifications to the Agilent Strand-Specific RNA Library Prep kit resulted in a new workflow called “RNA Direct” that generates RNA-Seq data with minimal ribosomal contamination and good sequencing coverage. Using RNA isolated from a set of matched samples including fresh frozen (FF) or formalin-fixed, paraffin-embedded (FFPE) from tumor/normal tissues we generated high-quality data using a protocol that does not require upfront ribosomal depletion or poly(A) selection. Using SureSelectXT RNA Direct protocol (RNA Direct) workflow, we found transcripts to be upregulated or downregulated to similar degrees with similar confidence levels in both the FF and FFPE samples, demonstrating the utility for meaningful gene expression studies with biobank samples of variable quality.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Halvardson J, Zaghlool A, Feuk L (2013) Exome RNA sequencing reveals rare and novel alternative transcripts. Nucleic Acids Res 41(1):e6
Cieslik M, Chugh R, Wu Y-M et al (2015) The use of exome capture RNA-Seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res 25(9):1372–1381
Kumar G, Denslow ND (2017) Gene expression profiling in fish toxicology: a review. Rev Environ Contam Toxicol 241:1–38. Review. PMID: 27464848
Cui Y, Paules RS (2010) Use of transcriptomics in understanding mechanisms of drug-induced toxicity. Pharmacogenomics 11(4):573–585. https://doi.org/10.2217/pgs.10.37. Review. PMID: 20350139
Schroeder A et al (2006) The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7:3. PMID: 16448564
“Evaluating RNA quality from FFPE samples” Illumina publication number 470-2014-001
“SureSelectXT RNA direct protocol provides simultaneous transcriptome enrichment and ribosomal depletion of FFPE RNA” Agilent publication number 5991-8119EN
SureSelectXT RNA Direct for preparation of strand-specific sequencing libraries from high-quality or FFPE-derived RNA samples for the Illumina Platform Version A0 July 2017 or newer version (p/n G9691-90050)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Jones, J.C., Siebold, A.P., Livi, C.B., Lucas, A.B. (2018). SureSelectXT RNA Direct: A Technique for Expression Analysis Through Sequencing of Target-Enriched FFPE Total RNA. In: Raghavachari, N., Garcia-Reyero, N. (eds) Gene Expression Analysis. Methods in Molecular Biology, vol 1783. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7834-2_4
Download citation
DOI: https://doi.org/10.1007/978-1-4939-7834-2_4
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-7833-5
Online ISBN: 978-1-4939-7834-2
eBook Packages: Springer Protocols