Abstract
Ethylene (Et) plays a remarkable role in regulating plant signaling networks involved in responding to multiple biotic and abiotic stresses. Jasmonic (JA) and salicylic (SA) acids are also highly implicated in this scenario. In the last decade, understanding of this complicated puzzle of plant-stress regulation has progressed considerably. This chapter contains the recent progress in the plant adaptation to the most important and investigated hostile environments (e.g., O2-shortage, microorganisms infection, O3, and freezing) and the role of Et in protecting plants from pathogens. It is also highlighted that Et acts via a complex signaling pathway leading to the activation of Ethylene Response Factor (EtRF) genes which represent one of the largest transcription factor families in the plant kingdom. Likewise, other notable components of the Et signal transduction network are also included in this chapter. The contribution of all these different signals is discussed within the context of their role in plant-stress adaptation.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Salicylic Acid
- Necrotrophic Pathogen
- Ethylene Response Factor
- Systemic Acquire Resistance
- Aerenchyma Formation
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
When plants colonized the terrestrial ecosystems (some 475 million years ago), they had to acquire a number of organs necessary to keep erectile (i.e., root system, stem, and especially cell walls lignin enriched) (Kendrick and Crane 1997; Peter and Neale 2004; Martone et al. 2009). Likewise, terrestrial plants also had to develop a leaf system able to carry out both photobiosynthesis (i.e., carbohydrate and energy biosynthesis) and transpiration (i.e., gas exchange and generation of a force that allows the ascent and distribution of water and nutrients from the soil). However, the colonization process also caused serious problems of stress as a result of the transition from aquatic, motile ancestors into terrestrial, sessile organisms (Martone et al. 2009). Thus, the lack of mobility resulted in a complicated process of adaptation to the environment and the acquisition of defense mechanisms against diseases and predators (Ausubel 2005). In order to avoid a progressive disappearance, plants improved their physiological plasticity and developed a complicated set of signaling networks. These networks are tightly regulated by hormones that allow plants to survive by protecting them against biotic and abiotic stresses (Robert-Seilaniantz et al. 2011). Ethylene (Et), in combination with hormones such as jasmonic (JA) and salicylic (SA) acids, is one of the main players involved in the resistance and susceptibility to bacterial, fungal, and nematode pathogens (Adie et al. 2007; Kazan and Manners 2008; León-Reyes et al. 2009, 2010; Lin et al. 2009). The biosynthesis, transport, and accumulation of the above-mentioned hormones trigger a cascade of signaling pathways involved in plant defense. Et and JA signaling pathways are activated in response to necrotrophic plant pathogens; whereas salicylic acid (SA) play a major role during the triggering of plant defenses toward biotrophic pathogens (reviewed in Glazebrook 2005; Thaler et al. 2012). In general, SA and JA/Et defensive signaling pathways have been demonstrated to be mutually antagonistic (van Loon et al. 2006; Adie et al. 2007; Pieterse et al. 2012). Recently, it was demonstrated that both SA- and JA-dependent disease resistance is inhibited by a simultaneously reduced red:far light ratio (De Wit et al. 2013). In addition, it seems fairly clear that: (1) Et production plays a role in plant responses to flooding, salinity, drought, and several contaminant agents (e.g., ozone); and (2) plant growth-promoting rhizobacteria (PGPR) can improve plant tolerance to drought, salinity, and metal toxicity (Haas and Defago 2005; Lugtenberg and Kamilova 2009; Barreto-Figueiredo et al. 2011; Hol et al. 2013), although the role of Et in this puzzle is not fully decoded. This chapter aims to give an overview on the role of Et in the defense mechanisms of land plants against different types of environmental stresses.
2 Updated Overview of the Plant Hormone Ethylene
Et is the simplest plant hormone. Zhong and Burns (2003) showed that 7 % of the 6,000 investigated Arabidopsis genes were regulated by Et. During the plants life cycle, Et regulates key processes such as root hair development, flowering, climateric fruit ripening, seed dormancy, and germination (for review, see Czarny et al. 2006; Delseny et al. 2008; Matilla and Matilla-Vázquez 2008; García et al. 2010). In general, with the exception of lateral root initiation and fruit ripening (see flooding below), elevated levels of Et are deleterious to plant health and growth. Likewise, Et is also involved in environmental stress signaling upon wounding and during the interaction with pathogen and non-pathogen microorganisms (Pieterse et al. 2007, 2012; Verk et al. 2009). Consequently, the biosynthesis and perception of Et must be tightly controlled within the plant. The biosynthesis of Et begins with the transformation of methionine (Met), a scarce amino acid in plants, to S-adenosyl-methionine (SAM). This conversion is catalyzed by the SAM synthase (Peleman et al. 1989). SAM synthases are not specific to the Met cycle (Yang Cycle) since SAM also serves as substrate for several reactions, including cell transmethylations. Subsequently, the 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS; S-adenosyl-l-Met methylthioadenosine-lyase) catalyzes the rate-limiting step in Et biosynthesis by converting SAM into ACC and 5′-methyl-thioadenosine (MTA), which regenerates Met in several steps (Bradford 2008) (Fig. 7.1, left). Besides plants, the Yang Cycle is also found in bacteria, archeae, and animals and it is well known that in higher plants it is tightly regulated (Rzewuski and Sauter 2008). The ACS gene was first cloned from Cucurbita pepo (Sato and Theologis 1989) and then significant efforts were conducted to study this key ACS multigene family. All ACS members are under strict regulatory control and the abundance of ACS proteins is closely correlated with the level of Et production in most plant tissues. Furthermore, various members of the ACS gene family were found to be differentially expressed in response to developmental and environmental triggers (Tsuchisaka and Theologis 2004a, b). The ACS family includes 6 members in rice (Rzewuski and Sauter 2008) and 12 members in Arabidopsis, of which only 9 appear to be implicated in Et biosynthesis (Yamagami et al. 2003; Vandenbussche et al. 2006; Vandenbussche and Van der Straeten 2007; Lin et al. 2009). Three types of ACS have been defined based on their C-terminal regions, which are involved in enzyme stability. Five of the ACS genes and their expression patterns were described previously in deepwater rice internodes since at least two of them are sequentially induced during submergence (Rzewuski and Sauter 2008). Since OsACS5 expression is induced within 60 min of submergence, this family member might be responsible for the early increase in ACS activity. By contrast, OsACS1 expression is enhanced within 6 h of submergence. It has also been suggested that OsACS1 together with OsACS5 contributes to sustain Et production during long submergence (Zarembinski and Theologis 1997; van der Straeten et al. 2001). On the other hand, several MAPKs were found to regulate ACS activity (Broekaert et al. 2006; Schweighofer and Meskiene 2008). Thus, the phosphorylation of ACS2 and ACS6 by the MAPK MPK6 results in an increased production of Et (Christians et al. 2009). These phosphorylations also protect ACS2 and ACS6 from recognition and breakdown by the 26S proteasome pathway (Wang et al. 2004).
The last step of Et biosynthesis is catalyzed by ACC oxidase (ACO). In this metabolizing ACC reaction cyanoformic acid is also formed which is spontaneously degraded to cyanide (HCN) (Yip and Yang 1988). The HCN must be rapidly metabolized to keep its concentration below toxic levels. The molecular bases for HCN detoxification were recently studied in plants (Yi et al. 2012). The main HCN detoxification process described to date is catalyzed by β-cyanoalanine synthase (CAS), a pyridoxal phosphate-dependent enzyme that converts cysteine and HCN to HS and β-cyanoalanine. In Arabidopsis, the CAS gene family is composed of three members (Watanabe et al. 2008). The most abundant CAS protein (CYS-C1) is in the mitochondria, whereas CYS-D1 and DYS-D2 are found in the cytosol (Watanabe et al. 2008). Mitochondrial CAS is essential for formation of root hairs in Arabidopsis (García et al. 2010). HCN enhances the resistance of N. tabacum and Arabidopsis leaves to TMV and turnip vein clearing virus (TVCV), respectively (Wong et al. 2002). Likewise, it has also been proposed that HCN and Et are responsible for the resistance of young rice plants to blast fungus (Magnaporthe grisea) infection. In this fungus resistance mechanism, the induced OsACS2 and OsACO7 contributed specially (Iwai et al. 2006). On the other hand, plant pretreatment with KCN relieved stress induced by oxidative damage, and plainly induced the alternative oxidase (AOX) activity and Et production, proving a new fangled role of HCN against environmental stress (Xu et al. 2012).
In tomato and Arabidopsis ACO families are composed of four and six members, respectively (Babula et al. 2006; Lin et al. 2009). By contrast, in the rice genome six ACO members were found through computational analysis. Thus, in rice seedlings: (1) the highest expression of OsACO1 was found in the very young growing internodes (i.e., OsACO1 was induced after 4 h and at least up to 24 h of submergence; Mekhedov and Kende 1996); and (3) the expression of OsACO2 and OsACO3 were induced by auxin and Et, respectively, in a dose-dependent way (Chae et al. 2000). Taken together, Et biosynthesis is heavily regulated, including transcriptional and post-transcriptional control of the key enzymes (i.e., ACS and ACO). The presence of the enzyme ACC deaminase (ACCD), involved in the degradation of ACC to ammonia and α-ketobutyrate, is common in soil bacteria (Fig. 7.2), including biocontrol bacterial strains (Glick et al. 2007; Chen et al. 2013; Roca et al. 2013). ACC is a frequent component of seeds, roots, and leaves exudates (Glick et al. 2007) and bacteria can act as a sink of ACC, lowering Et levels in the plant. As a consequence, plant growth can be promoted and some of the potentially deleterious consequences of high Et concentrations under environmental stresses (e.g., flooding, heavy metals, salinity, drought, and microorganisms attack) may be reduced (Glick et al. 2007; Gamalero and Glick 2012; Stearns et al. 2012). Interestingly, several plant-associated bacteria have a positive effect over the Et levels in the plants that they are colonizing. Thus, (1) some pathovars of the plant-pathogen Pseudomonas syringae have the ability to synthesize Et both in vitro and in vivo (Weingart and Volksch 1997; Sato et al. 1997); (2) the Pseudomonas fluorescens root colonization trigger an increase of ACO activity in vivo (Hase et al. 2003); (3) the expression amounts of ACO1 and ACO2 are up-regulated by the infection of Botrytis cinerea (Adie et al. 2007); and (4) the transcriptional activation of ACO genes in tomato has been demonstrated in response to P. syringae infection (Weingart et al. 2001; Cohn and Martin 2005). Data on Et, JA, and SA production seems to conclude that a highly and tightly regulated Et biosynthesis may be used by pathogens Et producers to bypass defenses (Adie et al. 2007).
Model explaining the role of plant growth promoting rhyzobacteria (PGPR) in generating plant growth under general stress conditions (left) and particularly under flooding (right). In the left model, PGPR synthesize and secrete IAA. Bacterial IAA, together with the IAA synthesized by the root, induce ACS transcription and consequently the production of ACC. A percentage of this Et precursor can be degraded by root-associated bacteria causing a notable decrease in the biosynthesis of Et. The remaining ACC is exported to the plant shoot where the ACC oxidase (ACO) catalyzes the synthesis of Et, triggering plant growth. In the right model, flooding is the environmental factor that induces ACS expression. The role of the ACC deaminase (ACCD) in both models is evident
The Et signaling pathway is well established in Arabidopsis (de la Torre et al. 2006; Stepanova and Alonso 2009; Yoo et al. 2009). Thus, this gaseous hormone is sensed by receptors located in the endoplasmic reticulum. In the Arabidopsis there are five receptors (ETR1, ERS1, ETR2, ERS2, and EIN4), all of them with an active kinase domain (Stepanova and Alonso 2009; Yoo et al. 2009). The receptors operate as negative sensors of Et signaling and interact with Constitutive Triple Response 1 (CTR1), an Raf-like protein kinase (Fig. 7.1, left). In the absence of Et, CTR1 has a negative regulatory function, actively suppressing the Et signaling pathway. Upon Et-receptor binding, CTR1 is no longer capable of repressing Ethylene Insensitive 2 (EIN2) which is a transmembrane protein with homology to NRAMP metal ion transporters. EIN2 acts as a positive regulator of the Et responses. Et destabilizes the F-box proteins called ETP1 and ETP2, stabilizing EIN2 and promoting downstream effects (Qiao et al. 2009). EIN2 prevents the binding of the key Et Response Factors (EtRFs) EIN3 and its homolog EIN3-like 1 (EIL1) to EBF1 and EBF2 (EIN3 binding F-box proteins 1 and 2) which are part of an SCF E3 ligase complex (SCFEBF1/2) (An et al. 2010). Consequently, EBF1 and EBF2 are down-regulated by Et, suggesting that this gaseous hormone stabilizes EIN3/EIL1 by promoting EBF1 and EBF2 degradation by the proteasome complex. Thus, EIN3 (a short-lived transcription factor (TF) with five homologs in the Arabidopsis genome) and EIL1 are no longer degraded through the 26S proteasome pathway and induce transcription of EBF1 and EBF2 (Guo and Ecker 2003; Potuschak et al. 2003; Binder et al. 2007; Konishi and Yanagisawa 2008). When the Et levels decrease or Et is absent, EIN3 is ubiquitinated by SCFEBF1/EBF2 and degraded by the 26S proteasome. All this process is under control of EIN5, a 5′ → 3′ exoribonuclease that acts downstream of CTR1 (Fig. 7.1, left). In the presence of Et, EIN5 promotes the EBF1 and EBF2-mRNA decay, which allows the accumulation of EIN3 (Olmedo et al. 2006). In short, EIN3 is: (1) stabilized by Et; (2) phosphorylated by an MAPK cascade which can be activated by CTR1; (3) accumulated in nuclei after the increase in the Et levels with the subsequent binding to the promoter of EBF2; and finally, (4) together with EIL1, regulates the expression of target genes such as EtRF1, which encodes the transcription factor Et-Response Element Binding Protein (AP2/EREBP) involved in plant defense against necrotrophic pathogens (Glazebrook 2005; Verk et al. 2009; Zhao et al. 2012). EtRF1 and AtMYC2 are two notable regulators of Et–JA interactions in defense. However, AtMYC2 works in the opposite way to EtRF1 (for more information, see Adie et al. 2007). Interestingly, genes encoding group-VII EtRFs (Ethylene Response Factors) are up-regulated under anaerobic stress in several plant species (Nakano et al. 2006; Bailey-Serres et al. 2012).
Finally, it is especially important to note that during a stress process: (1) the Et action mode is modulated by the concentration of the hormone rather than by its presence (Pierik et al. 2006); (2) Et, SA, and JA signaling pathways, individually or in crosstalk, play significant roles in the physiology of stress in land plants (Wasternack 2007; Thaler et al. 2012); (3) during resistance to necrotrophic pathogens, Et synergistically with JA plays a key role, as demonstrated by genetic approaches (Grant and Jones 2009; Pieterse et al. 2012); and (iv) ACC-JA conjugation may be fundamental for the Et-JA crosstalk regulation (Wasternack 2007; Fonseca et al. 2009).
3 Crosstalk Between Oxygen Deficient Stress and Ethylene Biosynthesis and Signaling
O2 is the final electron acceptor in the mitochondrial respiratory chain. In soil, and more specifically in the rhizosphere, O2 concentrations can be limiting (hypoxia) or absent (anoxia). The decrease of O2 diffusion capacity in the soil (e.g., compact structure, water logging, and deep flooding) limits its availability for the root (Dat et al. 2004). Thus, the O2 shortage in the soil generates a partial pressure around radical system incapable to oxygenate in the root the machinery of respiratory ATP biosynthesis. Additionally, the consumption of O2 by aerobic rhizosphere microorganisms can further aggravate the root stress. Indirect and direct sensing of O2 status may be responsible for the acclimatization responses that extend survival under O2 deprivation (Bailey-Serres and Chang 2005). For this reason, plants can adapt to this energy crisis by promoting anaerobic metabolism and thus increase substrate-level ATP production (Magneschi and Perata 2009).
Rice (O. sativa) is a model plant for the study of metabolic control under O2 limiting conditions since this semiaquatic organism is well adapted to a partially flooded environment. However, abrupt flooding can cause sharp submergence by imposing, among other factors, a complex stress due to a 103-fold reduction in the diffusion of O2 and CO2. The growth of deep water rice in wetlands is adapted to gradual flooding by means of acceleration in the elongation of submerged internodes to keep aerial tissues above the air–water environment. When sudden submerged, deepwater and most lowland varieties accelerate internode and/or leaf elongation to avoid the flooding. By contrast, lowland varieties tolerant to submergence save complete submergence through a constraint in shoot elongation and carbohydrate spending, thereby conserving energy reserves to restarting development upon desubmergence (Fig. 7.3). Consequently, an immediate response must be triggered by the plant in order not to block energy biosynthesis (Geingenberger 2003; Bailey-Serres et al. 2012). Thus, almost 50 genes responding to O2-shortage, including EtRFs, were identified in several species such as Arabidopsis, rice, cotton, and poplar (Mustroph et al. 2010). Recent reports contain excellent updates on the molecular biology of O2-shortage response (Mustroph et al. 2010; Bailey-Serres et al. 2012; Licausi 2011, 2012).
3.1 Role of Ethylene Response Factors Under Low-Oxygen Stress
A large quantity of microarray data for Arabidopsis and rice under low-O2 stress (i.e., anoxia and hypoxia) are available, and these experiments have revealed much about plant responses to low O2 (Licausi et al. 2010; Mustroph et al. 2010; Lee et al. 2011; Licausi 2012). For example, EtRFs are TFs unique to plants that bind specifically to TAAGAGCCGCC (GCC box) sequences found in the promoter regions of Et Response (EtR) genes (e.g., Hookless1). EtRFs are ubiquitous in the plant kingdom and their functional implications have been studied in a wide range of processes including response to biotic and abiotic stresses (for more information, see Pirrello et al. 2012). The EtRF family is a large gene family of TFs which is part of the APETALA2 (AP2)/EtRF superfamily. AP2 is one of the largest families of TFs in plants, including three different sub-families which are characterized by the number of EtRF domains and by having either one or two AP2 DNA-binding domains. The EtRF, also known as the Et-Responsive Element-Binding Protein (EtREBP) family, has one AP2 domain, the RAV family has two domains (i.e., AP2 and B3), and the AP2 family has two AP2 domains (Nakano et al. 2006; Romanel et al. 2009). In Arabidopsis and rice, the EtRF family comprises about one hundred members which are categorized into ten clades. Clade VII has an MCGGAI/L highly conserved motif at its NH2-terminal (Nakano et al. 2006). In all rice varieties studied, a sub-group VIIb exists where all members lack this NH2-terminal motif. On the other hand, a major QTL responsible for tolerance to submergence, Submergence1 (SUB1; located in chromosome 9), was identified in varieties of lowland indica rice (Fukao et al. 2006; Xu et al. 2012). This SUB1 locus consists of a clade of three sub-group VIIb genes (OsSUB1A, OsSUB1B, and OsSUB1C genes), but the SUB1A is present only in indica and not japonica cultivars. OsSUB1C acts downstream of OsSUB1A (Fukao et al. 2006).
The expression of an Arabidopsis clade VII gene, AtRAP2.2, is induced by Et in shoots but not in roots (Hinz et al. 2010). RAP2.2 protein only affects to the induction of genes linked to sugar metabolism, fermentation, and Et biosynthesis (Hinz et al. 2010). Unlike rice, Arabidopsis possesses five genes within group VII, including HYPOXIA-RESPONSIVE1 (HRE1) and HRE2. The plants overexpressing HRE1 and HRE2 showed an increased tolerance to anoxia, whereas the hre1hre2 double mutant showed reduced tolerance (Licausi et al. 2010). A further study showed that in the presence of exogenous ACC transgenic seedlings with silenced HRE1 displayed exaggerated apical hook curvatures. These results indicate a negative role of HRE1 in the Et responses (Yang et al. 2011). HRE1 and HRE2 shows a strong up-regulation under O2 depletion, mediated by both Et-dependent and Et-independent signals (Licausi et al. 2010; Yang et al. 2011). Like SUB1A, HRE1 transcript accumulation is induced by Et, which synergistically increases its rise during O2 stress (Yang et al. 2011). Not long ago, another member of the AP2/Etr2 family named Octedecanoic-Responsive Arabidopsis59 (ORA59) was found to be as the more important integrator of the JA and Et signaling pathways. ORA59 is induced and synergistically activated by JA and Et.
Et also induces the gene expression of alcohol dehydrogenase (ADH1) in Arabidopsis (Peng et al. 2001, 2005). Ethanolic fermentation through ADH1 activity contributes substantially to low-O2 stress adaptation. For this reason, an adh1 null mutant showed lower survival when exposed to low-O2 pressure (Ellis et al. 1999). Likewise, the pyruvate decarboxylases (PDC1 and PDC2) overexpression in Arabidopsis results in improved survival under low-O2 conditions (Ismond et al. 2003). EtRFs are also involved in several developmental processes such as zygotic embryogenesis (Riechmann and Meyerowitz 1998) and abiotic and biotic stress responses (Fujimoto et al. 2000; Sakuma et al. 2002).
Finally, the degradation of clade VII-EtRF proteins is carried out by the N-end rule pathway (i.e., N-erp; Hinz et al. 2010; Gibbs et al. 2011; Bailey-Serres et al. 2012). More specifically, all five Arabidopsis VII-EtRFs proteins are N-end rule substrates. N-erp is a pathway to degrade proteins that relates the in vivo stability of a specific protein to the nature of its N-terminal. These N-terminal destabilizing residues are known as N-degrons (Varshavsky 2011). In eukaryotes, N-erp is a part of the ubiquitine (Ub) system (Graciet and Wellmer 2010).
3.2 Crosstalk Between Low-O2 and Ethylene Under Submergence
Many investigations have demonstrated the involvement of Et in O2-shortage responses (i.e., flooding and submergence). In contrast to flooding avoidance, which involves increased Et and enhanced stem elongation rates to permit the plant to have access to atmospheric O2 (Kende et al. 1998), submergence tolerance is the result of an efficient reduction in the consumption of carbohydrates and an ethanolic fermentation-dependent metabolism, together with a reduced production of Et and restricted cell elongation (Jackson and Ram 2003). Careful research in rice and a wetland dicot, marsh dock (Rumex palustris), pointed out that Et accumulation in submerged organs triggers a hormonal signaling pathway that cause the reduction of the antagonism between gibberellins (GA) and abscisic acid (ABA) which is usually responsible for the restriction of the internodal cell elongation. In submerged parts, the restriction of internodal elongation is achieved via a decreased responsiveness to GA arising from elevated levels of DELLA proteins that repress GA-induced growth (Fukao and Bailey-Serres 2008). SNORKEL (SK) 1 and 2 and SUB1A (EtRFs that confers prolonged tolerance to submergence in deepwater rice) genes are involved in the above signaling cascade (Hattori et al. 2009; Bailey-Serres and Voesenek 2010). The deepwater rice adaptation to flooding is the result of its ability to elongate the cell internodes. These internodes possess hollow structures which prevent plant drowning allowing gas exchange with the atmosphere. The internode elongation response in deepwater rice is regulated by Et (Hattori et al. 2009). Many physiological and molecular studies have shown that Et, GA, and ABA signaling are implicated in the elongation response. However, most of the gene(s) involved in this trait needs to be identified. Thus, the Hattori´s group found for the first time that the EtRFs-encoding genes SK1 and SK2 trigger deepwater response. Consequently, the deepwater rice requires SK1 and SK2 to extend the hollow stem to the water surface through the elongation of its stem internodes (Hattori et al. 2009). Therefore, under these deepwater conditions, Et accumulates and induces expression of SK1 and SK2 whose products triggers notable internode elongation via GA (Hattori et al. 2009).
As indicated above (section “Role of Ethylene Response Factors Under Low-Oxygen Stress”), several EtRF proteins from the major QTL SUB1 were demonstrated to have a main role in submergence tolerance in rice (Xu et al. 2006). Both flooding and submergence are controlled by SUB1A, SUB1B, and SUB1C. However, since the expression of SUB1A-1 confers submergence tolerance to submergence intolerant rice plants, SUB1A is thought to be the key gene in this SUB1 gene cluster (Xu et al. 2006). Some key features of SUB1A-1 are described below. SUB1A-1 overexpression in japonica rice, a flooding-sensitive cultivar, resulted in an enhanced ADH1 expression and tolerance to flooding (Fukao et al. 2006; Xu et al. 2006). Several authors have proposed that the conferred submergence tolerance is the result of a complex signaling pathway that reduces carbohydrate consumption and growth elongation (Fukao et al. 2006; Xu et al. 2006; Perata and Voesenek 2007; Jung et al. 2010). SUB1A-1 transcripts, as with SK1 and SK2, are Et-induced. Additionally, (1) SUB1A-1 boosts the accumulation of SLENDER RICE 1 (SLR1) and SLENDER RICE-LIKE 1 (SLRL1), two negative regulators of GA responses; and (2) SUB1A-1 protein ultimately limits Et biosynthesis (Fukao et al. 2006; Fukao and Bailey-Serres 2008). Other effects induced by SUB1A-1 were described by Bailey-Serres et al. (2012). All together, SUB1A-1 seems to be included in an appropriate point in the signaling pathway belonging to submergence response. Thus, SUB1A-1 maintains cell viability and prevents plant growth during submergence stress. Furthermore, during a subsequent recovery period (i.e., reoxygenation), SUB1A-1 is also involved in homeostasis restoration. The reduced elongation response is only beneficial when the submergence is deep and/or relatively short lasting. However, when the submergence is prolonged but relatively shallow floods, several plant species have been shown to elongate their stems in a hormonal-dependent manner. Thus, the accumulated Et inhibits ABA biosynthesis and increases its degradation resulting in reduced levels of ABA (Benschop et al. 2005, 2006; Saika et al. 2007). The decline of ABA levels results in the release of the repression of GA biosynthesis promoting the increase of the concentration of bioactive GA in the submerged tissues. Additionally, in response to Et and submergence, the sensitivity to GA is also enhanced, through yet unknown mechanisms. SK1 and SK2 genes, belonging to the same APETALA2/EtRF subfamily as the SUB1A-1 gene, play a role in rice elongation when submerged (Hattori et al. 2009). Although it is not known whether the SK genes interfere with GA biosynthesis or action, it has been demonstrated that they act upstream of GA. A rapid underwater elongation requires carbon and energy, and, therefore, depends on the accessibility to nonstructural carbohydrates. Chen et al. (2010) shown that the translocation of newly fixed carbon to the elongation tissues and the mobilization of starch can both be induced under submergence conditions (Chen et al. 2010). Model explaining the relationship between Et, ABA, and GA in submergence adaptation process of rice is indicated in Fig. 7.3.
SUB1A perhaps can represses cell elongation though an involving expansin-A, increase in ethanolic fermentation via control of ADH gene expression, and a decrease in carbohydrate consumption, among other metabolic factors (Bailey-Serres and Voesenek 2010). Strikingly, SUB1A represses SUB1C which acts in an antagonistic way by promoting GA-induced carbohydrate breakdown and cell elongation. Both SUB1A and SUB1C are induced by Et. However, since SUB1A responds to Et at concentrations two orders the magnitude lower than SUB1C, is expected to be induced earlier. Therefore, in the presence of SUB1A, a delay in the induction of the expression of SUB1C during submergence is observed (for more information see Rzewuski and Sauter 2008). Although it is clear that several hormones, cell wall loosening proteins and carbohydrates are required for the elongation response, nowadays is poorly understood which part of the signal transduction pathway may cause the differences within and among naturally occurring species. In contrast to wild species, more research has been done in cultivated rice varieties to explain the variation in underwater elongation.
Recently, Chen et al. (2010) suggested that, under submergence conditions, the variation in the elongation rate of the petioles of the wetland plant Rumex palustris is controlled by an Et-regulated pathway that alters the dynamics of endogenous ABA levels in the petioles. This variation in the endogenous ABA concentration affects the responsiveness to GA and consequently the underwater petiole elongation rate. In this wetland species, the stimulation or inhibition of the underwater elongation is controlled by the AP2/EtRF genes (Voesenek and Bailey-Serres 2009). The slow elongating varieties maintain relatively high levels of ABA, which then results in a limited GA responsiveness and thus reduced growth rate. The effect of ABA on GA in the model species R. palustris suggests a novel role of ABA regulating GA. Notoriously, if we compare this study with previous research investigating the role of Et and ABA under submergence conditions in the fast and slow elongating species R. palustris and R. acetosa, respectively, the results strongly indicate that differences between and within species in petiole elongation induced by flooding are controlled by the same switch point(s) and pathway(s), i.e., by regulating the levels of ABA and the subsequent GA responses (Benschop et al. 2005; Chen et al. 2010). It may be hypothesized that the inter- and intra-species genotypic variation in wetland plant species is the result of the strong selective force exerted by flooding stress.
3.3 Ethylene and Flooding
To survive flooding, many plant species have evolved by developing new adaptive traits (Bailey-Serres and Voesenek 2010; Bailey-Serres et al. 2012). The privatization of O2 to the roots is the main consequence of flooding. Flooding together with salinity, dryness, and temperature is an important generator of abiotic stress and significantly affects distribution of plants in terrestrial environments. Shortage (i.e., hypoxia; [O2] < 50 mmol m–3) or absence (i.e., anoxia) of O2 in waterlogged environments generates different responses in root systems (Matilla and Rodríguez-Gacio 2013). Under flooding, gases diffuse 104-fold slower. Thus, within the first 60 min of flooding, a decline from 20.8 to 7.9 kPa in the partial pressure of O2 was observed, which continues to decrease to 1 kPa after 24 h. Under low O2 conditions, soil microorganisms are the main consumers of the available O2 and several toxic compounds may accumulate in the rhizosphere. The O2 consumption by soil microorganisms generates a strong stress around the roots. Some plants (e.g., rice) may remain temporarily in soils with low O2 levels and show a positive response to Et and enhanced tissue sensitivity to GAs (Knaap et al. 1996). Therefore, survival of rice upon a great increase of the water level depends on the fast elongation of the stem, which is Et-regulated. OsACS1 alone, or in combination with OsACS5, maintains Et production during submergence (van der Straeten et al. 2001; Rzewuski and Sauter 2008). It was hypothesized the increase in the OsACS expression, together with the increase in the activity of OsACS due to the escape of OsACS1 from OsEOL-mediated degradation, result in a rising of Et production within the first hours of submergence (Yoshida et al. 2006). The appearance of aerenchyma vessels (i.e., soft tissues), which allow O2 exchange from the aerial parts to the root tissues and adventitious roots, was an evolutionary key for the flooding adaptation (Vartapetian and Jackson 1997; Watkin et al. 1998; Bacanammwo and Purcell 1999; Gunawardena et al. 2001; Aschi-Smiti et al. 2004). Aerenchyma formation occurs through two different processes: schizogeny and lysigeny. Schizogenous aerenchyma is characteristic of Rumex spp. and involves reorganization of the cell wall (CW) and cell separation. However, in plant such as Arabidopsis or rice, programmed cell death (PCD) is responsible for the formation of lysigenous aerenchyma. Many of the adaptive growth responses occurring in roots under hypoxic conditions, including aerenchyma formation, occur in response to Et which is stored by physical trapping in flooding soil solution and submerged parts of plants at concentrations of 103 mm3 dm−3 (Voesenek et al. 2006). Therefore the application of Et induces the aerenchyma formation in hypoxic maize roots, while the presence of Et inhibitors repress its appearance (Dat et al. 2004).
The expression level of genes responsible for Et biosynthesis is up-regulated under flooding conditions (van der Straeten et al. 1997; Peng et al. 2005). Thus, in root tissues ACO activity is inhibited (Voesenek et al. 1993) and ACS activated (Van Der Straeten et al. 2001; Rieu et al. 2005), generating high levels of ACC (Geisler-Lee et al. 2010). Notably, since ACC is a mobile molecule does not necessarily require to be produced at sites where Et acts. Under flooding, ACC synthesized in plant roots is transported via the xylem to enable the biosynthesis of Et in the distant tissues (Finlayson et al. 1991). Therefore, the ACC must be transported to the next aerobic zones (i.e., shoots) for its conversion into Et. In tomato plants, English et al. (1995) showed that ACO activity regulates the Et production in response to flooding. On the other hand, during flooding of Rumex palustris, Et biosynthesis seems to be limited at the level of ACO activity rather than by ACS (Voesenek et al. 1993). However a portion of ACC biosynthesized by the roots is translocated to the rhizosphere and become available to bacteria possessing ACCD (see above, section “Updated Overview of the Plant Hormone Ethylene”) (Fig. 7.2). If bacteria with ACCD are not abundant in the rhizosphere, the ACC is mostly translocated to the oxygenated upper parts of the plant for subsequent transformation to Et (Grichko and Glick 2001). Interestingly, aerenchyma formation does not always require Et. In some species such as Arabidopsis, constitutive lysigenous aerenchyma is formed in response to Et and H2O2 signaling (Mühlenbock et al. 2007). In support of the latter, the involvement of ROS, Ca2+ signaling, and CW metabolism in aerenchyma formation was recently demonstrated under waterlogged conditions (Rajhi et al. 2011).
Finally, to summarize rice adaptation to flooding: (1) the triggered Et biosynthesis and accumulation leads to an increase in bioactive GA and appearance of PCD; (2) PCD of epidermal cells facilitates emergence of adventitious roots at the nodes of the submerged stems, while GA induces the internodal growth; (3) Et prevents ABA biosynthesis and consequently the GA action on growth and PCD.
3.4 Crosstalk Between Low-O2 and Ethylene in Seeds
The production of seeds is crucial and represents the main strategy that allows most plants species to maintain their genetic diversity, survive, and spread. Before germination is triggered, viable seeds can overcome long periods of severe desiccation and dormancy (Iglesias-Fernández et al. 2011; Graeber et al. 2012). Indeed, one of the key milestones during plant evolution has been the acquisition of desiccation tolerance (Linkies et al. 2010). Under desiccation conditions, the seed undergoes strong metabolic and hormonal readjustments, such as an increase in dehydrin and ABA levels (Rodríguez-Gacio et al. 2009; Leprince and Buitink 2010). During seed development and early imbibition, the internal high metabolic activity and the outer seed layers (i.e., seeds coats) prevent O2 diffusion. Likewise, this hypoxic environment inside the seed causes an ATP deficiency (Borisjuk and Rolletschek 2009). Hence, the seed needs to develop strategies to reduce or prevent O2 restriction besides an ability to adjust its endogenous levels of O2 as well as O2 demands. For this, the seed requires mechanisms for O2-sensing and O2-dependent regulatory systems (Bailey-Serres and Chang 2005; Borisjuk and Rolletschek 2009). Although the O2 sensors have not been definitely identified, in Arabidopsis seedlings, two independent research groups have recently demonstrated that one branch of the Ub-dependent N-end rule pathway functions as a mechanism for sensing O2 (Gibbs et al. 2011; Licausi 2011; Licausi et al. 2011). Additionally, an increasing amount of data supports the leading role for the non-symbiotic hemoglobins/NO (nsHbs/NO) cycle in O2-sensing (Sairam et al. 2009; Siddiqui et al. 2010; Matilla and Rodríguez-Gacio 2013). However, it has not yet been successfully demonstrated whether Et biosynthesis and signaling are involved in triggering processes of hypoxia in seeds. The down-regulation of the nsHbs1 biosynthesis in Hordeum vulgare (barley) enhanced the production of Et in Zea mays (maize) suspension cells during hypoxia (Manach-Little et al. 2005). On the other hand, studies in Gossypium hirsutum showed that GhnsHb1 expression is up-regulated by Et, SA, and JA, suggesting that GhnsHb1 may be involved in defensive mechanisms (Qu et al. 2006).
4 Ethylene and Plant Defense Against Microorganisms
Land plants are anchored to the soil and therefore the root system is in close contact with the neighboring soil environment (Darrah and Roose 2007). The release of nutrients in the form of root exudates to the rhizosphere (Loyola-Vargas et al. 2007; Newman and Römheld 2007; Uren 2007; Badri and Vivanco 2009) results in a highly active and dense population of microorganisms. In fact, bacterial population densities in the rhizosphere can reach 1–2 orders of magnitude higher than in the bulk soil (Molina et al. 2000; Morgan et al. 2005). The root exudation occurs through root hairs and both the apex and young parts of roots (Newman and Römheld 2007; Uren 2007) and influences microbial root colonization (Lugtenberg and Bloemberg 2004; Gamalero et al. 2005; Watt et al. 2006). At the same time, rhizosphere colonizing microorganisms can directly alter the metabolism and development of the root system (Ahemad and Khan 2011; Berendsen et al. 2012).
The presence of rhizosphere microorganisms can affect the root exudate properties due to an active degradation of its components (Jones et al. 2003). Furthermore, rhizosphere microorganisms can also increase the exudation levels and alter the root exudates composition, facilitate the availability of some soil nutrients and promote the plant growth (Phillips et al. 2004; Rosas et al. 2006; van Loon 2007; Lugtenberg and Kamilova 2009; Matilla et al. 2010). Additionally, plant-associated microorganisms can synthesize plant hormones such as cytokinins, GA, and auxins (Preston 2004; Vessey 2003; Ahemad and Khan 2011; Roca et al. 2013) besides releasing Et (Freebairn and Buddenhagen 1964; Weingart and Volksch 1997; Sato et al. 1997). Microorganisms use two different Et biosynthetic pathways, both different from that of higher plants (see above). Thus, most of these microorganisms produce small traces of the hormone via the 2-keto-4-methylthiobutyric acid (KMBA) pathway, in which the NADH:Fe(III)EDTA oxidoreductase generates hydroxyl radicals from molecular O2 (Fukuda et al. 1989; Nagahama et al. 1992). However, several microorganisms can synthesize Et using 2-oxoglutarate as precursor via an Et-forming enzyme (Weingart and Volksch 1997).
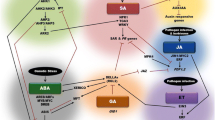
During evolution, plants have acquired a complex system of defense mechanisms that protect them against plant-pathogenic fungi, oomycetes, and bacteria, besides viruses and nematodes (Bari and Jones 2009). Successful plant pathogens can interfere or block the plant immune system whereas beneficial plant–microorganisms associations can promote plant growth and help to overcome different environmental stresses. However, beneficial microorganisms are firstly recognized as potential pathogens and the plants can react to their presence by activating an immune response (Pieterse et al. 2012). Thus, the recognition of pathogen- or microbe-associated molecular patterns (PAMP/MAPS) by the plant can also trigger the so-called effector-triggered immunity (De Vleesschauwer and Höfte 2009). Found mostly in plant-associated bacteria, PAMP/MAPS are bacterial determinants such as flagella, lipopolysaccharides, siderophores, and antibiotics, amongst others (reviewed by Bakker et al. 2007; De Vleesschauwer and Höfte 2009; Vlot et al. 2009). Recently, it was shown that microbial elicitors and JA differentially modulates the plant’s innate immune response (Flury et al. 2013). Plant pathogen infection may result in the induction of systemic acquired resistance (SAR), a broad spectrum, and long-lasting disease resistance. SAR is generally involved in the protection against (hemi-)biotrophic pathogens (Glazebrook 2005) and its induction requires the accumulation of SA. Moreover, SAR-induced plants show increased expression of pathogenesis-related (PR) genes (Durrant and Dong 2004; Vlot et al. 2009; Fu and Dong 2013). On the other hand, the plant root colonization by certain non-pathogenic PGPRs can suppress disease by triggering systemic induced resistance (ISR). ISR is phenotypically similar to SAR but it is dependent of the Et and JA signaling pathways (van Loon and Bakker 2005; De Vleesschauwer and Höfte 2009) (Fig. 7.4). In general, ISR is associated with defense against necrotrophic pathogens and herbivorous (Glazebrook 2005; Pieterse et al. 2012) and is not associated with an enhanced expression of PR genes (van Loon and Bakker 2005; De Vleesschauwer and Höfte 2009). Interestingly, the ISR induced by the rhizobacteria Pseudomonas fluorescens WCS417r is not associated with the endogenous increase of the JA and Et, suggesting that enhanced hormonal sensitivity causes this improved defense (Pieterse et al. 2000; De Vleesschauwer and Höfte 2009 and references therein). PGPR-mediated ISR has been shown to be efficient against a broad range of plant pathogens on both monocotyledonous and dicotyledonous species (reviewed by Bakker et al. 2007; De Vleesschauwer and Höfte 2009) and it is well known that for its induction an effective colonization of the rhizosphere is required (Raaijmakers et al. 1995). Both SAR and ISR signaling pathways have been shown to be dependent on the transcriptional activator NPR1 (Non-expresser of Pathogenesis-Related; Pieterse et al. 1998, 2007; Niu et al. 2011; Zhang et al. 2012) (Fig. 7.4).
Elicitation of induced systemic resistance (ISR) and systemic acquired resistance (SAR) transduction pathways in Arabidopsis thaliana. (a) Simplified model for triggering of SAR and ISR. etr1 (ET receptor mutant 1 plants); jar1 (JA response 1 mutant); NahG (SA non-accumulating transgenic plants); npr1 (non-expressor of PR genes 1 mutant plants). (b) Quantification of ISR and SAR in Arabidopsis plants infected with P. syringae pv. tomato DC3000. ISR was induced by inoculating plant roots with the rhizobacterium P. fluorescens WCS417r. SAR was triggered by infiltrating plant leaves with an avirulent variant of P. syringae pv. tomato. Disease index represents the percentage of leaves showing symptoms relative to the control plants. Wt: wild type; C: non-treated plants. Adapted from Pieterse et al. (1998) with permission of Dr. Pieterse
4.1 Involvement of Ethylene in Pathogenic Infections
In ISR-triggered plants no defense mechanism is activated before the recognition of a pathogen. However, the plant tissues are sensitized to react faster and strongly in response to the pathogen, a phenomenon known as “priming” (Verhagen et al. 2004; Conrath 2009). For example, experiments with endophytic biocontrol strain Enterobacter radicincitans DSM 16656 demonstrated that this bacterium is capable of inducing priming via SA or JA/Et signaling pathways to protect plants against potential pathogen attack (Brock et al. 2012) (Fig. 7.4). Importantly, primed plants show a wide spectrum of resistance with low impact on the plant fitness (i.e., plant growth and seed production) (Van Hulten et al. 2006). A number of studies show that priming: (1) often depends on the induced disease resistance key regulator Non-expresser of Pathogenesis-Related genes (NPR1) (León-Reyes et al. 2009); and (2) is an evolutionary advantage over constitutive activation of defense response (Van Hulten et al. 2006; Conrath 2009).
A hypothesis on the involvement of the Et signaling in the plant defense mechanisms in the presence or absence of a pathogen is shown in Fig. 7.1. It has long been known that Et can act positively and negatively on plant immunity (van Loon et al. 2006). Thus, pathogen attack activates Et production in many plants (Broekaert et al. 2006; van Loon et al. 2006 and references therein) and rhizobacteria-mediated ISR requires responsiveness to Et and JA (van Wees et al. 2008; Pieterse et al. 2007). Unfortunately, the role of Et during the plant–pathogen interaction has remained secondary and deserves more attention. Thus, after the infection, plants often respond with a rapid rate of Et biosynthesis (Iwai et al. 2006; van Loon et al. 2006 and references therein). Pathogenic infection triggers a rapid and low Et biosynthesis from pre-existing ACC in affected tissues. This first Et wave may be a protective response by the plant (van Loon et al. 2006). Subsequently, the activation of the transcription of the ACS genes to generate a net biosynthesis of Et immediate precursor and then a highly elevated ACO activity provokes a second wave of hormone (Iwai et al. 2006; van Loon et al. 2006 and references therein). If the pathogenic attack is ongoing, autocatalytic biosynthesis of Et takes place. This remarkable process is highly damaging for the infected plant. Therefore, it is logical to suppose that (1) the inhibition of the biosynthesis of Et decreases the severity of infection; and (2) transgenic plants with high expression of ACCD are strongly protected against some pathogenic attacks (Czarny et al. 2006; Glick et al. 2007).
The ISR model system Arabidopsis-Pseudomonas fluorescens WCS417r is one of the best characterized (Pieterse et al. 2007; De Vleesschauwer and Höfte 2009 and references therein). In this model, the Arabidopsis mutants etr1 (ET-response) and jar1 (JA-response) were unable to trigger resistance against the pathogen bacteria P. syringae after colonization with P. fluorescens WCS417r (Pieterse et al. 1998). Investigation with other mutants in Et signaling concluded that the establishment of ISR requires an intact Et signaling pathway (Ton et al. 2002a). Particularly interesting results emerged from the study of the eir1 mutant, insensitive to Et in the roots but not in the shoots. Arabidopsis eir1 plants were unable to show ISR after root colonization by the rhizobacteria WCS417r. However, eir1 mutants exhibited ISR when the strain WCS417r was infiltrated into the leaves, suggesting the importance of responsiveness to Et at the site of application (Knoester et al. 1999). Interestingly, in Arabidopsis, etr1 plants failed to exhibit ISR after treatment with ACC or JA. However, jar1 plants were able to response to JA but not to ACC suggesting that JA pathway acts upstream of Et pathway in the signaling cascade (Pieterse et al. 1998).
It is interesting to point that the locus ISR1, encoding a key component of the Et signal transduction pathway, is required for both ISR and basal resistance in Arabidopsis (Ton et al. 1999, 2001, 2002b). Likewise, the endogenous Et levels are crucial for the development and fine-tuning of appropriate defense responses (Zhao et al. 2012, and references therein). The importance of Et content in plant defense responses may have led to the development of Et-producing pathogens. These evolved pathogens might interfere with the Et plant status altering or preventing the defense response to their benefit.
As described previously, Et alone or in combination with other hormones is involved in determining the most appropriate defensive response. However, the function of Et in plant defense is complex and highly regulated. This is reflected in the enumeration of Et-associated mutants and their susceptibility to phytopathogens (van Loon et al. 2006). For example, although ACS expression is poorly understood during pathogenesis, recent results indicate that the rice OsEDR1 (Enhanced Disease Resistance 1; ortholog of Arabidopsis EDR1) is a positive regulator of Et biosynthesis. Thus, the expression of the ACS gene family was suppressed in OsEDR1-defective mutants resulting in rice plants more resistant against the biotrophic pathogen Xanthomonas oryzae pv. oryzae (Shen et al. 2011). The TFs EIL1 and EIN3 regulate the expression of the Et transcriptional activator ERF1. Likewise, ERF1 regulates EtR and Et defense-related genes (e.g., Pathogenesis-Related gene 3 (PR-3) and Plant Defensin 1.2) playing a role in the defense against necrotrophic pathogens (Berrocal-Lobo and Molina 2004; Adie et al. 2007). In Arabidopsis, Et appears to act antagonistically in SA signaling. Thus, it was demonstrated that EIL1 and EIN3 repress SA biosynthesis by binding to the isochorismate synthase 1 promoter, a well-known SA biosynthetic gene (Robert-Seilaniantz et al. 2011; Pieterse et al. 2012). Conversely, Et potentiated the response of Arabidopsis plants to SA, resulting in a increased expression of PR-1, an SA-responsive gene (De Vos et al. 2006). Moreover, in tobacco (Nicotiana tabacum) Et was shown to be key player for the establishment of SA-dependent SAR against TMV (León-Reyes et al. 2009 and references therein).
Considerable research in recent years has demonstrated that Et regulates the expression of defensive genes such as PR-2 (β-1, 3-glucanases), PR-3 (chitinases), and PR-12 (plant defensin factors) (van Loon et al. 2006). However, Et works as a component of a tangled network of signaling compounds including SA, JA, and ABA. Likewise, in different plant species the presence of the GCC box (see section “Role of Ethylene Response Factors Under Low-Oxygen Stress”) was demonstrated to be essential, and sometimes sufficient, for the regulation of the expression PR genes by EtRFs (Adie et al. 2007). The EtRFs–GCC binding can also take place in promoters of EtR genes not involved in pathogenesis (e.g., Hookless1), evidencing a wider role for GCC box in the transcriptional regulation by Et. On the other hand, EtRF family members can activate or repress concrete defense pathways, often with opposite effects, resulting in susceptibility or resistance to the attacking pathogens (Berrocal-Lobo and Molina 2004; McGrath et al. 2005; Ham et al. 2006). Other examples of the involvement of Et in plant defense are listed below. In Arabidopsis, Et has also been involved in both local and systemic defensive responses against the necrotrophic fungus Alternaria brassicicola. Et, but not SA or JA, was capable of inducing the expression of the Arabidopsis secreted lipase GLIP1, which shows antifungal activity against A. brassisicola (Oh et al. 2005). More recently, the elicitation of systemic resistance was shown to not significantly alter the structure community of rhizosphere bacteria (Doornbos et al. 2011). Referring to aggressive pathogens, the necrotrophic fungus Botrytis cinerea is one of the most stressful and destructive (Williamson et al. 2007). Et, synergistically with JA, plays a key role during resistance to necrotrophic pathogens (van Loon et al. 2006; Grant and Jones 2009). In a recent study, Zhang et al. (2012) found that the mutation of the Arabidopsis mediator complex subunit 16 (MED16) blocks the expression of several Et and JA response genes compromising, consequently, the plant defenses against necrotrophic pathogens such as B. cinerea and A. brassicicola. Furthermore, studies with Arabidopsis have shown that the ein2 and the ein3eil1 double mutant, both Et-insensitive, are more susceptible to B. cinerea (Alonso et al. 2003). Several EtRFs (e.g., ORA59, RAP2.2, and EtRF1) have been also recognized as remarkable regulators in the Botrytis resistance (Nakano et al. 2006; Wehner et al. 2011; Zhao et al. 2012). Moreover, ectopic expression of EtRF1 and ORA59 enhanced resistance of Arabidopsis to B. cinerea, Fusarium oxysporum, and Plectosphaerella cucumerina (Berrocal-Lobo and Molina 2004; Pré et al. 2008). Taken together with the RAP2.2 function in low-O2 tolerance (see section “Role of Ethylene Response Factors Under Low-Oxygen Stress”), the Zhao group’s data suggested that RAP2.2 (1) may act as a global regulatory protein in the Et signaling pathway and could play a dual role in the low-O2 tolerance and Botrytis resistance; and (2) might serve as a global TF involved in the regulation of the Et signaling pathway and as node in the crosstalk signaling between biotic and abiotic stress responses (Zhao et al. 2012). Recently, it has been shown that EtRF6 is a notable regulator of biotic stress defense. Thus, EtRF6 controls the ROS-responsive genes expression after activation by MPK3/MPK6 (Wang et al. 2013). Likewise, ERF6 plays a dual role under stress as it activates both stress tolerance and growth inhibition, and both roles take play independently from each other (Dubois et al. 2013).
4.2 Non-pathogenic Infections and Induced Ethylene Production
As described above, different biotic and abiotic stresses can cause an imbalance in the Et production of land plants and the increased level of gaseous phytohormone can inhibit the overall plant growth or the length of specific organs including roots (Bleecker and Kende 2000; Mayak et al. 2004; De la Torre et al. 2006; Matilla and Matilla-Vázquez 2008). Et and JA have been shown to be required for the establishment of a broad-spectrum ISR response, stressing the crucial modulating role of Et in plant defense (van Wees et al. 2008). Thus, Et and JA are indispensable for the development of ISR in leaves after root colonization by beneficial microorganisms such as Piriformospora indica (Verma et al. 1998) and P. fluorescens (van der Ent et al. 2009). The fungus P. indica colonizes plant roots and promotes Arabidopsis growth and seed production. Interestingly, the growth of Arabidopsis Et-related mutants etr1, ein2, and ein3eil1 was not promoted by the P. indica, although the roots were more colonized by the fungus (Camehl et al. 2010). Conversely, the overexpression of EtRF1 reduced P. indica colonization and constitutively activated plant defense. Camehl et al. (2010) suggested that the Et homeostasis is required to balance fungal colonization and defense responses. Recent studies have also demonstrated that P. indica induces ACC biosynthesis (Khatabi et al. 2012). The ability to inhibit the Et biosynthesis without the necessity of applying exogenous inhibitors has allowed the study of the accurate role of Et in multiple stress and developmental-related phenomena. Thus, the heterologous expression of the Pseudomonas ACCD gene in tomato plants showed to greatly decrease the production of Et (Klee et al. 1991). No apparent vegetative phenotypic abnormalities were detected in these tomato transgenic plants. However, there were notable alterations in the reproductive phase (i.e., several weeks delayed fruit ripening). After these early results, the ACCD was considered as a marker for the Et role in many stress and developmental processes. Interestingly, degradation of ACC in tomato inhibits Et biosynthesis but does not prevent the ability of fruits to sense Et and no ripening defects were observed in transgenic fruits exposed to Et (Klee et al. 1991). On the other hand, during the symbiotic association between rhizobia and legumes, the exogenous application of Et inhibits the formation and functioning of radical nodules. As an example, a Medicago truncatula Et-insensitive mutant showed increased nodulation by its symbiont Sinorhizobium meliloti (Penmetsa and Cook 1997). Additionally, the results of Stearns et al. (2012) support the possibility of a direct connexion between Et and auxin response, and evidenced the stress-reducing benefits of ACCD-expressing PGPRs (Fig. 7.2). Thus, some ACCD-encoding rhizobial strains can decrease Et production in the plant and therefore enhance the formation of nodules. This increased nodulation was enhanced when ACCD-containing PGPRs and rhizobial strains were co-inoculated (Baby et al. 2011). Soil bacteria expressing ACCD reduce the level of Et and confer resistance and growth of plant under various stresses (Glick et al. 1998, 2007) including flooding and pathogen attack (Wang et al. 2000; Farwell et al. 2007; see section “Updated Overview of the Plant Hormone Ethylene”). It has been hypothesized that under conditions of stress, the root excretes the majority of ACC to the rhizosphere where it is degraded by the ACCD of appropriate bacteria (e.g., Pseudomonas sp.; Zahir et al. 2009). Therefore, rhizobacteria with ACCD activity have the ability to reduce Et production in roots and promote plant growth (e.g., root elongation) under several stress conditions (Siddikee et al. 2011; Chen et al. 2013) (Fig. 7.2). For example, in vitro experiments showed that ACCD-producing PGPRs enhanced the salt tolerance of important crops such as canola (Cheng et al. 2007), tomato (Mayak et al. 2004), and wheat (Zahir et al. 2009). Much work is still required to transfer these results to field conditions in order to gain insight on how microorganisms induce ACC biosynthesis in plant roots. However, some progress has already been made in this regard (Ma et al. 2004; Gamalero et al. 2008; Gamalero and Glick 2012).
5 The Relationship Between Ethylene and Other Environmental Stress-Inducing Factors
5.1 Ozone
Ozone (O3) is a highly unstable and reactive allotrope of O2. O3 is a common constituent of troposphere, with powerful oxidizing properties and the most phytotoxic air pollutant affecting plants, causing damage to the photosynthetic apparatus (Ashmore 2005; Wittig et al. 2009). Surface O3 concentrations (i.e., >60 nL L−1) have been shown to negatively affect the yields of crops (Fiscus et al. 2005). Et production is (1) the quickest and most commonly observed response to O3 (Kangasjärvi et al. 2005), including in many important crop plants (Wilkinson and Davies 2009); (2) highly correlated with O3 injury (Tamaoki et al. 2003); and (3) clearly associated with the induction of Hypersensitive Response (HR) and PCD (Kangasjärvi et al. 2005; Overmyer et al. 2003, 2005). On the other hand, in some species it was demonstrated the prominent role of JA in the O3–Et signaling pathway (Tamaoki et al. 2003; Grantz et al. 2010).
Rice, a moderately O3-sensitive crop species, has significant reductions in its yields (~15–20 %) due to elevated O3 levels (Shi et al. 2009). Moreover, O3 also induces a quick stomatal closure response (Wittig et al. 2007; Wilkinson and Davies 2009). ABA is considered the main regulator of stomatal functioning in plants and induces stomatal closure via a network of chemical messengers (Acharya and Assmann 2008) and Et has been shown to antagonize the stomatal response to ABA (Tanaka et al. 2006). Thus, plants pretreated with 1-methylcyclopropene (1-MCP), an Et perception antagonist, were able to close the stomata normally in response to ABA (Wilkinson and Davies 2009). On the other hand, when O3 penetrates the plant leaf through the stomata, it is quickly transformed to ROS (e.g., O2 – anion and H2O2) in the apoplast (Baier et al. 2005). Subsequently, in Arabidopsis, the H2O2 production in guard cells as a consequence of oxidative stress of O3 causes stomatal closure in an Et-dependent manner (Matilla-Vázquez and Matilla 2012; and references therein). In this process, Et also induces the stomatal closure stimulating the production of H2O2 by the NADPH-oxidase AtRbohF (Matilla-Vázquez and Matilla 2012). For more detailed information about the O3 harmful effects on stomata movements, see Wilkinson and Davies (2010).
As indicated above (section “Cross-Talk Between Oxygen Deficient Stress and Ethylene Biosynthesis and Signaling”), when the root system is subject to stresses like flooding, the ACC is transported from there to the oxygenated parts (e.g., shoots) and transformed in Et by ACO. However, to our knowledge, studies on spatial alterations of ACC content and Et production in response to O3 still remain to be performed. Several mutants and accessions of Arabidopsis described as O3-sensitive have now been demonstrated that overproduce Et (Kangasjärvi et al. 2005), and Arabidopsis mutants insensitive to Et are O3-tolerant. Recently, (1) an essential JA–Et interaction was found to be mediated by JA-Zim domain (JAZ). These JAZ proteins repress the transcription of JA-responsive genes and interact with TFs involved in mediating responses to Et (Wager and Browse 2012); and (2) O3 surface levels induce plant physiology responses in Gossypium barbadense with no increase in the production of Et (Grantz et al. 2010; Grantz and Vu 2012). However, when the plants were exposed to high O3 levels, Et biosynthesis was induced and further enhanced in MeJA-treated plants (Grantz and Vu 2012). In G. barbadense, the application of MeJA as an anti-ozonant has been proposed.
5.2 Freezing
Although Et regulates several specific aspects of plant responses against biotic and abiotic stress (sections “Cross-Talk Between Oxygen Deficient Stress and Ethylene Biosynthesis and Signaling” and “Ethylene and Plant Defense Against Microorganisms”), their definite role in freezing stress remains unclear (Zhang and Huang 2010 and references therein). In general, high levels of Et production are associated with chilling sensitivity (see Morgan and Drew (1997) for review of earlier literature). Nevertheless, the TFs known as C-repeat Binding Factor (CBF), belonging to the AP2/ERF superfamily, are involved in the well-understood cold signaling pathway (CBF/DREB) transcriptional regulatory cascade. Recent results in Arabidopsis demonstrated the negative effect of Et biosynthesis and signaling over the plant freezing tolerance by repressing type-A Arabidopsis Response Regulators (ARR) genes and the cold-inducible CBFs (Shi et al. 2012). Namely, ETR1 and EIN4, in contrast to EIN2 and EIN3/EIL1, have positive roles during the modulation of the plant adaptations to freezing. Diverse and contradictory implications of Et biosynthesis in chilling sensitivity were previously shown in maize, mung bean, tomato, cucumber, and tobacco plants (more information in Shi et al. 2012).
6 Conclusions and Future Perspective
At present, there is no doubt about the critical role of Et in plant defense strategies against biotic and abiotic stresses. Et participates in a highly complex and tightly regulated signaling network that also includes crosstalk with JA, SA, GA, and ABA signaling pathways. In order to obtain goods and services orientated to the development of modern agriculture, the knowledge of all these plant signaling networks has undergone a strong progress during the last decade. As a result, the number of biocontrol and biotechnological strategies designed to improve plant responses to stressful environmental cues, such as low O2, freezing, and pathogens, is growing exponentially. It seems beyond doubt that the level of endogenous Et is critical for the establishment and adjustment of appropriate plant responses, and that these processes require tight spatial and temporal regulation of Et biosynthesis. A major research priority to improve the understanding of the Et signaling at molecular level was the identification of transcriptional networks that regulate the synthesis of developmental modulators. Thereby, functional analysis of the large ERF family is helping to characterize how Et coordinates plant adaptive responses to stress. Ultimately, unscrambling how plants alter their microbiome and the mechanisms by which plant-associated microorganisms control plant health will provide an excellent opportunity to enhance crop productivity and quality. However, the molecular mechanisms by which rhizosphere microorganisms are recognized to subsequently activate Et-mediated responses are still poorly understood.
Due to ET action is included in a plant hormone network, it is indispensable to unravel the ET crosstalk with SA-, JA-, and ABA-depeudent signaling pathways. The result of this extensive study is to understand the plant response to a particular type of stress. This biotechnology challenge will require the characterization and contribution of the molecular components involved in this tangled network. To fill this complicated puzzle, molecular platforms as microarrays, protein–protein interactions, knock-out gene collection, or RNA-seq facilities must be utilized to this aim without ruling out new -omics technologies.
References
Acharya BR, Assmann SM (2008) Hormone interactions in stomatal function. Plant Mol Biol 69:451–462
Adie B, Chico JM, Rubio-Somoza I, Solano R (2007) Modulation of plant defenses by ethylene. J Plant Growth Regul 26:160–177
Ahemad M, Khan MS (2011) Assessment of plant growth promoting activities of rhizobacterium Pseudomonas putida under insecticide-stress. Microbiol J 1:54–64
Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci U S A 100:2992–2997
An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, Zhang S, Ecker JR, Guo H (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-like1 is mediated by proteasomal degradation of EIN3 binding f-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22:2384–2401
Aschi-Smiti S, Chaïbi W, Brouquisse R, Bérénice-Ricard B, Saglio P (2004) Assessment of enzyme induction and aerenchyma formation as mechanisms for flooding tolerance in Trifolium subterraneum ‘Park’. Ann Bot 91:195–204
Ashmore MR (2005) Assessing the future global impacts of ozone on vegetation. Plant Cell Environ 28:949–964
Ausubel FM (2005) Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 6:973–979
Babula D, Misztal LH, Jakubowicz M, Kaczmarek M, Nowak W, Sadowski J (2006) Genes involved in biobiosynthesis and signalisation of ethylene in Brassica oleracea and Arabidopsis thaliana: identification and genome comparative mapping of specific gene homologues. Theor Appl Genet 112:410–420
Baby S, Muhammad I, Muhammad A, Azeem K (2011) Manipulation of ethylene biosynthesis in roots through bacterial ACC deaminase for improving nodulation in legumes. Crit Rev Plant Sci 30:279–291
Bacanammwo M, Purcell LC (1999) Soybean root morphological and anatomical traits associated with acclimation to flooding. Crop Sci 39:143–149
Badri DV, Vivanco JM (2009) Regulation and function of root exudates. Plant Cell Environ 32:666–681
Baier M, Kandlbinder A, Golldack D, Dietz KJ (2005) Oxidative stress and ozone: perception, signaling and response. Plant Cell Environ 28:1012–1020
Bailey-Serres J, Chang R (2005) Sensing and signaling in response to oxygen deprivation in plants and other organisms. Ann Bot 96:507–518
Bailey-Serres J, Voesenek LACJ (2010) Life in the balance: a signaling network controlling survival of flooding. Curr Opin Plant Biol 13:489–494
Bailey-Serres J, Fukao T, Gibbs DJ, Holdsworth MJ, Lee SC, Licausi F, Perata P, Voesenek LACJ, van Dongen JT (2012) Making sense of low oxygen sensing. Trends Plant Sci 17:129–138
Bakker PAHM, Pieterse CMJ, Van Loon LC (2007) Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology 97:239–243
Bari R, Jones J (2009) Role of plant hormones in plant defense responses. Plant Mol Biol 69:473–488
Barreto-Figueiredo MC, Seldin L, Araujo FF, Ramos-Mariano RL (2011) Plant growth promoting rhizobacteria: fundamentals and applications. In: Maheshwari DK (ed) Plant growth and health promoting bacteria, vol 18. Springer-Verlag, Berlin, pp 21–43
Benschop JJ, Jackson MB, Guhl K, Vreeburg RAM, Croker SJ, Peeters AJM, Voesenek LACJ (2005) Contrasting interactions between ethylene and abscisic acid in Rumex species differing in submergence tolerance. Plant J 44:756–768
Benschop JJ, Bou J, Peeters AJM, Wagemaker N, Guhl K, Ward D, Hedden P, Moritz T, Voesenek LACJ (2006) Long-term submergence-induced elongation in Rumex palustris requires ABA-dependent biobiosynthesis of GA1. Plant Physiol 141:1644–1652
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 8:478–486
Berrocal-Lobo M, Molina A (2004) Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol Plant Microbe Interact 17:763–770
Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD (2007) The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19:509–523
Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16:1–18
Borisjuk L, Rolletschek H (2009) The oxygen status of the developing seed. New Phytol 182:17–30
Bradford KJ (2008) Shang Fa Yang: Pioneer in plant ethylene biochemistry. Plant Sci 175:2–7
Brock AK, Berger B, Mewis I, Ruppel S (2012) Impact of the PGPB Enterobacter radicincitans DSM 16656 on growth, glucosinolate profile, and immune responses of Arabidopsis thaliana. Microb Ecol. doi:10.1007/s00248-012-0146-3
Broekaert WF, Delauré SL, De Bolle MFC, Cammue BPA (2006) The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol 44:393–416
Camehl I, Sherameti I, Venus Y, Bethke G, Varma A, Lee J, Oelmüller R (2010) Ethylene signaling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytol 185:1062–1073
Chae HS, Cho YG, Park MY, Lee MC, Eun MY, Kang BG, Kim WT (2000) Hormonal cross-talk between auxin and ethylene differentially regulates the expression of two members of the 1-aminocyclopropane-1-carboxylate oxidase gene family in rice (Oryza sativa L.). Plant Cell Physiol 41:354–362
Chen X, Pierik R, Peeters AJM, Visser EJW, Huber H, de Kroon H, Voesenek LACJ (2010) Endogenous abscisic acid as a key switch for natural variation in flooding-induced shoot elongation. Plant Physiol 154:969–977
Chen L, Dodd IC, Theobald JC, Belimov AA, Davies WJ (2013) The rhizobacterium Variovorax paradoxus 5C-2, containing ACC deaminase, promotes growth and development of Arabidopsis thaliana via an ethylene-dependent pathway. J Exp Bot 64:1565–1573
Cheng Z, Park E, Glick BR (2007) 1-Aminocyclopropane-1-carboxylate deaminase from Pseudomonas putida UW4 facilitates the growth of canola in the presence of salt. Can J Microbiol 53:912–918
Christians MJ, Gingerich DJ, Hansen M, Binder BM, Kieber JJ, Vierstra RD (2009) The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J 57:332–345
Cohn JR, Martin GB (2005) Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J 44:139–154
Conrath U (2009) Priming of induced plant defense responses. Adv Bot Res 51:362–395
Czarny JZ, Grichko VP, Glick BR (2006) Genetic modulation of ethylene biosynthesis and signaling in plants. Biotechnol Adv 24:410–419
Darrah PR, Roose T (2007) Modeling the rhizosphere. In: Picton R, Varanini Z, Nannipieri P (eds) The Rizosphere: biochemistry and organic substances at the soil-plant interface. CRC Press, New York, pp 331–370
Dat JF, Capelli N, Folzer H, Bourgeade P, Badot P-M (2004) Sensing and signaling during plant flooding. Plant Physiol Biochem 42:273–282
De la Torre F, Rodríguez-Gacio MC, Matilla AJ (2006) How ethylene works in the reproductive organs in higher plants. A signaling update from third milennium. Plant Signal Behav 1:231–242
De Vleesschauwer D, Höfte M (2009) Rhizobacteria-induced systemic resistance. In: Van Loon LC (ed) Adv Bot Res, vol 51. Academic Press Ltd-Elsevier Science Ltd, London, pp 223–281
De Vos M, Van Zaanen W, Koornneef A, Korzelius JP, Dicke M, Van Loon LC, Pieterse CMJ (2006) Herbivore-induced resistance against microbial pathogens in Arabidopsis. Plant Physiol 142:352–363
De Wit M, Spoel SH, Sánchez-Pérez GF, Gommers CM, Pieterse CM et al (2013) Perception of low red:far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. doi:10.1111/tpj.12203
Delseny M, Charng Y, Wang L-C (2008) Ethylene biology. Plant Sci 175:1–196
Doornbos RF, Geraats BPJ, Kuramae EE, Van Loon LC, Bakker PHM (2011) Effects of jasmonic acid, ethylene, and salicylic acid signaling on the rhizosphere bacterial community of Arabidopsis thaliana. Mol Plant Microbe Interact 24:395–407
Dubois M, Skirycz A, Claeys H, Maleux K et al (2013) The ethylene response factor 6 acts as central regulator of leaf growth under water limiting conditions in Arabidopsis thaliana. Plant Physiol. doi:10.1104/pp. 113.216341
Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42:185–209
Ellis MH, Dennis ES, Peacock WJ (1999) Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol 119:57–64
English PJ, Lycett GW, Roberts JA, Jackson MB (1995) Increased 1-aminocyclopropane-1-carboxylic acid oxidase activity in shoots of flooded tomato plants raises ethylene production to physiologically active levels. Plant Physiol 109:1435–1440
Farwell AJ, Vesely S, Nero V, McCormack K, Rodríguez H, McCormack K, Shah S, Dixon DG, Glick BR (2007) Tolerance of transgenic canola (Bassica napus) amended with ACC deaminase-containing plant growth-promoting bacteria to flooding stress at a metal-contaminated field site. Environ Pollut 147:540–545
Finlayson SA, Foster KR, Reid DM (1991) Transport and metabolism of 1- aminocyclopropane-1-carboxylic acid in sunflower (Helianthus annuus L.) seedlings. Plant Physiol 96:1360–1367
Fiscus EL, Booker FL, Burkey KO (2005) Crop responses to ozone: uptake, modes of action, carbon assimilation and partitioning. Plant Cell Environ 28:997–1011
Flury P, Klauser D, Schulze B, Boller T, Bartels S (2013) The anticipation of danger: microbe-associated molecular pattern perception enhances AtPep-triggered oxidative burst. Plant Physiol 161:2023–2035
Fonseca S, Chico JM, Solano R (2009) The jasmonate pathway: the ligand, the receptor and the core signaling module. Curr Opin Plant Biol 12:539–547
Freebairn HT, Buddenhagen IW (1964) Ethylene production by Pseudomonas solanacearum. Nature 202:313–314
Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol. doi:10.1146/annurev-arplant-042811-105606
Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12:393–404
Fukao T, Bailey-Serres J (2008) Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellins responses in rice. Proc Natl Acad Sci U S A 105:16814–16819
Fukao T, Xu K, Ronald PC, Bailey-Serres J (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18:2021–2034
Fukuda H, Takahashi M, Fujii T, Tazaki M, Ogawa T (1989) An NADH:Fe(III)EDTA oxidoreductase from Cryptococcus albidus: an enzyme involved in ethylene production in vivo? FEMS Microbiol Lett 60:107–112
Gamalero E, Glick BR (2012) Ethylene and abiotic stress tolerance in plants. In: Ahmad P, Prasad MNV (eds) Environmental adaptations and stress tolerance of plants in the era of climatie change. Springer, New York, pp 395–412
Gamalero E, Lingua G, Tombolini R, Avidano L, Pivato B, Berta G (2005) Colonization of tomato root seedling by pseudomonas fluorescens 92rkG5: spatio-temporal dynamics, localization, organization, viability, and culturability. Microbiol Ecol 50:289–297
Gamalero E, Berta G, Massa N, Glick BR, Lingua G (2008) Synergistic interactions between the ACC deaminase-producing bacterium Pseudomanas putida UW4 and the AM fungus Gigaspora rosea positively affect cucumber plant growth. FEMS Microbiol Ecol 64:459–467
García I, Castellano JM, Vioque B, Solano R, Gotor C, Romero LC (2010) Mitochondrial β-cyanoalanine synthase is essential for root hair formation in Arabidopsis thaliana W. Plant Cell 22:3268–3279
Geingenberger P (2003) Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6:247–256
Geisler-Lee J, Caldwell C, Gallie DR (2010) Expression of the ethylene biosynthetic machinery in maize roots is regulated in response to hypoxia. J Exp Bot 61:857–871
Gibbs DJ, Lee SC-H, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, Holdsworth MJ (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 497:415–418
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Glick BR, Penrose DM, Li J (1998) A model for the lowering of plant ethylene concentrations by plant growth promoting bacteria. J Theor Biol 190:63–68
Glick BR, Cheng Z, Czarny J, Duan J (2007) Promotion of plant growth by ACC deaminase-producing soil bacteria. Eur J Plant Pathol 119:329–339
Graciet E, Wellmer F (2010) The plant N-end rule pathway: structure and functions. Trend Plant Sci 15:447–453
Graeber K, Nakabayashu K, Miatton E, Leubner-Metger G, Soppe WJJ (2012) Molecular mechanisms of seed dormancy. Plant Cell Environ 35:1769–1786
Grant MR, Jones JDG (2009) Hormone (dis)harmony moulds plant health and disease. Science 324:750–752
Grantz DA, Vu H-B (2012) Root and shoot gas exchange respond additively to moderate ozone and methyl jasmonate without induction of ethylene: ethylene is induced at higher O3 concentrations. J Exp Bot 63:4303–4313
Grantz DA, Vu H-B, Aguilar C, Rea MA (2010) No interaction between methyl jasmonate and ozone in Pima cotton: growth and allocation respond independently to both. Plant Cell Environ 33:717–728
Grichko VP, Glick BR (2001) Ethylene and flooding stress in plants. Plant Physiol Biochem 39:1–9
Gunawardena A, Pearce DM, Jackson MB, Hawes CR, Evans DE (2001) Characterization of programmed cell death during aerenchyma formation induced by ethylene or hypoxia in roots of maize (Zea mays L.). Planta 212:205–214
Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115:667–677
Haas D, Defago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 17:307–319
Ham BK, Park JM, Lee SB, Kim MJ, Lee IJ, Kim K-J, Kwon CS, Paek KH (2006) Tobacco Tsip1, a DnaJ-type Zn finger protein, is recruited to and potentiates Tsi1-mediated transcriptional activation. Plant Cell 18:2005–2020
Hase S, Van Pelt JA, Van Loon LC, Pieterse CMJ (2003) Colonization of Arabidopsis roots by Pseudomonas fluorescens primes the plant to produce higher levels of ethylene upon pathogen infection. Physiol Mol Plant Pathol 62:219–226
Hattori Y, Nagai K, Furukawa S, Song X-J, Kawano R, Sakakibara H, Jianzhong Wu J, Matsumoto T, Yoshimura A, Kitano H, Matsuoka M, Mori H, Ashikari M (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allows rice to adapt to deep water. Nature 460:1026–1030
Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R (2010) Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153:757–772
Hol WH, Bezemer TM, Biere A (2013) Getting the ecology into interactions between plants and the plant growth-promoting bacterium Pseudomonas fluorescens. Front Plant Sci 4:81. doi:10.3389/fpls.2013.00081
Iglesias-Fernández R, Rodríguez-Gacio MC, Matilla AJ (2011) Progress in research on dry after ripening. Seed Sci Res 21:69–80
Ismond KP, Dolferus R, de Pauw M, Dennis ES, Good AG (2003) Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol 132:1292–1302
Iwai T, Miyasaka A, Seo S, Ohashi Y (2006) Contribution of ethylene biobiosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol 142:1202–1215
Jackson MB, Ram PC (2003) Physiological and molecular basis of susceptibility and tolerance of rice plants to complete submergence. Ann Bot 91:227–241
Jones DL, Dennis PG, Owen AG, Van Hees PAW (2003) Organic acid behavior in soils-misconceptions and knowledge gaps. Plant Soil 248:31–41
Jung K-H, Seo Y-S, Walia H, Cao P, Fukao T, Canlas PE, Amonpant F, Bailey-Serres J, Ronald PC (2010) The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol 152:1674–1692
Kangasjärvi J, Jaspers P, Kollist H (2005) Signaling and cell death in ozone-exposed plants. Plant Cell Environ 28:1021–1036
Kazan K, Manners JM (2008) Jasmonate signaling: toward an integrated view. Plant Physiol 146:1459–1468
Kende H, van der Knaap E, Cho H-T (1998) Deepwater rice: a model plant to study stem elongation. Plant Physiol 118:1105–1110
Kendrick P, Crane PR (1997) The origin and early evolution of plants on land. Nature 389:33–39
Khatabi B, Molitor A, Lindermayr C, Pfiffi S, Durner J, von Wettstein D, Kogel K-H, Schäfer P (2012) Ethylene supports colonization of plant roots by the mutualistic fungus Piriformospora indica. PLoS One 7:e35502
Klee HL, Hayford MB, Kretzmer KA, Barry GF, Kishore GM (1991) Control of ethylene biosynthesis by expression of a bacterial enzyme in transgenic tomato plants. Plant Cell 3:1187–1193
Knaap E, Sauter M, Wilford R, Kende H (1996) Identification of a gibberellin-induced receptor-like kinase in deepwater rice. Plant Physiol 112:1397–1401
Knoester M, Pieterse CM, Bol JF, Van Loon LC (1999) Systemic resistance in Arabidopsis induced by rhizobacteria requires ethylene-dependent signaling at the site of application. Mol Plant Microbe Interact 12:720–727
Konishi M, Yanagisawa S (2008) Ethylene signaling in Arabidopsis involves feedback regulation by an elaborate control of EBF2 expression by EIN3. Plant J 55:821–831
Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LACJ, Bailey-Serres J (2011) Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol 190:457–471
León-Reyes A, Steven H, Spoel SH, De Lange ES, Abe H, Kobayashi M, Tsuda S, Millenaar FF, Welschen RAM, Ritsema T, Pieterse CMJ (2009) Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol 149:1797–1809
León-Reyes A, Du Y, Koornneef A, Proietti S, Körbes AP, Memelink J, Pieterse CMJ, Ritsema T (2010) Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol Plant Microbe Interact 23:187–197
Leprince O, Buitink J (2010) Desiccation tolerance: from genomics to the field. Plant Sci 179:554–564
Licausi F (2011) Regulation of the molecular response to oxygen limitations in plants. New Phytol 190:550–555
Licausi F (2012) Molecular element of low-oxygen signaling in plants. Physiol Plant. doi:10.1111/ppl.12011
Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62:302–315
Licausi F, Weits DA, Pant BD, Scheible W-R, Geigenberger P, van Dongen JT (2011) Hypoxia responsive gene expression is mediated by various subsets of transcription factors and mRNAs that are determined by the actual oxygen availability. New Phytol 190:442–456
Lin Z, Zhong S, Grierson D (2009) Recent advances in ethylene research. J Exp Bot 60:3311–3336
Linkies A, Graeber K, Knight CA, Leubner-Metzger G (2010) The evolution of seeds. New Phytol 186:817–831
Loyola-Vargas VM, Broeckling CD, Badri D, Vivanco JM (2007) Effect of transporters on the secretion of phytochemicals by the roots of Arabidospis thaliana. Planta 225:301–310
Lugtenberg BJ, Bloemberg GV (2004) Life in the rhizosphere. In: Ramos JL (ed) Pseudomonas: genomics, life style and molecular arquitecture, vol I. Kluwer Academic/Plenum Publishers, New York, pp 403–430
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Ma W, Charles TC, Glick BR (2004) Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Sinorhizobium meliloti increases its ability to nodulate alfalfa. Appl Environ Microbiol 70:5891–5897
Magneschi L, Perata P (2009) Rice germination and seedling growth in the absence of oxygen. Ann Bot 103:181–196
Manach-Little N, Igamberdiev AU, Hill RD (2005) Hemoglobin expression affects ethylene production in maize cell cultures. Plant Physiol Biochem 43:485–489
Martone PT, Estévez JM, Lu F, Ruel K, Denny MW, Somerville C, Ralph J (2009) Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr Biol 19:169–175
Matilla AJ, Matilla-Vázquez MA (2008) Involvement of ethylene in seed physiology. Plant Sci 175:87–97
Matilla-Vázquez MA, Matilla AJ (2012) Role of H2O2 as signaling molecule in plan5. In: Ahmad P, Prassad MNV (eds) Environmental Adaptations and stress Tolerance of Plan5 in the Era of climate change. Springer, New York, pp 361–380
Matilla AJ, Rodríguez-Gacio MC (2013) Non-symbiotic hemoglobins in the life of seeds. Phytochemistry 87:7–15
Matilla MA, Ramos JL, Bakker PAHM, Doornbos R, Badri DV, Vivanco JM, Ramos-González MI (2010) Pseudomonas putida KT2440 causes induced systemic resistance and changes in Arabidopsis root exudation. Environ Microbiol Rep 2:381–388
Mayak S, Tirosh T, Glick BR (2004) Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol Biochem 42:565–572
McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible W-R, Udvardi MK, Kazan K (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139:949–959
Mekhedov SL, Kende H (1996) Submergence enhances expression of a gene encoding 1-aminocyclopropane-1-carboxylate oxidase in deepwater rice. Plant Cell Physiol 37:531–537
Molina LA, Ramos C, Duque E, Ronchel MC, García JM, Wyke L, Ramos JL (2000) Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol Biochem 32:315–321
Morgan PW, Drew MC (1997) Ethylene and plant responses to stress. Physiol Plant 100:620–630
Morgan JAW, Bending GD, White PJ (2005) Biological costs and benefits to plant-microbe interactions in the rhizosphere. J Exp Bot 56:1729–1739
Mühlenbock P, Plaszczyca M, Plaszczyca M, Mellerowicz E, Karpinski S (2007) Lysigenous aerenchyma formation in Arabidopsis is controlled by LESION SIMULATING DISEASE1. Plant Cell 19:3819–3830
Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152:1484–1500
Nagahama K, Ogawa T, Fujii T, Fukuda H (1992) Classification of ethylene-producing bacteria in terms of biosynthetic pathways to ethylene. J Ferment Bioeng 73:1–5
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Newman G, Römheld V (2007) The release of root exudates as affected by the plant physiology status. In: Picton R, Varanini Z, Nannipieri P (eds) The rhizosphere: biochemistry and organic substances at the soil-plant interface. CRC Press, New York, pp 23–72
Niu DD, Liu HX, Jiang CH, Wang YP, Wang QY, Jin HL, Guo JH (2011) The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol Plant Microbe Interact 24:533–542
Oh IS, Park AR, Bae MS, Kwon SJ, Kim YS, Lee JE, Kang NY, Lee S, Cheong H, Park OK (2005) Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17:2832–2847
Olmedo G, Guo H, Gregory BD, Saeid D, Nourizadeh SD, Aguilar-Henonin L, Li H, An F, Guzman P, Ecker JR (2006) ETHYLENE-INSENSITIVE5 encodes a 5′ → 3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc Natl Acad Sci U S A 103:13286–13293
Overmyer K, Brosché M, Kangasjärvi J (2003) Reactive oxygen species and hormonal control of cell death. Trend Plant Sci 8:335–342
Overmyer K, Brosche M, Pellinen R, Kuittenen T, Tuominen H, Ahlfors R, Keinänen M, Saarma M, Scheel D, Kangasjärvi J (2005) Ozone-induced programmed cell death in the Arabidopsis radical-induced cell death 1 mutant. Plant Physiol 137:1092–1104
Peleman J, Boerjan W, Engler G, Seurinck J, Botterman J, Alliotte T, van Montagu M, Inzé D (1989) Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. Plant Cell 1:81–93
Peng H-P, Chan C-S, Shih M-C, Yang SF (2001) Signaling events in the hypoxic induction of alcohol dehydrogenase gene in Arabidopsis. Plant Physiol 126:742–749
Peng H-P, Lin T-Y, Wang N-N, Shih M-C (2005) Differential expression of genes encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis during hypoxia. Plant Mol Biol 58:15–25
Penmetsa RV, Cook DR (1997) A legume ethylene-insensitive mutant hyper infected by its rhizobial symbiont. Science 275:527–530
Perata P, Voesenek LA (2007) Submergence tolerance in rice requires Sub1A, an ethylene-response-factor-like gene. Trends Plant Sci 12:43–46
Peter G, Neale D (2004) Molecular basis for the evolution of xylem lignification. Curr Opin Plant Biol 7:737–742
Phillips DA, Fox TC, King MD, Bhuvaneswari TV, Teubner LR (2004) Microbial products trigger amino acid exudation from plant roots. Plant Physiol 136:2887–2894
Pierik R, Tholen D, Poorter H, Visser EJ, Voesenek LA (2006) The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci 11:176–183
Pieterse CMJ, Van Wees SCM, Van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10:1571–1580
Pieterse CMJ, Van Pelt JA, Ton J, Parchmann S, Mueller MJ, Buchala AJ, Métraux J-C, van Loon LC (2000) Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is no accompanied by an increase in their production. Physiol Mol Plant Pathol 57:123–134
Pieterse CMJ, Van der Ent S, Van Pelt JA, Van Loon LC (2007) Ramina A et al (eds) Advances in plant ethylene research: proceeding of the 7th international symposium of the plant hormone ethylene. p 325–331
Pieterse CMJ, Van der Does D, Zamioudis C, León-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521
Pirrello J, Prassad N, Zhang W, Chen K, Mila I, Zouine M, Latché A, Pech JC, Ohme-Takagi M, Regad F, Bouzayen M (2012) Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol 12:190
Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115:679–689
Pré M, Atallah M, Champion A, De Vos M, Pieterse CMJ, Memelink J (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147:1347–1357
Preston GM (2004) Plant perceptions of plant growth-promoting Pseudomonas. Philos Trans R Soc Lond 359:907–918
Qiao H, Chang KN, Yazaki J, Ecker JR (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23:512–521
Qu Z-L, Zhong N-Q, Wang H-Y, Chen A-P, Jian G-L, Xia G-X (2006) Ectopic expression of the cotton non-symbiotic hemoglobin gene GhHb1 triggers defense responses and increases disease tolerance in Arabidopsis. Plant Cell Physiol 47:1058–1068
Raaijmakers JM, Leeman M, Van Oorschot MMP, Van der Sluis I, Schippers B, Bakker PAHM (1995) Dose–response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathology 85:1075–1081
Rajhi I, Yamauchi T, Takahashi H, Nishiuchi S, Shiono K, Watanabe R, Mliki A, Nagamura Y, Tsutsumi N, Nishizawa NK, Nakazono M (2011) Identification of genes expressed in maize root cortical cells during lysigenous aerenchyma formation using laser microdissection and microarray analyses. New Phytol 190:351–368
Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379:633–646
Rieu I, Cristescu SM, Harren FJM, Huibers W, Voesenek LACJ, Mariani C, Vriezen WH (2005) RP-ACS1, a flooding-induced 1-aminocyclopropane-1-carboxylate synthase gene of Rumex palustris, is involved in rhythmic ethylene production. J Exp Bot 56:841–849
Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49:317–343
Roca A, Pizarro-Tobías P, Udaondo Z, Fernández M, Matilla MA, Molina-Henares MA, Molina L, Segura A, Duque E, Ramos JL (2013) Analysis of the plant growth-promoting properties encoded by the genome of the rhizobacterium Pseudomonas putida BIRD-1. Environ Microbiol 15:780–794
Rodríguez-Gacio MC, Matilla-Vázquez MA, Matilla AJ (2009) Seed dormancy and ABA signaling: the breakthrough goes on. Plant Signal Behav 4:1035–1857
Romanel EAC, Schrago CG, Couñago RM, Russo CAM, Alves-Ferreira M (2009) Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One 4:e5791
Rosas SB, Andre JA, Rovera M, Correa NS (2006) Phosphate-solubilizing Pseudomonas putida can influence the rhizobia-legume symbiosis. Soil Biol Biochem 38:3502–3505
Rzewuski G, Sauter M (2008) Ethylene biosynthesis and signaling in rice. Plant Sci 175:32–42
Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S, Jikumaru Y, Fujimoto M, Arikawa T, Takahashi H, Ando M, Arimura S-I, Miyao A, Hirochika H, Kamiya Y, Tsutsumi N, Nambara E, Nakazono M (2007) Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8´-hydroxylase in rice. Plant Cell Physiol 48:287–298
Sairam RK, Kumutha D, Ezhilmathi K (2009) Waterlogging tolerance: nonsymbiotic haemoglobin–nitric oxide homeostasis and antioxidants. Curr Sci 96:674–682
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Sato T, Theologis A (1989) Cloning the mRNA encoding 1- aminocyclopropane-1-carboxylate synthase, the key enzyme for ethylene biosynthesis in plants. Proc Natl Acad Sci U S A 86:6621–6625
Sato M, Watanabe K, Yazawa M, Takikawa Y, Nishiyama K (1997) Detection of new ethylene-producing bacteria, Pseudomonas syringae pvs. cannabina and sesami, by PCR amplification of genes for the ethylene-forming enzyme. Phytopathology 87:1192–1196
Schweighofer A, Meskiene I (2008) Regulation of stress hormones jasmonates and ethylene by MAPK pathways in plants. Mol Biosyst 4:799–803
Shen X, Liu H, Yuang B, Li X, Xu C, Wang S (2011) OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis: rice bacterial resistance. Plant Cell Environ 34:179–191
Shi G, Yang L, Wang Y, Kobayashi K, Zhu J, Tang H, Pan S, Chen T, Liu G, Wang Y (2009) Impact of elevated ozone concentration on yield of four Chinese rice cultivars under open air field condition. Agric Ecosyst Environ 131:178–184
Shi Y, Tian S, Hou L, Huang X, Zhang X, Guo H, Yang S (2012) Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and Type-A ARR genes in Arabidopsis. Plant Cell 24:2578–25952
Siddikee MA, Glick BR, Chauhan PS, Yim WJ, Sa T (2011) Enhancement of growth and salt tolerance of red pepper seedlings (Capsicum annuum L.) by regulating stress ethylene biosynthesis with halotolerant bacteria containing 1-aminocyclopropane-1-carboxylic acid deaminase activity. Plant Physiol Biochem 49:427–434
Siddiqui MH, Al-Whaibi MH, Basalah MO (2010) Role of nitric oxide in tolerance of plants to abiotic stress. Protoplasm 248:447–455
Stearns JC, Woody OZ, McConkey BJ, Glick BR (2012) Effects of bacterial ACC deaminase on Brassica napus gene expression. Mol Plant Microbe Interact 25:668–676
Stepanova AN, Alonso JM (2009) Ethylene signaling and response: where different regulatory modules meet. Curr Opin Plant Biol 12:548–555
Tamaoki M, Nakajima N, Kubo A, Aono M, Matsuyama T, Saji H (2003) Transcriptome analysis of O3-exposed Arabidopsis reveals that multiple signal pathways act mutually antagonistically to induce gene expression. Plant Mol Biol 53:443–456
Tanaka Y, Sano T, Tamaoki M, Nakajima N, Kondo N, Hasezawa S (2006) Cytokinin and auxin inhibit abscisic acid-induced stomatal closure by enhancing ethylene production in Arabidopsis. J Exp Bot 57:2259–2266
Thaler JS, Humphrey PT, Whiteman NK (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17:260–270
Ton J, Pieterse CMJ, Van Loon LC (1999) identification of a locus in Arabidopsis controlling both the expression of rhizobacteria-mediated induced systemic resistence (ISR) and basal resistence against Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 12:911–918
Ton J, Davison S, Van Wees SCM, Van Loon LC, Pieterse CMJ (2001) The Arabidopsis ISR1 locus controlling rhizobacteria-mediated induced systemic resistance is involved in ethylene signaling. Plant Physiol 125:652–661
Ton J, de Vos M, Robben C, Buchala A, Metraux JP, van Loon LC, Pieterse CMJ (2002a) Characterization of Arabidopsis enhanced disease susceptibility mutants that are affected in systemically induced resistance. Plant J 29:11–21
Ton J, Van Pelt JA, Van Loon LC, Pieterse CMJ (2002b) The Arabidopsis ISR1 locus is required for rhizobacteria mediated induced systemic resistance against different pathogens. Plant Biol 4:221–227
Tsuchisaka A, Theologis A (2004a) Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol 136:2982–3000
Tsuchisaka A, Theologis A (2004b) Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc Natl Acad Sci U S A 101:2275–2280
Uren NC (2007) Types, amount and possible functions of compounds released into rhizosphere by soil-grown plants. In: Picton R, Varanini Z, Nannipieri P (eds) The rhizosphere: biochemistry and organic substances at the soil-plant interface. CRC Press, New York, pp 1–21
van der Ent S, Van Wees S, Pieterse CMJ (2009) Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70:1581–1588
van der Straeten D, Anuntalabhochai S, Van Caeneghem W, Zhou Z, Gielen J, Van Montagu M (1997) Expression of three members of the ACC synthase gene family in deepwater rice by submergence, wounding and hormonal treatments. Plant Sci 124:79–87
van der Straeten D, Zhou Z, Prinsen E, Van Onckelen HA, van Montagu MC (2001) A comparative molecular physiological study of submergence response in lowland and deepwater rice. Plant Physiol 125:955–968
van Hulten M, Pelser M, Van Loon LC, Pieterse CMJ, Ton J (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci U S A 103:5602–5607
van Loon LC (2007) Plant responses to plant growth-promoting rhizobacteria. Eur J Plant Pathol 119:243–254
van Loon LC, Bakker PAHM (2005) Induced systemic resistance as a mechanism of disease suppression by rhizobacteria. In: Siddiqui ZA (ed) PGPR: biocontrol and biofertilization. Springer, Dordrecht, pp 39–66
van Loon LC, Geraats BP, Linthorst HJ (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11:184–191
van Wees SCM, Van der Ent S, Pieterse CMJ (2008) Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol 11:443–448
Vandenbussche F, Van der Straeten D (2007) Cross-talk of multiple signals controlling the plant phenotype. J Plant Growth Regul 26:176–187
Vandenbussche F, Vriezen WH, Van Der Straeten D (2006) Ethylene biosynthesis and signaling: a puzzle yet to be completed. In: Hedden P, Thomas SG (eds) Plant hormone signaling, vol 24. Blackwell Publishing, Oxford, pp 125–145
Varshavsky A (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci 20:298–1345
Vartapetian BB, Jackson MB (1997) Plant adaptations to anaerobic stress. Ann Bot 79:3–20
Verhagen BWM, Glazebrook J, Zhu T, Chang H-S, Van Loon LC, Pieterse CMJ (2004) The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol Plant Microbe Interact 17:895–908
Verk MC, Gatz C, Linthorst HJM (2009) Transcriptional regulation of plant defense responses. Adv Bot Res 51:397–438
Verma S, Varma A, Rexer KH, Hassel A, Kost G, Sarbhoy A, Bisen P, Bütehorn B, Franker P (1998) Piriformospora indica, a new root-colonizing fungus. Mycologia 90:896–903
Vessey JK (2003) Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255:571–586
Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47:177–206
Voesenek LACJ, Bailey-Serres J (2009) Genetics of high-rise rice. Nature 460:959–960
Voesenek LACJ, Banga M, Thier RH et al (1993) Submergence-induced ethylene synthesis, entrapment, and growth in two plant species with contrasting flooding resistances. Plant Physiol 103:783–791
Voesenek LACJ, Colmer TD, Pierik R, Millenaar FF, Peeters AJM (2006) How plants cope with complete submergence. New Phytol 170:213–226
Wager A, Browse J (2012) Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front Plant Sci 3:41
Wang C, Knill E, Glick BR, Defago G (2000) Effect of transferring 1-aminocyclopropoane-1-carboxylic acid (ACC) deaminase genes into Pseudomonas fluorescens strain CH40 and its gacA derivative CHA96 on their growth-promoting and disease-suppressive capacities. Can J Microbiol 46:898–907
Wang KLC, Yoshida H, Lurin C, Ecker JR (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428:945–995
Wang P, Du Y, Zhao X, Miao Y, Son C-P (2013) The MPK6-ERF6-ROS-responsive cis-acting element7/GCC box complex modulates oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiol 161:1392–1408
Wasternack C (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot 100:681–697
Watanabe M, Kusano M, Oikawa A, Fukushima A, Noji M, Saito K (2008) Physiological roles of the β-substituted alanine synthase gene family in Arabidopsis. Plant Physiol 146:310–320
Watkin ELJ, Campbell CJ, Greenway H (1998) Root development and aerenchyma formation in two wheat cultivars and one Triticale cultivar grown in stagnant agar and aerated nutrient solution. Ann Bot 81:349–354
Watt M, Hugenholtz P, White R, Vinall K (2006) Numbers and locations of native bacteria on field-grown wheat roots quantified by fluorescence in situ hybridization (FISH). Environ Microbiol 8:871–884
Wehner N, Hartmann L, Ehlert A, Bottner S, Oñate-Sánchez L, Droge-Laser W (2011) High-throughput protoplast transactivation (PTA) system for the analysis of Arabidopsis transcription factor function. Plant J 68:560–569
Weingart H, Volksch B (1997) Ethylene production by Pseudomonas syringae Pathovars in vitro and in planta. Appl Environ Microbiol 63:156–161
Weingart H, Ullrich H, Geider K, Volksch B (2001) The role of ethylene production in virulence of Pseudomonas syringae pvs. glycinea and phaseolicola. Phytopathology 91:511–518
Wilkinson S, Davies WJ (2009) Ozone suppresses soil drying and abscisic acid (ABA)-induced stomatal closure via an ethylene-dependent mechanism. Plant Cell Environ 32:949–959
Wilkinson S, Davies WJ (2010) Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant Cell Environ 33:510–525
Williamson B, Tudzynsk B, Tudzynski P, van Kan JAL (2007) Botrytis cinerea: the cause of grey mould disease. Mol Plant Pathol 8:561–580
Wittig VE, Ainsworth EA, Long SP (2007) To what extent do current and projected increases in surface ozone affect photosynthesis and stomatal conductance of trees? A meta-analytic review of the last three decades of experiments. Plant Cell Environ 30:1150–1162
Wittig VE, Ainsworth EA, Naidu SL, Karnosky DF, Long SP (2009) Quantifying the impact of current and future tropospheric ozone on tree biomass, growth, physiology and biochemistry: a quantitative meta-analysis. Glob Change Biol 15:396–424
Wong CE, Carson RAJ, Carr JP (2002) Chemically induced virus resistance in Arabidopsis thaliana is independent of pathogenesis-related protein expression and the NPR1 gene. Mol Plant Microbe Interact 15:75–81
Xu KN, Xu X, Fukao T, Canlas P, Maghirang-Rodríguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442:705–708
Xu F, Zhang D-W, Zhu F, Tang H, Xin LV, Cheng J, Xie H-F, Lin H-H (2012) A novel role for cyanide in the control of cucumber (Cucumis sativus L.) seedlings response to environmental stress. Plant Cell Environ 35:1983–1997
Yamagami T, Tsuchisaka A, Yamada K, Haddon WF, Harden LA, Theologis A (2003) Biochemical diversity among the 1- amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J Biol Chem 278:49102–49112
Yang CY, Fu-Chiun Hsu F-C, Li JP, Wang NN, Shih M-C (2011) The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis thaliana. Plant Physiol 156:202–212
Yi M, Juergens M, Jez JM (2012) Structure of soybean β-cyanoalanine synthase and the molecular basis for cyanide detoxification in plants. Plant Cell 24:2696–2706
Yip WK, Yang SF (1988) Cyanide metabolism in relation to ethylene production in plant tissues. Plant Physiol 88:473–476
Yoo SD, Cho YH, Sheen J (2009) Emerging connections in the ethylene signaling network. Trends Plant Sci 14:270–279
Yoshida H, Wang KLC, Chang CM, Mori K, Uchida E, Ecker JR (2006) The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. Plant Mol Biol 62:427–437
Zahir AZ, Ghani U, Naveed M, Nadeem SM, Asghar HN (2009) Comparative effectiveness of Pseudomonas and Serratia sp. containing ACC-deaminase for improving growth and yield of wheat (Triticum aestivum L.) under salts tressed conditions. Arch Microbiol 191:415–424
Zarembinski TI, Theologis A (1997) Expression characteristics of Os-ACS1 and Os-ACS2, two members of the 1-aminocyclopropane-1-carboxylate synthase gene family in rice (Oryza sativa L. cv. Habiganj Aman II) during partial submergence. Plant Mol Biol 33:71–77
Zhang Z, Huang R (2010) Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol Biol 73:241–249
Zhang X, Wang C, Zhang Y, Sun Y, Mou Z (2012) The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24:4294–4309
Zhao Y, Wei T, Yin K-Q, Chen Z, Gu H, Qu L-J, Qin G (2012) Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol 195:450–460
Zhong GV, Burns JK (2003) Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol Biol 53:117–131
Acknowledgments
This work was financially supported by Ministerio de Ciencia e Innovación (MICINN, Spain) Grant CGL2009-11425. M.A.M-V was supported by the EU Marie-Curie Intra-European Fellowship for Career Development (FP7-PEOPLE-2011-IEF) grant number 298003. The authors wish to apologize to all those scientists whose manuscripts have not been directly mentioned. The authors thank Dr. J. Ludwig-Müller for providing Fig. 7.1. We wish to thank Dr. J.C. Mortimer (Department of Biochemistry, Hopkins Building, Tennis Court Road, Cambridge CB2 1QW, UK) for critical reading of the manuscript and the language polishing.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Matilla-Vázquez, M.A., Matilla, A.J. (2014). Ethylene: Role in Plants Under Environmental Stress. In: Ahmad, P., Wani, M. (eds) Physiological Mechanisms and Adaptation Strategies in Plants Under Changing Environment. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-8600-8_7
Download citation
DOI: https://doi.org/10.1007/978-1-4614-8600-8_7
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-8599-5
Online ISBN: 978-1-4614-8600-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)