Abstract
Bacteriophages with contractile tails epitomize the concepts of “virus” and “phage” for many because the tails of these phages undergo a large conformational change – resembling the action of a syringe – upon the attachment to the host cell. The contractile tails belong to the recently recognized class of “contractile systems,” which includes phage tails, their close relatives R-type pyocins, the bacterial type VI secretion system, and the virulence cassette of Photorhabdus. Their function is to deliver large proteins and/or DNA into the cytoplasm of a bacterial or eukaryotic cell. The structure of the core components of all contractile tail-like systems is conserved, but the corresponding genes have diverged to such a degree that the common ancestry can no longer be easily detected at the level of amino acid sequence. At present, it is unclear, whether the contractile systems originated in bacteria or in phages. This chapter describes the structure and function of phage contractile tails and compares them with other phage tails and with other known contractile systems.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
The host-cell attachment organelle of bacteriophages – the tail – is a complex macromolecular machine, which is responsible for host-cell recognition, attachment, and cell envelope penetration. The tail forms a channel connecting the phage capsid with the host cell during infection. Phage genomic DNA and proteins, which are packaged in the capsid, are translocated into the host-cell cytoplasm through this channel.
At present, all tailed phages (the order Caudovirales) are divided into three families based on the tail morphology: Myoviridae (long contractile tail), Siphoviridae (long noncontractile tail), and Podoviridae (short noncontractile tail) (Ackermann 2003). This classification is based on the visual appearance of the tail and does not take into account the life cycle of the phage, its genome replication strategy, or any other phage–host interactions, which occur inside the cell. Nevertheless, bacteriophages with contractile tails on average have larger genomes than other bacteriophages. In fact, giant Myoviridae bacteriophages represent some of the most complex viruses with genomes as large as ∼700 kbp (Serwer et al. 2007). Phages with contractile tails and large genomes are strictly lytic, i.e., infection leads to host-cell lysis and phage genome does not integrate into the host-cell chromosome. All known phages with contractile tails have double-stranded DNA genome.
A contractile tail consists of the baseplate, the central tube (or the core), the external contractile sheath, and the terminator complex (Fig. 5.1) (Leiman et al. 2010). The baseplate carries host-cell-binding proteins [also called receptor-binding proteins (RBPs)] and coordinates host-cell attachment with sheath contraction. The tail terminator complex attaches the tail to the phage capsid via a head-to-tail connector protein, a cone- or mushroom-shaped dodecamer, which is positioned in one of the 12 pentameric vertices of the capsid (Fig. 5.1).
Structure of a phage with a contractile tail. (a) Schematic showing major components of a contractile tail phage. RBP stands for receptor-binding proteins. (b, c) CryoEM reconstructions of phage T4 with the tail in the extended and contracted conformations (Fokine et al. 2004; Leiman et al. 2004; Kostyuchenko et al. 2005)
Upon binding to the host-cell surface with the RBPs, the baseplate changes its conformation triggering sheath contraction. The sheath contracts to about half or more of its original length, which moves the entire phage particle closer to the cell surface and drives the rigid internal tail tube through the cell envelope (Fig. 5.1). Depending on the phage species, the tube might extend to several hundreds of angstroms from the plane of the baseplate.
Phage tails have evolved to effectively breach all the constituent layers of the bacterial host-cell envelope. The extended external polysaccharides (if present) are both recognized and locally digested by RBPs, which possess an enzymatic activity specific to these sugars (Leiman et al. 2007; Walter et al. 2008). This allows the phage to bind to the cell’s outer membrane, which is breached by the tail tube upon contraction of the tail sheath. The peptidoglycan layer is disrupted with the help of a glycosidase, which is also part of the tail, but its location in the tail and, possibly, number of such molecules in the tail varies in different phages. The tube is then able to interact with the cytoplasmic membrane.
The final step of the infection is the translocation of DNA and proteins from the phage capsid into the host-cell cytoplasm. The tail is thought of as a passive conduit in this process because it does not appear to contain a source of energy necessary to make it an active participant of this translocation event. A very detailed description of phage genome delivery into the host cell is given in a different chapter of this book (see Chap. 7 in this book).
The tail of bacteriophage T4 is by far the best-studied contractile system (Coombs and Arisaka 1994; Leiman et al. 2010). The T4 tail composition, assembly, structure, and conformational changes on attachment to the host cell are fairly well understood (Coombs and Arisaka 1994; Leiman et al. 2010). The structure and function of phage P2 tail (the other “model” contractile tail) have been studied to a much lesser extent (Kahn et al. 1991). These model systems are used for gene function prediction and assignment in all newly characterized contractile tail-like systems.
The accumulating genomic and protein structure data suggest that the contractile tail systems contain a set of conserved genes and have a common ancestor. Furthermore, many genes are shared with noncontractile tails also pointing to the common ancestry. A very compelling and plausible hypothesis stating that the contractile tails have likely evolved from noncontractile tails is beautifully presented in Chap. 6. Thus, this chapter will not discuss such evolutionary relationships, but instead will summarize the currently available data and emphasize common features of contractile tail-like systems.
2 Contractile Tail Structure
Contractile tails have an average diameter of ∼220 Å but vary greatly in length from ∼1,000 to ∼4,500 Å. Despite its relatively large size, the organization of a contractile tail is fairly simple because its major part is a repetitive structure composed of two proteins – the internal tube and the external contractile sheath. Most of the tail’s complexity is concentrated in the baseplate, which is the control center coordinating the host attachment process. The sheath and the tube represent two major bands on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) and serve as the definitive markers of a contractile sheath-like structure.
All known contractile phages have sheaths with molecular weights (MWs) between ∼40 and ∼80 kDa with the mean shifted to the lower values. A sheath with a MW of ∼45 kDa is the most widespread among currently sequenced phages. The sheath proteins of two model phages, P2 and T4, have MWs of 43 and 71 kDa, respectively. On the other side, known tube proteins display less variation in their size. Their MWs are within the 15–21-kDa interval (18.5 kDa for T4 and 19.1 kDa for P2). The pair of sheath and tail tube genes also serves as a genetic marker of a contractile tail because the two genes are adjacent to each other with the sheath gene preceding the tube one and are transcribed from the sheath gene promoter.
2.1 Structure of the Sheath
The structure of the T4 sheath, the model object for all contractile sheath-like systems, was studied by several generations of structural biologists (reviewed in Coombs and Arisaka 1994; Leiman et al. 2010). However, a major breakthrough happened only recently, when the crystal structure of the T4 sheath protein gp18 (gene product 18) was solved (Aksyuk et al. 2009a). Full-length recombinant gp18 forms polysheaths of various lengths, which are not suitable for crystallization (King 1968). These polysheaths contain defects (missing subunits and other irregularities) making them difficult to analyze to high resolution because averaging techniques cannot be fully exploited. A large body of mutagenesis work went into producing a gp18 mutant, which would not form a polymeric structure (Kuznetsova et al. 1998; Poglazov et al. 1999). The result of this work is a mutant called gp18M constituting about three-fourth of the full-length molecule (residues 1–510 out of 659) with one amino acid substitution Arg510 → Pro.
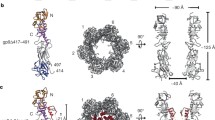
Gp18 consists of four domains. Twenty N-terminal and ∼160 C-terminal residues form Domain I, which is not present in the gp18M crystal structure (the domains are renumbered compared to Aksyuk et al. 2009a) to make the nomenclature consistent with other sheath proteins). Domain II (residues 21–87 and 346–510) consists of a β-sheet with five parallel and one antiparallel β-strands plus six α-helices, which surround the β-sheet. Domain III (residues 88–97 and 189–345) is a two-layer β-sandwich, flanked by four small α-helices. Domain IV (residues 98–188) is a six-stranded β-barrel plus an α-helix. Together domains III and IV form the protease resistant fragment of the sheath protein (Fig. 5.2).
Structure of the sheath protein. T4 gp18 (PDB ID 3FOA) and DSY3957 (PDB ID 3HXL) are shown as ribbon diagrams in the top and bottom panel, respectively. Domains I, II, III, and IV are colored purple, blue, green, and red, respectively. Domain I is not present in the crystals structure of T4 sheath protein gp18 and is shown as a purple rectangle. The domain-swapped N-terminal arm, which is donated to Domain I by a different subunit belonging to the same strand of the sheath, is colored yellow. The N-terminal arm belonging to the polypeptide chain of DSY3957 (PDB ID 3HXL) shown in the lower panel is colored in orange. In the crystal structure, it is donated to Domain I of a molecule related by crystal symmetry (not shown)
The overall topology of the gp18 polypeptide chain is quite remarkable. Domain IV is “inserted” between residues 97 and 189, which belong to a loop connecting two β-strands of Domain III (Figs. 5.2 and 5.3). The latter is inserted between residues 87 and 346 into a loop of Domain II. Domain II, in turn, is an insertion into Domain I.
Domainal organization of sheath proteins. The color code as in Fig. 5.2 Domains I, II, III, and IV are colored purple, blue, green, and red, respectively. The domain-swapped N-terminal arm is colored yellow. The orientation of the sheath subunit relative to the tail axis is shown below each diagram
T4 tail has been studied by cryo-electron microscopy (cryoEM) in the extended and contracted conformations (Leiman et al. 2004; Kostyuchenko et al. 2005). The crystal structure of gp18M fits (as a rigid body) into the cryoEM map of the sheath in both conformations. Domain I interacts with the tail tube and forms the internal core of the sheath. Domains II and III are partially exposed to solution. Domain IV forms the sheath’s external surface.
T4 sheath is composed of 138 gp18 subunits, which are arranged in a six-start helix (Leiman et al. 2004). Each of the six strands comprising the helix contains 23 gp18 subunits (Leiman et al. 2004). The sheath is 240 Å wide and 925 Å long. The pitch and twist of the helix are 40.6 and 17.2 Å, respectively (Kostyuchenko et al. 2005) (Fig. 5.4). Each gp18 subunit interacts with four neighbors: two related by the sixfold symmetry (interstrand contacts) and two within the same strand (intrastrand contacts). The interactions within the strand govern the connectivity and helical interpretation of the sheath structure because the surface of the intrastrand subunit contact is five times that of the interstrand (Kostyuchenko et al. 2005).
Structure of T4 sheath in the extended and contracted states as determined by cryoEM (Leiman et al. 2004; Kostyuchenko et al. 2005; Aksyuk et al. 2009a). Three of the six gp18 helices are extracted from the complete tail structure, which is shown in the extended (a) and contracted (b) conformations. Each strand is colored in a distinct color (pink, blue, and green). The successive layers of gp18 subunits are numbered 1, 2, 3, 4, and 5 with the layer numbered 1 being closest to the baseplate. The middle panels of (a) and (b) show the arrangement of Domains II trough IV in three neighboring strands. The rightmost panels show the interactions of Domains I from the same three strands (note, that these domains maintain connectivity in the extended and contracted states). A simplistic representation of how Domains II through IV belonging to the three neighboring strands change their interactions in the extended and contracted states is also shown in the middle panels
The contracted T4 sheath is 330 Å wide and 420 Å long. It is also a six-start helix with a 16.4 Å pitch and 32.9° twist (Leiman et al. 2004). Compared to the side-by-side arrangement of the gp18 strands in the extended sheath, the strands in the contracted sheath are interdigitated (Fig. 5.4). The axial compression or contraction is accomplished by sliding one layer of the sheath subunits into the next one, which is accompanied by the increase in the sheath’s diameter. Upon sheath contraction, each of gp18 subunits moves by ∼50 Å away from the tail’s axis and tilts by ∼45° (Aksyuk et al. 2009a).
In the process of sheath contraction, gp18 subunits move as rigid bodies without refolding or significant domain shifts (Aksyuk et al. 2009a). Domains I of the gp18 subunits belonging to the same strand maintain the connectivity throughout the contraction process, thus preserving the helical structure of the sheath, whereas Domains II through IV form new interactions in the contracted state (Fig. 5.4). Upon contraction, the contact area between gp18 molecules increases by about four times (Aksyuk et al. 2009a).
2.2 Evolution of Sheath Proteins
The topology of gp18 polypeptide chain alone suggests that Domain I is likely the oldest part of the sheath structure (Figs. 5.2 and 5.3). Domains II, III, and IV were “picked up” over the period of evolution of the sheath to its current structure. This assumption is strengthened by comparing the primary and tertiary structure of gp18 with its homologs.
Due to the diversity of phage tail sheath sequences, only relatively close relatives of phage T4 can be identified using the gp18 amino acid sequence. Mapping the amino acid sequence conservation onto the gp18M structure shows that Domains I and II are the most conserved parts of the structure.
In addition to gp18, crystal structures of two other sheath proteins are currently available at the Protein Data Bank (PDB, http://www.pdb.org) under the deposition codes (IDs) 3HXL and 3LML (Figs. 5.2 and 5.3). They represent the complete products of genes dsy3957 and lin1278 from two different prophages infecting Gram-positive Desulfitobacterium hafniense and Listeria innocua, respectively (note that T4 is a Gram-negative Escherichia coli lytic phage). DSY3957 (3HXL) and LIN1278 (3LML) have very similar folds and show 43% sequence identity and a root mean square deviation (RMSD) of 2.8 Å upon superposition with 81% of residues participating in the alignment. The proteins consist of three domains connected by flexible linkers. The domains are shifted and rotated relative to each other, thus elevating the RMSD value, but the matching domains have virtually identical folds.
A careful inspection of the structure of DSY3957 and LIN1278 shows that folds of two of their three domains are similar to those of gp18 Domains II and III even though there is no significant sequence similarity (17% or lower overall) (Table 5.1) (Fig. 5.3). Domains II of the three proteins are very similar in size (∼220 residues), whereas Domain III of gp18 is almost twice as big as that in the other two proteins (∼160 residues vs. ∼90 residues) because it contains many small and large insertions, one of which is Domain IV (90 additional residues). The third domain of DSY3957 and LIN1278 corresponds to Domain I of gp18, which is not present in the gp18M structure. HHpred analysis shows that Domains I of the three proteins have a 100% probability to have the same fold (Table 5.2).
This analysis shows that gp18, DSY3957, and LIN1278 have a common ancestor. Furthermore, we can propose that Domains I, II, and III likely represent the smallest unit, which can form a contractile sheath-like structure. The common ancestry of the sheath proteins also suggests that all sheaths undergo similar reorganization during the contraction event with Domains I maintaining the connectivity throughout the process and Domains II and III forming new and more extensive intersubunit interactions in the contracted state compared to the extended one.
A possible explanation of how Domains I maintain their connectivity during the contraction event can be given with the help of the DSY3957 crystal structure and the T4 sheath cryoEM map. Domain I of DSY3957 is formed by residues 308–438 of one polypeptide chain and residues 1–25 of the symmetry-related molecule. In other words, one molecule donates its N-terminal arm to its crystallographic neighbor, where these residues become a part of a β-sheet within Domain I. By superimposing DSY3957 onto the structure of gp18 fitted into the T4 sheath cryoEM map, we find that the length of the donated N-terminal arm and the spatial orientation of Domains I stacked onto each other should allow such a domain swapping to take place within the sheath structure. Therefore, the entire inner part of the sheath is “cross-linked” by the chain-swapped Domains I. This property is likely universal for all contractile tail-like structures.
2.3 Structure of the Tube
The accumulated body of data does not allow to give a fully consistent description of the tail tube structure. Earlier studies of isolated T4 tubes suggested that their helical structure is identical to that of the sheath in the extended state. Furthermore, the sheath and the tube appear to contain the same number of subunits. The sheath polymerizes into the extended conformation only in the presence of both the baseplate and the tube. It was proposed that during tail assembly the tube serves as a scaffold for the sheath and that the latter adapts the helical symmetry of the tube.
These conclusions are difficult to reconcile considering the latest structural data. Subunits, comprising the tube, are not well resolved in the T4 tail cryoEM maps, making symmetry determination impossible. If the tube and the sheath had the same symmetry, tube subunits should be well resolved, similar to the sheath subunits, but this is not the case. Furthermore, as it is shown in Chap. 6 in this book, the contractile tails, the long noncontractile tails, and the bacterial type VI secretion system (T6SS) have a common ancestor. The T6SS tube protein (called Hcp) forms tubes with sixfold symmetry in the crystalline form and, possibly, in solution. However, in these tubes, Hcp hexamers are stacked onto each other without any twist, which is inconsistent with the helical symmetry of the sheath.
It is nevertheless possible that the tube and the sheath have the same helical symmetry. The subunits of the tube might be unresolved in the cryoEM maps due to the powerful Fourier ripple of negative density caused by the sheath. This ripple overlaps with the tube density and significantly deteriorates it.
An explanation – consistent with current data – for the tube and the sheath to have different symmetries also exists. The subunits of the sheath and the tube might come in register only at certain points of the elongated structure, and the tube would still serve as a scaffold for sheath polymerization. Yet another possibility is that sheath subunit polymerization into the extended conformation is determined and driven by the baseplate, and the tube per se is not required for polymerization. Instead, only the proteins associated with the tube’s end are necessary for stabilization of the sheath, which is known to depolymerize quickly without the tail capping complex.
2.4 The Simplest Contractile Tail-Like Structure
In phages with large genomes, the baseplate is a very complex organelle. For example, the baseplate of phage T4 consists of 12 proteins. Two additional proteins form a cylindrical structure, onto which the tail tube subunits are assembled and polymerized into the tube. The structure and position of nine constituent proteins (or their major domains) in T4 baseplate are known. This information is used to understand the function of proteins in other baseplates.
Phage genomes are diverse in general, but even close relatives display large variations in their RBPs, which emanate from the baseplate. Since the baseplate is responsible for interaction with the host-cell surface, it evolves faster compared to the rest of the genome because a small change in the baseplate structure might cause the phage to switch host specificity, and such an adaptability is in general beneficial for phage survival.
The diversity of baseplate genes of closely related phages increases toward the periphery culminating in the host-cell-binding regions of RBPs. This diversity pattern is even more pronounced in phages, which are related very distantly. Often, homologous proteins show no similarity at the amino acid sequence level, but the common ancestry could still be found at the level of protein structure. Unfortunately, structures of very few non-T4 baseplate proteins are currently available. One such pair of previously unknown homologs is gp27 (the central hub protein of the T4 baseplate) and gp44 of phage Mu. The two proteins have very similar folds but show only 9.6% sequence identity (Kondou et al. 2005). It is clear now that the baseplate hub proteins of contractile and noncontractile tails have a common ancestor (Sciara et al. 2010). This is described in detail in Chap. 6 in this book.
Despite the limited information on the structure of non-T4 baseplate proteins, it is possible to establish the relationship between the T4 proteins with known functions and their phage P2 and phage Mu homologs and extend this information to all other contractile tail-like systems. Furthermore, we can even attempt to describe the composition of a simplest contractile tail-like structure. The HHpred Web server (Soding 2005; Soding et al. 2005) and several crystal structures solved in the framework of the structural genomics initiative are very useful for this analysis.
The hub structure. The central part of a simplest contractile tail-like structure is formed by the homologs of T4 gp27 and gp5 proteins. The orthologs of these and other proteins mentioned below, which comprise the particles of P2 and Mu phages, are listed in Table 5.3 and shown in Fig. 5.5. Gp5 forms the central membrane-puncturing needle or spike in T4 baseplate. The N-terminal domain of gp5 is an OB-fold, and HHpred clearly shows that the N-terminal domain of P2 gpV and Mu gp45 is a similar OB-fold. The three OB-fold domains of gp5 trimer are inserted into the channel formed by the trimeric gp27 donut-like structure, whereas the C-terminal domain comprises the membrane-attacking spike. In T4 gp5, this domain is a very stable, intertwined trimeric β-helix, and a high β-structure content is predicted for the C-terminal domains of P2 gpV and Mu gp45 suggesting that they also might form a β-helix. In general, the N-terminal OB-fold domain and the C-terminal β-structure-rich domain is a common feature of all membrane-puncturing needles or spikes in phages of Gram-negative bacteria.
The trimeric gp5–gp27 hub forms the centerpiece of the otherwise sixfold symmetric baseplate. The gp27 trimer acts as a three to sixfold symmetry adaptor. One part of the gp27 trimer tightly wraps around the three N-terminal domains of gp5 trimer, whereas the other part is composed of six tandem domains with two domains per monomer, which results in 2 × 3 = sixfold symmetry. The six tandem tube domains of gp27 trimer serve as a starting point for polymerization of the tail initiator complex and the rest of the tail tube. The structure of gp27 trimer is conserved not only in contractile but also in noncontractile tails making the three to sixfold adjustment principle universal for all long tails.
One feature of T4 tail not found in other contractile tails is a lysozyme domain in their gp5–gp27 complex orthologs. In T4, the lysozyme domain is “inserted” between the OB-fold and the C-terminal β-helical domain of gp5. The gp5 lysozyme domain shows 43% sequence identity to T4L, the cytoplasmic T4 lysozyme encoded by gene e (Mosig et al. 1989). T4L and the gp5 lysozyme domain have very similar structures with conserved catalytic residues suggesting their common ancestry (Kanamaru et al. 2002). The likely function of gp5 lysozyme domain is to locally digest the peptidoglycan layer during infection to make an opening for the tube (Kanamaru et al. 2002). However, a phage carrying a mutant gp5 with only 10% of the wild-type activity is almost as infective as the wild-type phage, suggesting that the residual activity is likely sufficient for infection in the laboratory conditions (Kanamaru et al. 2005).
Nevertheless, the presence of the lysozyme domain in T4 gp5–gp27 complex remains somewhat enigmatic because even in close relatives of T4, such as, for example, in Pseudomonas aeruginosa phage KVP40, the gp5 ortholog does not contain a lysozyme domain (Rossmann et al. 2004). Gp5 orthologs of P2 and Mu do not contain a lysozyme domain either (Table 5.3). However, a lysozyme or glycosidase domain can be detected at the C-terminus of tape measure proteins in some contractile and noncontractile phages (Piuri and Hatfull 2006; Boulanger et al. 2008). Most probably, the C-terminal domain of the tape measure protein interacts with the ortholog of gp27 hub protein and is translocated into the periplasm during infection. In summary, a functional glycoside does not appear to be required for contractile tails under all infection conditions.
It is worth to mention that the gp27 ortholog of the contractile tail iodobacteriophage ϕPLPE (Leblanc et al. 2009) encoded by gene 58 contains a lysozyme domain at its C-terminus (goose-type lysozyme), whereas its gp5 ortholog, encoded by gene 59, is lysozyme domain-free. Assuming that gp58 and gp59 of ϕPLPE form a complex similar to T4 gp5–gp27, the position of the lysozyme domain of ϕPLPE gp58 in the complex and in the baseplate would match that of the gp5 lysozyme domain.
Orthologs of T4 gp5–gp27 complex are also found in phages infecting Gram-positive bacteria, but the membrane-interacting C-terminal domain of gp5 proteins in these phages is a coiled coil instead of the β-helix (e.g., gp98 and gp99 from the Listeria phage A511 are homologs of T4 gp27 and gp5, respectively).
The minimal baseplate. The structure of T4 baseplate shows that the hub is surrounded by three proteins – gp6, gp25, and gp53. The ortholog of gp25 can be identified in the genomes of P2 and Mu using sequence similarity alone. The crystal structure of a gp25 homolog from a Geobacter sulfurreducens prophage (GenBank NP_952040.1) was solved by structural genomics (PDB ID 2IA7). The fold of this protein is very similar to that of Domain I of the sheath protein, which forms the inner part of the sheath. It is likely that gp25 and Domain I interact in the baseplate and that Domain I and, possibly, the rest of the sheath originates from a gp25-like protein. In general, the gp25 ortholog is the most conserved protein (in terms of the amino acid sequence) in the baseplate.
The orthologs of T4 gp6 in the baseplates of P2 and Mu (gpJ and gp47, respectively) are predicted by HHpred with a very high confidence (97% probability), but the “fifth” baseplate protein (the other four being gp5, gp27, gp25, and gp6) is somewhat enigmatic. In P2 and Mu, this protein is gpI and gp48, respectively, but none of them shows any sequence similarity to each other or T4 gp53. Their only common trait is a similar size (∼180 residues). It is possible that the T4 ortholog of the “fifth” protein is a domain of gp7.
The position and function of the three conserved baseplate proteins (orthologs of T4 gp25, gp6, and gp53/gp7) can be derived from the structure of T4 baseplate (Kostyuchenko et al. 2003; Aksyuk et al. 2009 b). The ortholog of T4 gp25 protein is positioned near the hub and might interact with the sheath. There are six copies of gp25 in the baseplate in agreement with the sheath symmetry. The gp6 ortholog forms a major part of the “minimal” baseplate. Six gp6 ortholog dimers form a continuous ring enveloping the hub and the gp25 ortholog protein. Six copies of the “fifth” baseplate protein (T4 gp53/gp7 ortholog) bind to the periphery of the gp6 ring. Tail fibers bind to the “fifth” protein and, possibly, to gp6 ortholog.
RBPs: The tail fiber/tailspike and tail fiber chaperone. The tail fiber/tailspike is responsible for host-cell receptor binding. It is an essential part of a functional tail. All known phage fibers are trimers having folds with very complex topologies often dominated by intertwined β-helices and, less often, α-helical coiled coils (Steven et al. 1988; van Raaij et al. 2001; Thomassen et al. 2003; Walter et al. 2008; Bartual et al. 2010). The C-terminal domain of the fiber is responsible for receptor binding, and its N-terminal domains attach the fiber to the baseplate.
The folding and attachment of the fiber to the baseplate is governed by a phage-encoded chaperone, which is always positioned downstream to the fiber. In some phages, the chaperone is part of the same gene or might remain bound to the fiber after its attachment to the baseplate (Riede et al. 1985; Tetart et al. 1996). In the latter case, the chaperone participates in host-cell receptor recognition and binding. The chaperones show a certain level of specificity in attachment of the fiber to the baseplate, suggesting that the chaperone–fiber complex binds to the tail during the fiber attachment (Williams et al. 2008).
The tail tube initiator. T4 clearly possesses the tail tube initiator complex, which serves as a starting point for polymerization of the tail tube. The initiator complex is built onto the gp27 trimer. In T4, this complex contains gp48 and/or gp54 (Kostyuchenko et al. 2003). Gp54 shows a significant sequence similarity to T4 tail tube protein gp19 and tube terminator protein gp3. There is not enough data to identify tube initiator proteins in P2 or Mu, but since P2 gpU and Mu gp43 are tail components with unknown function and location, we can tentatively assign them the tube initiator function.
The tape measure protein. Similar to noncontractile tails, the length of the tail is controlled by the tape measure protein (Abuladze et al. 1994). In T4, the tape measure protein gp29 appears to participate in the formation of the hub and the gp48/gp54 tube initiation complex (Kikuchi and King 1975a, b, c, d). The tape measure proteins of contractile and noncontractile tails are most likely homologous and thus function in a similar manner. The structure and function of tape measure proteins are described in detail in Chap. 6 in this book. Briefly, several copies of the tape measure protein extend through the central channel of the tail tube as it is being built. In the fully assembled tube, the tape measure protein adapts an extended α-helical conformation, and its N-terminus interacts with the tail tube terminator and, possibly, with the sheath terminator protein. Some tape measure proteins contain an identifiable glycosidase domain at the C-terminus. High-resolution studies of T4 tail did not provide sufficient detail on whether the tube channel is occupied by the tape measure protein or phage genomic DNA extending from the capsid.
Interestingly, the contractile tail of iodobacteriophage ϕPLPE mentioned earlier is one of the shortest known. The sheath part of the tail is only 600 Å (Leblanc et al. 2009), and its putative tape measure protein of ϕPLPE (gp55) is only 392 residues long. It does not contain an identifiable peptidoglycan hydrolase domain and is likely to have an α-helical structure throughout the sequence (McGuffin et al. 2000). In the extended α-helical conformation, it can form a fiber with a length of ∼600 Å, which roughly corresponds to the length of the tail, but unlike other tape measure proteins ϕPLPE gp55 does not appear to form even a small globular domain at its termini. As the sheath proteins of all phages have a common core structure (see below), the pitch of the sheath helix is likely to be conserved and approximately equal to that found in T4, i.e., ∼40 Å. Thus, the sheath and, consequently, the tube of ϕPLPE contain approximately 15 × 6 = 90 subunits each.
The tail tube and the sheath. The structure of the tube and the sheath of phage T4 is described above. The helical parameters and the length of the sheath vary in different phages even though the core structure of the sheath protein (the three inner domains) is conserved. The length of the tail in different phages varies, and phages with larger genomes have longer tails on average.
The tail terminator complex. In contractile tail phages, the tail is capped by two proteins, one of which interacts with the tail tube, whereas the other one most probably interacts with both the tube and the sheath (Coombs and Arisaka 1994; Leiman et al. 2010). The tail tube capping protein is a homolog of the noncontractile tail terminator protein. The tube and sheath terminators in T4 are encoded by gp3 and gp15, respectively. It is not possible to identify the terminator proteins in neither P2 nor Mu using the sequence data alone, except for Mu gp37, which is clearly related to the noncontractile tail terminator proteins (see Chap. 6).
In phage T4, the tail terminator complex can adapt two conformations – closed and open – associated with the extended and contracted sheath conformations, respectively (Kostyuchenko et al. 2005). The conformation of the tail terminator complex controls the location of the leading end of DNA molecule in the tail. In the extended tail, the central channel of the terminator complex is closed, and DNA cannot exit the capsid. Sheath contraction causes the channel to open up, allowing the leading end of phage DNA to exit from the capsid and descent down the tail tube in preparation for ejection into the host cell.
3 Assembly of a Contractile Tail
Extensive studies of T4 tail morphogenesis showed that the baseplate is assembled first (Kikuchi and King 1975a, b, c, d). The tail tube and sheath are built starting from the baseplate. Without the tube or the baseplate, T4 sheath assembles into a long polymeric structure called polysheath, whose structure is similar to that of the contracted sheath (Moody 1967a, b, 1973). The tube and the sheath must be capped with the tail terminator complex, or they would depolymerize (King 1968; Coombs and Eiserling 1977; Tschopp et al. 1979). The fibers are the last proteins, which attach to the baseplate. T4, like many large phages, contains two set of tail fibers. The short tail fibers attach to the baseplate directly, whereas attachment of the long tail fibers requires the phage particle to assemble fully: the tail must be sheathed and bound to the DNA-filled capsid. Another set of fibers, called whiskers or fibritins, emanating from the head-to-tail junction, participates in the long tail fiber assembly. No other phage tail assembly was studied to such an extent, but because all contractile tails have a common ancestor, their assembly pathway is likely to resemble that of T4.
The assembly process is strictly ordered. If a protein is excluded from the process, the assembly intermediate preceding the missing protein will accumulate in the cell (Kikuchi and King 1975a, b, c, d). Association of baseplate, tube, or sheath subunits into the complete structure does not appear to require any chemical energy. Except for gp5, no protein in the tail appears to undergo maturation proteolysis upon incorporation into the tail complex (Coombs and Arisaka 1994; Leiman et al. 2010).
It is apparent, however, that the tail is originally assembled as a high-energy metastable structure, akin to an extended spring, which is locked in this conformation by noncovalent interactions and can contract when those interactions are disturbed. Exposure of tails to 3 M urea and heat, and abrupt pH changes disturb these interactions causing contraction (Coombs and Arisaka 1994). Furthermore, the sheath protein assembles into a contracted sheath-like structure (the polysheath) in the absence of the baseplate and/or the tail tube. The contracted conformation of the sheath must be a very low-energy configuration because it is resistant to 6 M urea and detergents.
Still, there must be a source of energy, which is used to build the extended sheath structure in the first place. It is possible that the process is kinetically driven. The formation of the sheath in the extended conformation occurs about 50 times faster than the formation of polysheaths and, therefore, is a preferred pathway due to a lower activation energy (Arisaka et al. 1979; Tschopp et al. 1979). The integration of the N-terminal arm of the sheath protein into the C-terminal domain of the next layer subunit in the sheath (Sect. 5.2.2) might also be an important factor driving the polymerization of the sheath. Because the arm is evolved to interact with the C-terminal domain, its free energy is higher when it is extended into the solvent. This characterizes the sheath subunit before it becomes part of the polymeric structure. Thus, sheath subunits have a high intersubunit affinity, and the baseplate–tail tube complex shifts the assembly process toward the extended structure.
T4 baseplate can exist in one of two conformations, which are coupled to two sheath states: the “hexagonal” conformation is associated with the extended sheath, whereas the “star” conformation is associated with the contracted sheath, which is found in T4 particle after attachment to the host cell (Coombs and Arisaka 1994; Kostyuchenko et al. 2003; Leiman et al. 2004). The free energy of the hexagonal conformation is higher than that of the star because hexagonal baseplates switch to stars after prolonged storage. There is no published data about the structure or conformational changes of simpler baseplates (e.g., from phages P2 or Mu), but the structure of a noncontractile phage baseplate in two conformations is available (Sciara et al. 2010). Upon host-cell binding, this baseplate undergoes a rearrangement resembling that of T4 baseplate, from which we can conclude that the conformation of the baseplate is coupled to host-cell binding and determines sheath conformation in all contractile tail phages.
4 Interaction of the Contractile Tail with the Host-Cell Envelope
The baseplate coordinates host recognition and attachment with sheath contraction, which is initiated at the baseplate and is propagated through its entire length of the sheath in a wavelike fashion (Simon and Anderson 1967a, b; Moody 1973; Aksyuk et al. 2009a). Tail fibers, which emanate from the baseplate, are the primary determinants of host-cell specificity (Tetart et al. 1998; Williams et al. 2008). The host-cell binding parts of the fibers (their C-terminal domains) are positioned as far as 1,000 Å from the baseplate in some phages. Successful receptor binding triggers the conformational change of the baseplate, which then leads to sheath contraction. Thus, the host-cell-binding signal must be transmitted through the length of the fiber to the baseplate. The transmission of the signal occurs by way of a change in the orientation of the fibers relative to the baseplate because the fibers are not known to undergo refolding upon receptor binding. When the phage particle is free in solution, the fibers do not have a fixed orientation and point roughly sideways or even toward the capsid. The fibers of the phage bound to the host-cell surface point toward it. We will describe two possible mechanisms, which explain how and why this change in fiber orientation happens only on the cell surface, but not when the phage is free in solution (Fig. 5.6). Notably, no chemical energy is used in fiber reorientation and baseplate triggering.
Breaching the host-cell envelope. (a–h) Show stages and substages of the infection process. Blue arrows indicate motions caused by solvent movement (Brownian, thermal, convection, etc.). Magenta arrows show the movement of the phage capsid towards the host-cell surface, which is driven by sheath contraction. (a) The phage is free in solution, and its fibers move freely around their most favorable position. (b) The phage binds to the host cell with one or two of its fibers, which serve(s) as a tether restricting the movements of the phage particle. (c) The phage is moved in such a way that the fiber is brought into a conformation from which it cannot switch back to the “free phage” fiber configuration. The fiber forms new interaction with the baseplate proteins, and the baseplate initiates its conformational change. (d) The phage continues its hula dance on the cell surface while being attached to it with the tail fiber tether. Eventually, the other fibers find their binding partners on the cell surface. (e) As in (c), the phage is moved in such a way that all of its fibers, which have bound to the cell surface receptors, point toward the host cell. The conformational change of the baseplate now proceeds. (f) The baseplate conformational change initiates contraction of the sheath, which drives the capsid and the tube toward the cell membrane. The outer cell membrane is punctured with the help of the baseplate central spike protein. (g) The spike dissociates from the tail tube tip, thus opening the tail tube channel. The tail-associated glycosidase creates a small opening in the peptidoglycan layer. (h) The tail tube interacts with the inner membrane, which is pushed toward it by the osmotic pressure of the cytoplasm in small region now lacking the peptidoglycan layer. Phage DNA is released into the cytoplasm
The first mechanism, which we favor, can be described as follows (Fig. 5.6). When the phage comes across a susceptible host cell, most probably only one or two of the fibers bind to the host-cell receptors. Thus, attached phage is now under the influence of all the solvent movements originating from cell swimming and other molecular motions of various sorts (Brownian, thermal, convective currents, etc.). The phage is tethered to the cell with its fiber(s), while it is constantly being shaken by the solvent. Eventually, the other fibers bind to the host-cell receptors, which lead to both, orienting the phage perpendicular to the host-cell membrane and, at the same time, causing all the fibers to be pointed toward the host-cell surface. When the fibers are in this configuration, new interactions are formed in the baseplate causing it to switch to a lower energy conformation, which unlocks the sheath allowing it to contract. The spontaneous tail contraction does not occur because the orientations of the fibers associated with the extended and contracted sheath conformations are separated by a significant free energy peak. This peak can be overcome only if the fibers have become immobilized on the cell surface and the phage particle is being jiggled around by the solvent. This mechanism is supported by structural investigations of phage T4 baseplate (Leiman et al. 2004; Kostyuchenko et al. 2005) and R-type pyocin (P. Leiman, unpublished data).
The other possible mechanism of how fiber reorientation can be coupled to sheath contraction upon attachment to the host-cell surface comes from the observation that many phages require divalent cations (most often calcium) for infection. Cell surface molecules, which include phage receptors (polysaccharides and proteins), bind those cations making their concentrations near the cell surface very high. The high concentration of the ions can cause the baseplate to change its conformation and extend the fibers to bind to the host-cell surface. The baseplate will then change its conformation and unlock the sheath. This mechanism is similar to that proposed for the Gram-positive Lactococcus lactis bacteriophage p2 (Sciara et al. 2010), which requires calcium for infection. However, many contractile tail phages have very week requirements for medium composition for successful infection and can infect without calcium or any other divalent ions. Furthermore, phage tails contract in 3 M urea but stay intact in calcium or other divalent cations (Leiman et al. 2004; Leblanc et al. 2009).
It is possible that both proposed mechanisms are in play. The fibers become pointed to the host-cell surface as a result of solvent-induced phage movement relative to the host-cell surface, but, at the same time, the baseplate is more predisposed to changing its conformation in the presence of “surface-associated” ions. It is possible that binding of fibers to the host receptors releases additional ions, thus increasing the local concentration even further.
As the sheath contracts, it moves the capsid closer to the cell surface making the tail tube protrude from below the plane of the baseplate. The baseplate central hub complex, onto which the tube is assembled initially and which consists of T4 gp5 and gp27 orthologs, is dislodged from the baseplate and now forms the tip of the tube. This complex is the membrane-piercing “device,” which is driven through the cell outer membrane by the energy of the sheath. The C-terminal domain of the T4 gp5 ortholog forms a membrane-piercing spike or needle, and its N-terminal domain plugs the opening of the gp27 ortholog trimer, which forms the rim of the tube. The gp5 ortholog must dissociate from the gp27 ortholog to open the tube channel to allow the phage DNA to exit through the tube. It is unclear whether the gp5 ortholog is used to disrupt both membranes and it eventually leaves the gp27 ortholog in the cytoplasm or it comes off in the periplasm. Furthermore, there is little data about the interaction of the tube with the inner membrane.
The large energy, which is stored in the condensed conformation of phage DNA packaged into the capsid, is not used in membrane penetration because the pyocins (DNA-free complexes) are able to disrupt the host-cell envelope and kill the cells (Scholl et al. 2009). Furthermore, sheath contraction and DNA ejection are not linked (Leiman et al. 2004). The contraction can be induced by subjecting the phage to moderate concentrations of urea or pH and/or osmotic shock. Phages with contracted tails are, however, fairly stable and do not release their DNA. Therefore, the cytoplasmic membrane must contain a specific receptor, which opens up the tube and triggers the release of DNA. This receptor must be at least as abundant as the phage receptor on the cell surface. It is possible that the tube channel opening might be triggered by certain lipids comprising the cytoplasmic membrane. Translocation of DNA into the host cell through the tube is the next step in the infection process. The physics of this process is a hot topic of debate in today’s biophysics. This is covered in Chap. 7 of this book.
5 Structure and Function of Other Contractile Tail-Like Systems
In the past few years, several systems, which appear to share their ancestry with contractile tail phages, have been characterized. Among those are the R-type pyocins, the bacterial type VI secretion system (T6SS), the Photorhabdus virulence cassette (PVC), and rhapidosomes.
5.1 R-Type Pyocins
Pyocins are proteinaceous (DNA-free) bactericidal agents of P. aeruginosa. They were first described by Francois Jacob as high molecular weight bacteriocins of P. aeruginosa (Jacob 1954), but similar bacteriocins were found in other Gram-negative and Gram-positive bacteria (Williams et al. 2008). Pyocin DNA resides silently in the bacterial genome until it is activated with a UV or mitomycin C treatment (Matsui et al. 1993). This treatment causes DNA damage and activates RecA, which degrades the PrtR protein, the repressor of PrtN, a positive transcription regulator of the pyocin gene cluster.
R-type pyocins are close relatives of phage P2 tail (Nakayama et al. 2000). The R-type pyocin particle consists of at least 11 proteins all of which are related to P2 tail genes listed in Table 5.3. R-type pyocin represents one of the smallest contractile tail-like systems. The tube/sheath capping proteins of R-type pyocins have significantly diverged (in terms of the primary structure) from their P2 orthologs.
A pyocin-lysogenized bacterium can produce up to 200 R-type pyocin particles that leave the host by causing its lysis. Each of the newly released particles can bind to a bacterium and form a pore in its envelope causing depolarization of the cytoplasmic membrane and resulting in host death. Like phage, pyocins are very potent bacteriocins, and one pyocin particle is enough to kill a bacterium. A pyocin particle can strike only once. Being DNA-free, it does not multiply. Unlike that for a phage, the “infection” of a cell by R-type pyocin does not cause cell lysis, which means that the cytoplasmic content is not spilled out into the milieu immediately upon cell death. The host specificity of pyocins and phages is determined by the tail fibers, and P. aeruginosa pyocins can be retargeted to kill E. coli by equipping the pyocin with a fiber from an E. coli phage (Scholl et al. 2009).
R-type pyocins are divided into five subtypes (R1 through R5), each of which has narrow host range (up to a single P. aeruginosa strain). P. aeruginosa cells, which carry a particular pyocin, are resistant to it. Thus, P. aeruginosa cells carrying a pyocin can suppress growth or completely eliminate other susceptible P. aeruginosa cells in a given environmental niche (Kohler et al. 2010).
5.2 Type VI Secretion System
The type VI secretion system (T6SS) has been characterized as a secretion system of Gram-negative bacteria relatively recently compared to the other five types (Mougous et al. 2006; Pukatzki et al. 2006). It is a remarkably versatile system: T6SS was shown to mediate the interaction between a T6SS-expressing bacterial cell, its eukaryotic host and other bacteria (reviewed in (Records 2011). T6SS manifests itself in an increased pathogenicity or determines it for some strains of Vibrio cholerae, P. aeruginosa, Edwardsiella tarda, E. coli, and other bacteria. T6SS can function as a mediator of competitive interbacterial interactions and as a regulator of a cooperative social behavior. T6SS is found in plant-associated bacteria, including both pathogens and symbionts (Records 2011).
T6SS genes are clustered in pathogenicity islands containing 15–20 open-reading frames and can be easily detected using a set of conserved genes as markers. It is found in about one quarter of all bacterial genomes currently present in the database, often in several copies per genome. In addition to the contiguous cluster, homologs of certain T6SS genes could be found in as many as 25 copies in different genomic loci. The T6SS genes are predicted to encode cytoplasmic, periplasmic, and membrane-associated proteins, ATPases, lipoproteins, and various substrates, which are typically recognized by virtue of their extracellular secretion (reviewed in Filloux et al. 2008). Secreted substrates lack recognizable signal sequences and appear in culture supernatant as unprocessed polypeptides. Inactivation of individual T6SS genes causes the secreted proteins to be trapped in either the bacterial cytoplasm or periplasm, depending on the mutation (Raskin et al. 2006). One of the curious characteristics of the T6SS is that some of the proteins required for the function of the apparatus are also extracellularly secreted by the system (Mougous et al. 2006; Raskin et al. 2006).
Several T6SS proteins show either structural or sequence similarity to phage tail proteins (Leiman et al. 2009). In particular, T6SS contains an ortholog of the tail tube protein called Hcp (the hemolysin-coregulated protein), an ortholog of the gp5–gp27 complex called VgrG (valine-glycine repeat protein G), and a T4 gp25-like protein. Furthermore, two T6SS proteins with MWs of ∼19 and ∼55 kDa form a structure virtually identical to T4 polysheath in low-resolution electron microscopy images. This structure can be disassembled by the T6SS-encoded AAA+ ATPase in the presence of ATP and Mg ions. The presence of Hcp and VgrG in the external medium is the hallmark of active T6SS. VgrG often carries the “effector” or the pathogenicity domain at its C-terminus. Using T6SS, bacteria are able to deliver this large (1,000+ residues) trimeric protein directly into the cytoplasm of the host cell. Considering the information outlined above, it was proposed that T6SS forms a large phage tail-like structure, which is able to extend from the cell surface to attack the host cell. Furthermore, this complex uses a contractile tail-like mechanism for translocation of large molecules, such as a VgrG trimer, into the host-cell cytoplasm.
There are several features of T6SS, which set it apart from contractile phage tails. First, only three of the five baseplate proteins listed in Table 5.3 are found in T6SS. The orthologs of T4 gp6 and gp53 (or P2 gpI) are not found in the T6SS gene cluster. Second, there is no tape measure protein. Third, T6SS sheath is formed by two proteins. Fourth, phage tail contraction is a one-time irreversible event, but T6SS sheath must act continuously, and thus, it must be able to reset itself.
Nevertheless, T6SS is clearly related to phage contractile tails (Leiman et al. 2009). Because the structure of T6SS machine is unknown, it is very difficult to create a description of its mechanism, which would explain all the peculiarities of its structure and function.
5.3 Photorhabdus Virulence Cassette and Rhapidosomes
The cytotoxic activity toward eukaryotic cells by Serratia entomophila and Photorhabdus species is attributed to the expression of a set of genes from the so-called PVC (Hurst et al. 2004; Yang et al. 2006; Hurst et al. 2007). These genes assemble into structures resembling R-type pyocin particles. Bioinformatic analysis of the PCV gene cluster shows that it contains two orthologs of the tail tube protein or Hcp (afp1 and afp5), three sheath orthologs (afp2, afp3, and afp4), a protein with a glycosidase activity (afp7), the T4 gp5–gp27 complex ortholog (afp8), a T4 gp25-like protein (afp9), a T4 gp6 ortholog (afp11), and an AAA+ ATPase (afp15). The PVC gene cluster appears to represent a link between the pyocins and T6SS. On the one hand, it contains several tube and sheath proteins and the T6SS-like ATPase, but on the other hand, PVC appears to contain the complete minimal baseplate and, most importantly, is secreted into the medium, a property, which has never been seen for T6SS, but instead is a common feature of R-type pyocins and phages. Thus, the PVC is likely a pyocin-like structure, adapted to kill eukaryotic cells in a similar one-off stroke.
The function and structure of rhapidosomes are quite unclear due to limited data (Yamamoto 1967). Rhapidosomes appear to represent pyocin-like structures produced by Photobacterium, Proteus, and Saprospira. Apparently, many environmental isolates use such objects to inhibit growth of other strains or closely related species in their ecological niche.
6 Conclusion
The analysis presented in this chapter shows that the presence of just two proteins, orthologs of T4 gp25 and gp27, in a phage-like cluster of genes is a required and sufficient condition to define this cluster as a contractile tail relative. Orthologs of T4 gp25, but not the sheath or tube proteins, are better conserved and can often be detected using sequence alignment alone. For example, T4 gp25 and P2 gpW are the only two proteins from T4 and P2 tails with a sequence identity of 26%, which is slightly above the “gray area” threshold (Haggard-Ljungquist et al. 1995). The orthologs of T4 gp27 are more diverse, but because of their characteristic and unique secondary structure and domain organization, they can be identified with help of HMM algorithms (Soding 2005; Soding et al. 2005) with a high degree of confidence even if they display no significant sequence identity to known gp27 orthologs.
Contractile tail-like systems are widespread in the biosphere. Remarkably, the contractile tail-like secretion systems can penetrate through both, the bacterial and eukaryotic cell envelopes, suggesting that the underlying principle of creating a membrane channel for subsequent protein/DNA translocation must be universal for bacteria and eukaryotes. However, the structure of many proteins, which are involved in creating this channel, and their fate during membrane penetration remain unknown at present.
References
Abuladze NK, Gingery M, Tsai J, Eiserling FA (1994) Tail length determination in bacteriophage T4. Virology 199:301–310
Ackermann HW (2003) Bacteriophage observations and evolution. Res Microbiol 154:245–251
Aksyuk AA, Leiman PG, Kurochkina LP, Shneider MM, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG (2009a) The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J 28:821–829
Aksyuk AA, Leiman PG, Shneider MM, Mesyanzhinov VV, Rossmann MG (2009b) The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate. Structure 17:800–808
Arisaka F, Tschopp J, Van Driel R, Engel J (1979) Reassembly of the bacteriophage T4 tail from the core-baseplate and the monomeric sheath protein P18: a co-operative association process. J Mol Biol 132:369–386
Bartual SG, Otero JM, Garcia-Doval C, Llamas-Saiz AL, Kahn R, Fox GC, van Raaij MJ (2010) Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc Natl Acad Sci USA 107:20287–20292
Boulanger P, Jacquot P, Plancon L, Chami M, Engel A, Parquet C, Herbeuval C, Letellier L (2008) Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J Biol Chem 283:13556–13564
Coombs DH, Arisaka F (1994) T4 tail structure and function. In: Karam JD (ed) Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, DC, pp 259–281
Coombs DH, Eiserling FA (1977) Studies on the structure, protein composition and assembly of the neck of bacteriophage T4. J Mol Biol 116:375–405
Filloux A, Hachani A, Bleves S (2008) The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154:1570–1583
Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG (2004) Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci USA 101:6003–6008
Haggard-Ljungquist E, Jacobsen E, Rishovd S, Six EW, Nilssen O, Sunshine MG, Lindqvist BH, Kim KJ, Barreiro V, Koonin EV, Calendar R (1995) Bacteriophage P2: genes involved in baseplate assembly. Virology 213:109–121
Hurst MR, Beard SS, Jackson TA, Jones SM (2007) Isolation and characterization of the Serratia entomophila antifeeding prophage. FEMS Microbiol Lett 270:42–48
Hurst MR, Glare TR, Jackson TA (2004) Cloning Serratia entomophila antifeeding genes – a putative defective prophage active against the grass grub Costelytra zealandica. J Bacteriol 186:5116–5128
Jacob F (1954) Biosynthèse induite et mode d’action d’une pyocin, antibiotique de Pseudomonas pyocyanea. Ann Inst Pasteur (Paris) 86:149–160
Kahn ML, Ziermann R, Deho G, Ow DW, Sunshine MG, Calendar R (1991) Bacteriophage P2 and P4. Methods Enzymol 204:264–280
Kanamaru S, Ishiwata Y, Suzuki T, Rossmann MG, Arisaka F (2005) Control of bacteriophage T4 tail lysozyme activity during the infection process. J Mol Biol 346:1013–1020
Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG (2002) Structure of the cell-puncturing device of bacteriophage T4. Nature 415:553–557
Kikuchi Y, King J (1975a) Assembly of the tail of bacteriophage T4. J Supramol Struct 3:24–38
Kikuchi Y, King J (1975b) Genetic control of bacteriophage T4 baseplate morphogenesis. I. Sequential assembly of the major precursor, in vivo and in vitro. J Mol Biol 99:645–672
Kikuchi Y, King J (1975c) Genetic control of bacteriophage T4 baseplate morphogenesis. II. Mutants unable to form the central part of the baseplate. J Mol Biol 99:673–694
Kikuchi Y, King J (1975d) Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J Mol Biol 99:695–716
King J (1968) Assembly of the tail of bacteriophage T4. J Mol Biol 32:231–262
Kohler T, Donner V, van Delden C (2010) Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J Bacteriol 192:1921–1928
Kondou Y, Kitazawa D, Takeda S, Tsuchiya Y, Yamashita E, Mizuguchi M, Kawano K, Tsukihara T (2005) Structure of the central hub of bacteriophage Mu baseplate determined by X-ray crystallography of gp44. J Mol Biol 352:976–985
Kostyuchenko VA, Chipman PR, Leiman PG, Arisaka F, Mesyanzhinov VV, Rossmann MG (2005) The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol 12:810–813
Kostyuchenko VA, Leiman PG, Chipman PR, Kanamaru S, van Raaij MJ, Arisaka F, Mesyanzhinov VV, Rossmann MG (2003) Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol 10:688–693
Kuznetsova TA, Efimov AV, Aijrich LG, Kireeva IY, Marusich EI, Cappuccinelli P, Fiori P, Rappelli P, Kurochkina LP, Poglazov BF, Mesyanzhinov VV (1998) Properties of recombinant bacteriophage T4 tail sheath protein and its deletion fragments. Biochemistry (Mosc) 63:702–709
Leblanc C, Caumont-Sarcos A, Comeau AM, Krisch HM (2009) Isolation and genomic characterization of the first phage infecting Iodobacteria: phi PLPE, a myovirus having a novel set of features. Environ Microbiol Rep 1:499–509
Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG (2010) Morphogenesis of the T4 tail and tail fibers. Virol J 7:355
Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA 106:4154–4159
Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Muhlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ (2007) The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J Mol Biol 371:836–849
Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG (2004) Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118:419–429
Matsui H, Sano Y, Ishihara H, Shinomiya T (1993) Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J Bacteriol 175:1257–1263
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16:404–405
Moody MF (1967a) Structure of the sheath of bacteriophage T4. I. Structure of the contracted sheath and polysheath. J Mol Biol 25:167–200
Moody MF (1967b) Structure of the sheath of bacteriophage T4. II. Rearrangement of the sheath subunits during contraction. J Mol Biol 25:201–208
Moody MF (1973) Sheath of bacteriophage T4. 3. Contraction mechanism deduced from partially contracted sheaths. J Mol Biol 80:613–635
Mosig G, Lin GW, Franklin J, Fan WH (1989) Functional relationships and structural determinants of two bacteriophage T4 lysozymes: a soluble (gene e) and a baseplate-associated (gene 5) protein. New Biol 1:171–179
Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530
Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T (2000) The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38:213–231
Piuri M, Hatfull GF (2006) A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol Microbiol 62:1569–1585
Poglazov BF, Efimov AV, Marco S, Carrascosa J, Kuznetsova TA, Aijrich LG, Kurochkina LP, Mesyanzhinov VV (1999) Polymerization of bacteriophage T4 tail sheath protein mutants truncated at the C-termini. J Struct Biol 127:224–230
Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA 103:1528–1533
Raskin DM, Seshadri R, Pukatzki SU, Mekalanos JJ (2006) Bacterial genomics and pathogen evolution. Cell 124:703–714
Records AR (2011) The type VI secretion system: a multi-purpose delivery system with a phage-like machinery. Mol Plant Microbe Interact 24(7):751–757
Riede I, Degen M, Henning U (1985) The receptor specificity of bacteriophages can be determined by a tail fiber modifying protein. EMBO J 4:2343–2346
Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG (2004) The bacteriophage T4 DNA injection machine. Curr Opin Struct Biol 14:171–180
Scholl D, Cooley M, Williams SR, Gebhart D, Martin D, Bates A, Mandrell R (2009) An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the food-borne pathogen Escherichia coli O157:H7. Antimicrob Agents Chemother 53:3074–3080
Sciara G, Bebeacua C, Bron P, Tremblay D, Ortiz-Lombardia M, Lichiere J, van Heel M, Campanacci V, Moineau S, Cambillau C (2010) Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc Natl Acad Sci USA 107:6852–6857
Serwer P, Hayes SJ, Thomas JA, Hardies SC (2007) Propagating the missing bacteriophages: a large bacteriophage in a new class. Virol J 4:21
Simon LD, Anderson TF (1967a) The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. I. Attachment and penetration. Virology 32:279–297
Simon LD, Anderson TF (1967b) The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. II. Structure and function of the baseplate. Virology 32:298–305
Soding J (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960
Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248
Steven AC, Trus BL, Maizel JV, Unser M, Parry DA, Wall JS, Hainfeld JF, Studier FW (1988) Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol 200:351–365
Tetart F, Desplats C, Krisch HM (1998) Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: recombination between conserved motifs swaps adhesin specificity. J Mol Biol 282:543–556
Tetart F, Repoila F, Monod C, Krisch HM (1996) Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J Mol Biol 258:726–731
Thomassen E, Gielen G, Schutz M, Schoehn G, Abrahams JP, Miller S, van Raaij MJ (2003) The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. J Mol Biol 331:361–373
Tschopp J, Arisaka F, van Driel R, Engel J (1979) Purification, characterization and reassembly of the bacteriophage T4D tail sheath protein P18. J Mol Biol 128:247–258
van Raaij MJ, Schoehn G, Burda MR, Miller S (2001) Crystal structure of a heat and protease-stable part of the bacteriophage T4 short tail fibre. J Mol Biol 314:1137–1146
Walter M, Fiedler C, Grassl R, Biebl M, Rachel R, Hermo-Parrado XL, Llamas-Saiz AL, Seckler R, Miller S, van Raaij MJ (2008) Structure of the receptor-binding protein of bacteriophage det7: a podoviral tail spike in a myovirus. J Virol 82:2265–2273
Williams SR, Gebhart D, Martin DW, Scholl D (2008) Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol 74:3868–3876
Yamamoto T (1967) Presence of Rhapidosomes in various species of bacteria and their morphological characteristics. J Bacteriol 94:1746–1756
Yang G, Dowling AJ, Gerike U, ffrench-Constant RH, Waterfield NR (2006) Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J Bacteriol 188:2254–2261
Acknowledgments
We would like to express our sincere thank you to Prof. Michael Rossmann and Prof. Venigalla Rao, the editors of this book, for choosing the perfect timing in organizing this volume. This book was a unique opportunity for many scientists to summarize and publish the information, which was understood by many in the field because it was presented at conferences, but has never been published. We are very grateful to Prof. Rao for his great efforts in persuading the contributors to produce interesting chapters and, at the same time, keeping the entire project to a reasonable deadline.
Discussions with Prof. Alan Davidson, Prof. Ian Molineux, and Dr. Anastasia Aksyuk were extremely useful in preparation of this manuscript. We are very grateful to Prof. Liang Tong for sharing the coordinates of one of the sheath proteins prior to the publication of the paper describing the crystal structure.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Leiman, P.G., Shneider, M.M. (2012). Contractile Tail Machines of Bacteriophages. In: Rossmann, M., Rao, V. (eds) Viral Molecular Machines. Advances in Experimental Medicine and Biology, vol 726. Springer, Boston, MA. https://doi.org/10.1007/978-1-4614-0980-9_5
Download citation
DOI: https://doi.org/10.1007/978-1-4614-0980-9_5
Published:
Publisher Name: Springer, Boston, MA
Print ISBN: 978-1-4614-0979-3
Online ISBN: 978-1-4614-0980-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)