Abstract
Large, tailed dsDNA-containing bacteriophage genomes are packaged to a conserved and high density (∼500 mg/ml), generally in ∼2.5-nm, duplex-to-duplex, spaced, organized DNA shells within icosahedral capsids. Phages with these condensate properties, however, differ markedly in their inner capsid structures: (1) those with a naked condensed DNA, (2) those with many dispersed unstructured proteins embedded within the DNA, (3) those with a small number of localized proteins, and (4) those with a reduced or DNA-free internal protein structure of substantial volume. The DNA is translocated and condensed by a high-force ATPase motor into a procapsid already containing the proteins that are to be ejected together with the DNA into the infected host. The condensed genome structure of a single-phage type is unlikely to be precisely determined and can change without loss of function to fit an altered capsid size or internal structure. Although no such single-phage condensed genome structure is known exactly, it is known that a single general structure is unlikely to apply to all such phages.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Viral genomes are enclosed within a capsid for protection, transfer, and infection of host cells. Most well-studied viruses with capsids of icosahedral, or similar, symmetry have genomes composed of either DNA or RNA [either single-stranded (ss) or double-stranded (ds) in both cases] that range in length from <4 to >1,200 kb. In some cases, the genome may consist of more than one nucleic acid segment. The dimensions of icosahedral capsids vary dramatically to accommodate such variations in nucleic acid content and may, in some viruses, be elongated, and thus depart from strict icosahedral symmetry. Despite wide variations in genome composition, there are numbers of virus components that are structurally conserved across the phylogenetic spectrum (Koonin et al. 2006). Notably, the translocation of DNA into an empty immature capsid (the procapsid) by a viral DNA-packaging machine is an ancient invention that is found in all kingdoms, and its mechanism appears to be highly conserved among many DNA viruses, including Herpes and related viruses (Salmon and Baines 1998; Sheaffer et al. 2001), as well as among most tailed dsDNA bacteriophages.
In most large (greater than ∼10 kb) dsDNA viruses/phages, ATP-driven high force-generating molecular motors package the genome to high density within procapsids by a broadly conserved molecular mechanism (Smith et al. 2001). Generally, these molecular motors are composed of two main components, a portal-containing procapsid complexed with a multimeric ATPase enzyme, the terminase (Black 1989; Johnson and Chiu 2007; Rao and Feiss 2008; Rao and Black 2010). The portal is situated at a unique capsid vertex and contains a channel, through which DNA enters into the procapsid, and through which DNA and protein are ejected out of the capsid, via the tail (attached to the portal) into the host. DNA is often packaged in conjunction with formation of the mature genome ends by nucleolytic cleavage from a replicative concatemer (hence the name terminase) within bacteriophage or Herpes infected cells; in phages packaging is by a two component terminase, or as a protein terminated linear dsDNA by a single protein-component-packaging ATPase (e.g., Phi29). The terminase protein(s) in most phages/viruses dissociate from the virion after packaging and are not part of the mature virion (i.e., are not structural proteins). In contrast to this, and illustrating the amazing ability of viruses to create variations upon a theme, is the tailless phage PRD1 and its relatives, in which an icosahedral protein capsid encloses a lipid bilayer membrane packed with dsDNA. The packaging proteins of these dsDNA phages are permanently incorporated into a special vertex of the capsid and utilize this ATP driven motor to achieve a packaging rate and local packing density comparable to that of the tailed dsDNA phages (Gowen et al. 2003; Karhu et al. 2007). There are, of course, other icosahedral capsid phages with quite different packaging strategies that are often found to contain less densely packed genomes; e.g., circular supercoiled dsDNA containing PMS2; single-stranded DNA phage PhiX174; three-segment dsRNA containing outer lipid bilayer phage Phi6; not to mention filamentous phages and numerous others; however, phages of these families, which are very different from the tailed dsDNA phages utilizing molecular motors to densely fill procapsids with linear dsDNA, will not be reviewed here.
Most, if not all, tailed dsDNA containing bacteriophage genomes are highly condensed by the ATP-driven terminase complex to a remarkably characteristic and conserved concentration of ∼500 mg/ml, near to a liquid crystalline DNA state. Phage DNA packaged within the icosahedral capsid is thus at a concentration at least fivefold higher than found in metaphase chromatin (Cerritelli et al. 1997). Capsid DNA condensates can be both packaged within the infected cell, and unpackaged upon delivery to infected bacteria, within several minutes or less. Nevertheless, despite such high mobility, the metabolically inactive condensed genomes are stable, infectious, and deliverable from the capsid after many years of storage (Ackermann et al. 2004).
The similar structural and functional properties of condensed genomes found among tailed dsDNA bacteriophages have been the subject of numerous reviews and structural models (e.g., Earnshaw and Harrison 1977; Black 1989). However, it has only recently been appreciated that the comparable condensed genomic DNA structures are actually found among tailed bacteriophages with fundamentally different inner capsid environments. Phages can be broadly categorized based on these inner capsid differences: (1) those containing only a naked condensed DNA within the capsid with few or no internal proteins (e.g., lambda, HK97); (2) those with more than a thousand dispersed unstructured proteins embedded within the DNA (e.g., T4 and other T-evens); (3) those with a small number of localized proteins (e.g., Phi29); and (4) those with a reduced, or DNA-free, internal structure composed of more than one type of protein that has a defined shape and volume and occupies an appreciable fraction of the total capsid volume (e.g., T7, PhiKZ). Examples of these capsid structure types can be seen in Fig. 21.1. As noted, despite such significant and fundamental internal structural differences, the degree of DNA compaction found among all four types of phages is comparable in the condensed DNA-containing portions in the capsid interior. Studies have shown that the capsid internal proteins, whether structured or unstructured, found together with the condensed DNA are an internal part of the DNA-free precursor procapsid and associate with the DNA only following packaging rather than being transferred into the procapsid together with the DNA. Indeed, proteins with high DNA-binding affinities found bound to the metabolically active cellular DNA are in fact actively excluded from being packaged together with the DNA. In all four types of bacteriophages, based on the T4 precedent, the capsid also most likely contains many small-molecule DNA counterions. These small molecules taken up into the procapsid can vary in composition due to mutational changes to the host which prevent normal polyamine synthesis but allow viral packaging with divalent cations, apparently without affecting the packaging or the overall packaged DNA structure (see Black 1989 for early references).
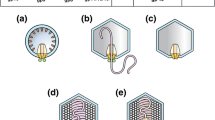
Examples of different internal capsid arrangements found in tailed dsDNA bacteriophages. Tails on capsids are not shown. Portal complex depicted by purple; 2.5-nm spaced DNA depicted by shades of blue. (a) lambda, HK97, and Phi29 have only a “naked” condensed DNA within their capsids; (b) the T4 capsid contains more than a thousand dispersed internal proteins (yellow) embedded within its DNA, whose copy numbers vary relative to capsid dimensions, as well as several localized proteins whose copy numbers remain unchanged in different-sized capsids (red), such as icosahedral capsids (T4i) or giant capsids (T4g); (c) the T7 capsid has an inner core composed of three different proteins (yellow); and (d) the PhiKZ capsid contains an extremely large inner protein body (purple)
In this review, emphasis will be placed on recent advances and discoveries made among these four classes of highly condensed genomes found in tailed bacteriophages, especially among the more recently discovered “environmental” phages. Experimental work bearing on structure and function will be emphasized over theoretical structural and biophysical modeling of the condensed DNA. How modeling of condensed capsid DNA structure relates to DNA packaging force and velocity, and to DNA ejection from the capsid have been discussed recently (Fuller et al. 2007; Rickgauer et al. 2008).
2 Naked DNA Condensate Structure in Phage Capsids
The exact three-dimensional structural properties of the packaged double-stranded DNA in bacteriophage capsids remain controversial. The packaged condensate structure found among the four different types of phage capsids shown in Fig. 21.1 displays an ordered ∼2.5-nm duplex-to-duplex DNA spacing that arises from the broadly conserved packaging motor mechanisms (Earnshaw and Casjens 1980). A number of models have been proposed to account for such broadly conserved structural features of these capsid condensates in which DNA is sometimes determined to be organized in concentric shells by X-ray scattering and cryo-EM (summarized in Mullaney and Black 1998; Comolli et al. 2008). However, definitive evidence to choose among these structural models (Fig. 21.2) appears to be lacking. In fact, despite some generally similar features such as the degree of packing density and the ordered 2.5-nm duplex spacings found among DNAs organized in shells, at least toward the periphery of the various phage capsids, other structural, chemical, and biological features of packaged DNA condensates found among a number of phages argue against a single unitary and deterministic packaged capsid DNA structure. Significant differences found among a number of packaged phage capsids are the following: Depending upon the phage capsid, DNA is oriented transverse (T7) (Cerritelli et al. 1997) or more nearly parallel (T4) (Earnshaw et al. 1978; Lepault et al. 1987) to the head-tail axis of the phage particle; the first DNA end packaged is the last end out (lambda, T7) (Sternberg and Weisberg 1975; Hartman et al. 1979) or the first end packaged is the first end out (T4) (Black and Silverman 1978), and is packaged to a comparable 500 mg/ml density despite being found in the capsid as a protein-free condensate or together with unstructured or DNA-free protein structures (Cerritelli et al. 1997; Mullaney and Black 1998). Moreover, functional DNA-containing “giant” phage capsids (Earnshaw et al. 1978) are packed with DNA to the same 2.5-nm spacing and final DNA density as normal capsids. In these giant phages, the DNA is clearly oriented parallel to the extended long axis of the giant capsids that can be more than tenfold longer than wild-type T4, while maintaining the same width as normal capsids. However, a phage with the unusual highly positively charged base a-putrescinylthymine packages DNA to ∼630 mg/ml, except when this modification is removed, showing that the DNA structure and charge critically determine packaging density (Scraba et al. 1983). Phages with unique sequence ends such as lambda significantly under-package deletion containing genomes of ∼75% normal length to yield active phages with looser than 2.5-nm of duplex-to-duplex overall spacing. On the other hand, headful packaging phages such as T4, SPP1, or P22 that lack a sequence-specific terminase cutting site package DNA to a conserved ∼500 mg/ml density irrespective of deletions because a headful cutting signal is communicated to the high-force packaging motor-terminase complex by the highly conserved portal DNA measuring device. Overall, functional fully packaged giant, more densely packaged positively charged base-containing DNA, and underpackaged short-genome phage capsids demonstrate how variable the capsid-condensate DNA structure can be among different phages.
Different models of packaged DNA structure. (a) The spool model containing longitudinally packaged DNA segments (Earnshaw et al. 1978); (b) spool or concentric shell models (Earnshaw and Harrison 1977; Cerritelli et al. 1997); (c) spiral-fold model (Black et al. 1985); (d) liquid crystal and hexagonal packing models (Lepault et al. 1987; Leforestier and Livolant 2010); (e) icosahedral-bend model which is the same as in (a) but sharp bends have been incorporated to follow the angular contours of the icosahedrons (Mullaney and Black 1998); (f) a model where the DNA forms 12 icosahedrally arranged pear-shaped rings was proposed for the large Bacillus phage G (Sun and Serwer 1997). This model is based on that proposed for lambda DNA (Witkiewicz and Schweiger 1985); (g) model for completion of genome packaging into shells in Phi29 based on a Monte Carlo simulation (Comolli et al. 2008); and (h) model for genome packaging with extensively knotted DNA that may arise following packaging (Marenduzzo et al. 2009). Nb. Capsid shell is not indicated in (b, f, g, and h)
DNA condensed within capsids of diverse size is often visualized by cryo-EM to be highly organized within shells with apparently gentle overall curvature in outer shells (Effantin et al. 2006; Xiang et al. 2006; Duda et al. 2009). An unresolved question is whether the DNA in such shells is uniformly bent in the wide outer shells even where contact is made with capsid angular icosahedral edges (e.g., Fig. 21.2e). Additional discontinuities in bending are expected in the inner shells near to the center of the icosahedron because packed DNA is observed to have essentially equal density in these portions of the capsid (Leforestier and Livolant 2010). This requires that DNA should be sharply bent below its persistence length in these regions. Overall, it appears uncertain whether DNA is arranged largely or exclusively with gentle bending (Fig. 21.2a, b), or with discontinuities – less gentle bends (Fig. 21.2e–g) or sharp kinks (Fig. 21.2c, d) – throughout the structure or only in those regions of the structure that apparently require this. Those DNA condensate models shown in Fig. 21.2 a–c, e, and g, whether or not they contain bends or kinks, would display DNA shells with generally similar overall structures in averaged cryo-EM reconstructions. It should be noted that DNA will readily undergo bending or kinking under certain constraints or interactions, even in the absence of high motor force, such as possibly with DNA-binding internal protein components of the capsid (see below) or domains of the main head protein itself which protrude into the capsid interior, as seen in the mature N4 capsid (Choi et al. 2008). Similarly, the T7 capsid apparently imparts angularity in portions of the outermost DNA shell (Agirrezabala et al. 2005).
Also, because of the highly energetic and rapid (up to 2,000 bp/s) packaging motor, it probably cannot be assumed a priori that minimum energy structures within the DNA must predominate in determining its structure. For example, it is frequently assumed in modeling (and other) studies that DNA is condensed within the capsid from outside to inside (Jiang et al. 2006; Petrov et al. 2007). However, in fact experimental evidence including ion etching of the intact fully packaged particle apparently establishes the opposite direction of packing condensation for phages T4 and lambda (Black and Silverman 1978; Black et al. 1985; Mendelson et al. 1992). EM observations of packaging intermediates in a number of phages also apparently show that the DNA is not initially preferentially packaged against the inner surface of the capsid, e.g., most recently in T3 (Fang et al. 2008).
It is unlikely that cryo-EM or other techniques presently have the resolution to resolve these questions of condensed DNA fine structure that requires that the entire length of an individual packaged duplex is resolved structurally at the nucleotide level. A number of techniques including Raman spectroscopy and solid-state NMR show that the condensed DNA is predominantly B form (Overman et al. 1998; Yu and Schaefer 2008). At the same time, evidence for some departure from uniform B-form structure comes from chemical evidence for kinks in the condensed DNA, in addition to known condensation below persistence length (see Black et al. 1985; Serwer 1986). Moreover, additional evidence against uniform B form comes from the observed hydrolysis of packaged T4 DNA in active phage particles by packaged staphylococcal nuclease whose enzymatic specificity is incompatible with B-form DNA cutting (see below). These experiments also show that many or a few molecules of the packaged nuclease can diffuse within the fully packaged DNA – condensed to ∼500 mg/ml DNA contains 75–85% (vol/vol) water – to reduce it more or less rapidly, varying with the copy number, to ∼160 bp limit digest lengths. Cryo-EM observations of such limit phage T4 head digests, however, show relatively small effects on DNA condensate structure. The observations also show the T4 DNA to be oriented differently from T7 in an elliptical coaxial spool whose long axes are oriented at approximately 30° tilt to the portal axis (N. Cheng, B. Mai, M.E. Cerritelli, L.W. Black, and A.C. Steven, personal communication).
A number of other fundamental properties of packaged DNA have recently been determined. It was known that one unique end of the packaged phage DNA descends through the portal to be injected into the infected host. Such a DNA duplex descending through the portal has recently been imaged by cryo-EM; the structure of the portal proximal DNA can also be visualized, and packaging is seen to change the portal structure (see below). Several studies had shown by classical electron microscopy that a single end is attached to the phage lambda (and other) tail(s) following disruption, thereby demonstrating that following packaging, but before ejection, DNA rearranges and descends through the portal and into the tail (Chattoraj and Inman 1974; Saigo and Uchida 1974; Thomas 1974; Saigo 1975). Recently, it has also been demonstrated by fluorescence correlation spectroscopy-Förster resonance energy transfer (FCS-FRET) and single-molecule FRET (smFRET) analysis of in vitro packaged Förster dye pair terminated 5- and 50-kb DNAs that the two DNA ends packaged into phage T4 capsids are held 80–90 Å apart; apparently both are within the portal channel of the T4 capsid (Ray et al. 2010). This structure suggests that a loop rather than an end of DNA is translocated by the T4 motor complex. Other less direct evidence (knotting and ion etching of DNA) is consistent with comparable colocalization of the two packaged DNA ends in other phages. However, colocalization of both packaged DNA ends such as in the phage T4 capsid also appears to place constraints on comparably packaged capsid DNA structures. How can this be reconciled with extensively knotted DNA derived from phage P4 capsids (Arsuaga et al. 2002)? DNA modeling suggests that the many knots found within DNA originating from the packaged phage P4 capsid would be delocalized and not interfere with DNA ejection from the capsid (see Fig. 21.2h) (Marenduzzo et al. 2009). However, if both P4 DNA ends are also anchored at the portal, it is not clear how extensive topological knotting could occur within the capsid; rather the knots may more likely arise following release and rearrangement of the condensate, or at least of its ends, from the native capsid structure.
3 Phage Capsids with DNA Condensed Together with Internal Proteins
In addition to the condensed genome, numbers of phage capsids contain a diversity of internal proteins, despite, as mentioned above, their DNA retaining conserved features, including a density of ∼500 mg/ml and displaying the characteristic 2.5-nm duplex-to-duplex spacing. These proteins vary in length, copy number, function, and localization within the capsid, illustrating not only the remarkable diversity that occurs between different phages, but also highlighting the fact that genome packaging occurs successfully despite numerous internal capsid conditions/states.
3.1 Phage Capsids with DNA Condensed Together with Dispersed Unstructured Proteins
The most intensively studied phage capsid internal proteins (IPs) are those of phage T4 and relatives. In T4, more than 1,000 molecules of three different small (∼8–20 kDa) proteins are packaged into DNA-free proheads by a capsid-targeting sequence (CTS) mechanism (Mullaney and Black 1996) (Fig. 21.3). IP incorporation into the procapsid is based on the affinity of a highly conserved consensus sequence found at the N-terminus of each IP (the CTS) that is responsible for incorporation of these proteins into the assembly and structure determining core of the procapsid. The affinity of the CTS for the core is such that it can be exploited to package diverse, nonphage proteins into the capsid (e.g., Fig. 21.3b, ii, iii, and iv). Processing of the assembly core by the morphogenetic protease (gp21) converts the essential shape-determining core protein (gp22) to small peptides, most of which exit the capsid, although a few of which are also retained within the mature capsid. Gp21 also modifies all of the IPs by removal of the 10–19-residue-long N-terminal CTS peptides. The internal proteins are unstructured and dispersed within the DNA condensate as shown by the ability of IP-nuclease fusion proteins to diffuse within and cut all the condensed DNA (Fig. 21.3c), IP protection from ion etching by the DNA (Black et al. 1985), and IP release and sedimentation together with packaged DNA released in condensed form by dissolution of the capsid (Zachary and Black 1991).
(a) Overview of the packaging of proteins by the T4 capsid targeting sequence (CTS) system: (i) capsid-size prohead core, minus the outer shell and DNA, is seen to assemble in vivo; (ii) completion of the capsid shell triggers cleavage of prohead proteins; (iii) small fragments of processed prohead proteins (including scaffold protein, CTS sequences, and protease) exit the capsid presumably via small holes in the shell prior to maturation–expansion; (iv) phage DNA is packaged into the capsid by the terminase; (v) mature T4 head. (b) Examples of proteins packaged into the mature T4 phage capsid: (i) structure of restriction endonuclease inhibitor IPI* (9 kDa) (the processed form of IPI) found in wild-type T4; examples of proteins packaged into the phage capsid using the phage-derived expression, packaging, and processing (PEPP); (ii) SN nuclease (15 kDa) (Chen et al. 2000); (iii) GFP (27 kDa) (3ADF) (Ebisawa et al. 2010); (iv) β-galactosidase (540-kDa tetramer) (1DP0) (Juers et al. 2000) (Nb. Structures not to scale). (c) Genomic DNA of recombinant T4 phage digested by SN nuclease within the capsid. Lane 1, lambda HindIII standard; lane 2, control DNA from recombinant T4[CTS▼IPIII▼SN], not treated with Ca2+; lanes 3–7, DNA from recombinant T4[CTS▼IPIII▼SN] incubated with Ca2+ for 1, 10, 30, 120 min, and 16 h, respectively (▼refers to a T4 protease processing site). Image reproduced from Mullaney and Black (1998)
Thus, it appears that these proteins fit within the conserved DNA condensate structure without loss of function. In addition, all three IPs can be eliminated from the capsid by mutation with only minor effects on procapsid assembly and shape and without significantly affecting the amount of DNA packaged within the capsid. Packaging of each IP is independent of the presence of the two other IPs, and in giant particles, the IP copy numbers per capsid increase with the available volume, presumably because the interior of the giant procapsid is seen to be filled with a giant scaffolding-core (composed of gp22) that also contains increased numbers of IPs (see Fig. 3a in Black et al. 1993).
The three internal proteins of T4 all have a net positive charge (all have predicted pI > 9) and are lysine rich relative to other T4 proteins, properties that might enhance their interaction with DNA. Solid-state NMR showed that there are specific IP-DNA contacts, and that these proteins could neutralize some (<5%) of the DNA charge balance (Yu and Schaefer 2008). Hence, this together with the fact that the small internal proteins are dispensable (Black et al. 1993) implies that their presence in the capsid is not essential for the stability of packaged DNA. This is supported by the fact that in other T-even phages the IPs are highly polymorphic and some of the internal proteins packaged by the CTS mechanism are small acidic or neutral proteins (Black et al. 1993; Repoila et al. 1994). Potentially, the main function of the IPs is to interact with the host in a manner that will enhance infection, as the T4 IPs are all injected into the host cell. Supporting this suggestion is the fact that one of them, IPI, protects the T4 DNA against a highly specific DNA modification dependent host restriction enzyme (Bair et al. 2007). Conditions or hosts in which the abundant IPII and IPIII are essential have not been found. The dimensions of IPI suggest it can be ejected through the portal and tail tube without unfolding thereby promoting immediate inhibition of its nuclease target in the infected host (Rifat et al. 2008). However, it appears probable that other such internal proteins (particularly larger ones) are ejected unfolded and are then refolded in the infected host. This is likely the case with E. coli phage P1 that may resemble the T-even phages with respect to packaged unstructured proteins, although little is known about their copy numbers and locations within the capsid. However, it is known that phage P1 injects two large and processed (60 and 200 kDa) anti-restriction endonuclease inhibitor proteins, dar A and dar B, to protect the injected DNA (Iida et al. 1987; Streiff et al. 1987).
That such permutations can occur was demonstrated by the packaging of β-galactosidase via an IPIII-β-galactosidase fusion and its subsequent ejection into, and activity within, the host cell (Fig. 21.3b, iv) (Hong and Black 1993). Physically, it is not feasible that packaged β-galactosidase could exit the capsid in its folded state. In addition, the appearance of the 540-kDa tetramer-dependent activity in the infected cell requires host chaperones. Hence, the ejection of the unfolded, or possibly semi-unfolded, protein must be followed by refolding and multimerization within the host (Hong and Black 1993). Possibly, unfolding of both unnatural and natural internal proteins, such as β-galactosidase and Alt, that are folded and active upon synthesis prior to encapsidation, is a secondary function of the high force-generating packaging motor and the resulting DNA pressure in the full capsid. GFP can also be packaged within viable T4 phage particles as CTS fusions and gradual formation of the fluorophore in mature phage shows the mobility of the encapsidated proteins; the IPIII-GFP fusions in DNA full heads, as opposed to CTS-deposited GFP, have fluorescent properties suggesting IPIII interacts specifically along the DNA to associate GFP monomers within the full head (Mullaney et al. 2000).
3.2 Phage Capsids with DNA Condensed with Localized Proteins
A number of unrelated phages contain proteins within their capsids that are injected into the host cell, ahead, or with, the phage DNA, but in contrast to the T4 IP proteins, these proteins have more specific locales, typically in close proximity to the portal complex. Examples of localized internal proteins are found in E. coli phages N4 and T4, and Bacillus phages Phi29 and SPP1. Despite having similar locales, these localized inner capsid proteins have a remarkably eclectic set of functions and several of them are apparently multifunctional. One localized protein that stands out for its unusualness is the N4 gp50, a virion RNA polymerase (vRNAP), the first RNAP to be identified as being packaged within the capsid of a tailed phage. Gp50 transcribes early phage promoters that are composed of four structural motifs at hairpin/stem structures (Gleghorn et al. 2008). The production of these structures and their binding by the vRNAP is assisted by host proteins (E. coli DNA gyrase and single-stranded DNA-binding protein) (Glucksmann et al. 1992; Gleghorn et al. 2008). The vRNAP is extremely large (∼380 kDa) relative to its mitochondrial and T7-like RNAPs homologues (Cermakian et al. 1996) and comprises three main domains, a central domain responsible for RNAP activity, an N-terminal domain that is required for injection of the first 500 bp of the genome and a C-terminal domain that is required for its incorporation into the head (Kazmierczak et al. 2002). About four copies of the N4 vRNAP are present per virion, and cryo-EM comparison of wild-type versus gp50-minus mutants indicate that the vRNAP is located at the base of an inner core, above the portal in such a manner that at least part of the vRNAP is posited close to the internal entrance to the portal channel (Choi et al. 2008).
In addition to its dispersed high-copy IP proteins, T4 also has several proteins, Alt and gp2, whose positions within the capsid are localized. Alt is a large protein (∼75 kDa) that is packaged into the capsids via its CTS. Alt is responsible for ribosylating host RNAP subunits (and other host proteins), resulting in preferential recognition of phage early promoters over host promoters (Depping et al. 2005). Alt is probably located above, possibly on, the portal complex as supported by the fact that in giant phages the copy number of Alt [about 40 copies (Black et al. 1993)] remains constant despite increases in the copy numbers of the three IP proteins proportional to increases in head size (Aebi et al. 1976; Bijlenga et al. 1976). Some finer details of this process remain to be clarified, such as why does Alt obtain such a very specific location within the head versus the dispersed locations of the IP proteins? Alt has the shortest CTS sequence, although comparable in sequence and also processing by the morphogenetic gp21 protease to the other IPs, leading to the question whether different portions of the Alt protein, other than the CTS sequence, may establish specific affinity for the portal protein.
N4 gp50 and Alt potentially have significant effect on the packaging of DNA and its final structure based on the volume they would occupy in the phage head, and the fact they are within the head prior to DNA packaging. Other proteins with specialized locations within the head potentially have less impact on the overall structure of packaged DNA as they are present in low copy number and/or have low molecular weights. However, their presence is likely to influence the structure of the packaged DNA in specific regions, particularly near or within the portal complex. Notable among such proteins are those that are noncovalently or covalently bound to the genome termini, such as T4 gp2 and Phi29 gp3, respectively. T4 gp2 prevents restriction of the phage genome in the host cell by exonuclease V (Lipinska et al. 1989) and likely also functions as a “genome plug” (see below) within the head as mutations in it produce unstable heads that leak DNA; however, the gp2 protein can be added to the already fully packaged head to complete active and stable phage particles (Wang et al. 2000). Since, as mentioned above, the ends of the T4 genome have been determined to be held 80–90 Å apart, it is likely that each copy of gp2 has a specialized location in close proximity to the portal. The main function of Phi29 gp3 is to act as a primer for DNA replication (Peñalva and Salas 1982; Watabe et al. 1983); however, it may also function as a genome plug (Xiang et al. 2006).
Genome plugs are proteins whose presence prevents the packaged genome from escaping through the portal vertex until the appropriate time for DNA ejection or descent into the tail tube. Genome plug proteins that are incorporated into the head are all likely to have some small effect on the structure of the packaged genome in the region close to where they reside. Due to the essential nature of such protein(s), all tailed phages are likely to have proteins with these functions, but as exemplified by T4 gp2 and Phi29 gp3, there is potentially a diversity of nonhomologous proteins that undertake this function. Conversely, it is also feasible that certain genome plug proteins may be conserved between related and even lesser-related phages. For instance, the minor capsid protein gp7 (NP_690662.1, also referred to as SPP1p011) of siphovirus SPP1 is a good candidate for a genome plug as it is present in one to two copies per virion, potentially injected into the host cell, and exhibits both DNA-binding capacity and affinity with the portal protein (Stiege et al. 2003). Homologues to gp7 exist in a wide diversity of phages (Stiege et al. 2003), as illustrated by its homology to the myovirus Mu gpF super family (cl10072) (e-30). However, for some potential genome plug proteins, such as T7 gp6.7 (9.2 kDa) (Kemp et al. 2005), determination of whether such a protein is widely conserved or not is complicated by their small size, which makes sequence comparisons difficult. It should be noted that the discussion above refers to genome plugs that are probably incorporated into the capsid, as this review focuses on DNA packaging and the factors that most immediately affect it. However, it should be noted that several phages, such as SPP1 and HK97, have neck or connector proteins that form rings under the portal structure which also function as genome plugs (and in these instances are homologous, belonging to large families) (Lhuillier et al. 2009; Cardarelli et al. 2010). Potentially, some phages, such as SPP1, have several proteins, in different locations, that participate in this function.
3.2.1 P22: A Localized Collection of Ejection Proteins?
The Salmonella podovirus P22 also potentially has localized proteins, one or more of which may act as a genome plug. These proteins, gps 7, 16, and 20 (whose genes are actually clustered), are known to be in the mature head and injected into the host cell, hence, they are referred to as pilot/injection or ejection proteins (Chang et al. 2006; Lander et al. 2006). Potentially, these ejection proteins are responsible for densities localized in the central channel of the portal, and/or in the channel of the tail hub (Chang et al. 2006); however, these densities observed by cryo-EM may, at least in part, be also due to the extended, large (12 × ∼80 kDa) portal structure itself whose crown projects well into the interior of the capsid (Tang et al. 2011). The proteins within the P22 head are of particular interest as its genome is packaged as a coaxial spool (Zhang et al. 2000; Chang et al. 2006; Lander et al. 2006), apparently in a similar manner to that of the podoviruses below (e.g., T7) whose heads contain inner cores which are likely to have a major influence on the structure of packaged DNA. However, unlike those phages the P22 capsid apparently does not contain a large core, except for the large portal mentioned. Large portal proteins have been indicated as a cause of coaxial spooling in simulation studies (Petrov et al. 2007). Hence, it will be interesting to determine the impacts of the P22 portal and its ejection proteins on DNA packaging and structure.
3.3 Phage Capsids with Multiprotein Internal Structures
The capsids of some podoviruses and myoviruses have been determined to contain multiprotein core structures within the densely packaged DNA. These structures are roughly cylindrical in shape, positioned above the connector complex, and typically occupy a considerable volume in the central region of the capsid, normally occupied by dsDNA in other phages, such that they are large enough to be visualized by normal electron microscopy. Their presence raises numbers of questions as to their function(s), particularly their impact on packaged DNA structure, and the packaging and ejection processes. Of the known core structures, that of the E. coli podovirus, T7, is a paradigm, illustrating that these structures can be morphologically complex and have functions of major consequence to various stages of the phage lifecycle.
The 510-Å-diameter T7 capsid contains a 265-Å-high and 175-Å-wide inner core that is surrounded by about six coaxial/concentric rings of dsDNA (Cerritelli et al. 2003a; Agirrezabala et al. 2005). The T7 core encloses a channel/cavity (varying in diameter from 35 to 110 Å) that is continuous through to the outer side of the capsid as a consequence of its docking onto the portal complex. In mature capsids, this inner channel contains a density attributed to a terminal segment of DNA (Agirrezabala et al. 2005). The core structure is not a simple cylinder, but is composed of three domains, each with a different symmetry 12-, 8-, and 4-fold (Cerritelli et al. 2003b; Agirrezabala et al. 2005). The 12-fold symmetrical domain most probably represents the connector complex, whereas the other two domains are composed of three proteins gps 14 (21 kDa), 15 (84 kDa), and 16 (144 kDa), present in 10–12, 8, and 4 copies, respectively (Cerritelli et al. 2003a; Agirrezabala et al. 2005; Kemp et al. 2005). The genes encoding these three proteins are clustered, and they are transcribed together (McAllister and Wu 1978).
As the T7 core structure extends so far into the capsid, it is likely to have a significant influence on the packaging of DNA into coaxial spools (Cerritelli et al. 1997), possibly preventing the formation of concentric spools (Petrov et al. 2007). However, the T7 core is not only important for the structure of DNA in the mature head, it most likely acts as a conduit during DNA packaging and/or release (as implied by the presence of DNA within it) and may assist the assembly of proheads into correct dimensions (Agirrezabala et al. 2005). Indeed, evidence supports the core being multifunctional, playing different and complex roles in different parts of the phage life cycle. Its component proteins, known to be essential and injected into the host cell (Garcia and Molineux 1996), have been proposed to create a channel that spans the host cell envelope (i.e., become an extensible tail of sorts), thereby enabling the phage DNA to enter the host cell and overcome the obstacle posed by the short tail of the mature virion which prevents it from reaching the host cytoplasm (Molineux 2001). Support for this includes electron microscopy of partially emptied bacteriophage T7 capsids showing a needle-like extension, about twice the length of the normal T7 tail (Serwer et al. 2008), the N-terminal domain of gp16 has transglycosylase activity (Moak and Molineux 2000; Moak and Molineux 2004), and membrane localization experiments have shown that gp14 localizes to the outer membrane, whereas gp15 and gp16 are found predominantly in both soluble and cytoplasmic membrane-bound forms. Notably, recent studies have demonstrated that a DNA-gp15-gp16 motor is responsible for the translocation of the first 850 bp of the T7 genome into the host cell (Chang et al. 2010a). Another interesting feature of the three T7 core proteins is that their injection into the host implies that they would need to almost completely disassemble to be able to exit the head via the narrow connector channel (Kemp et al. 2005). Hence, the T7 core structure is likely to be intriguing, not only for its morphogenesis, but also for its disaggregation pathway.
Core structures have been identified in other podoviruses, such as Prochlorococcus phage P-SSP7, another member of the subfamily Autographivirinae to which T7 belongs, as well as in podoviruses that are more distantly related to T7 and belong to genera outside of its subfamily, such as Salmonella phage Epsilon15 and E. coli phage N4 (Lavigne et al. 2008). While the cores of these phages are not as well described as the T7 core, there is increasing evidence that they share a number of characteristics. Notably, the DNAs of all these phages, or at least the most outermost strands that have been resolved, have coaxial spools, or hexagonal packing (Leforestier and Livolant 2010) a formation that must most certainly be affected by the presence of the core in each phage. The core structures of these phages are all likely to contain an inner channel in which a terminal segment of DNA resides, either based on the observation of densities interpreted as DNA in Epsilon15 (Jiang et al. 2006) and P-SSP7 (Liu et al. 2010) or the presence of cylindrical structure as for N4. Interestingly, the extreme terminus of the DNA that descends into the portal in P-SSP7 is split into single-stranded DNA (Liu et al. 2010). The core proteins of Epsilon15 and P-SSP7 are also probably injected into the host cell, based on the absence of core density in empty particles (Chang et al. 2010b; Liu et al. 2010). In addition, Epsilon15 particles adsorbed to host cells gain a tubular density in the host periplasm (in conjunction with the disappearance of the core), suggesting that, as in T7, the Epsilon15 core protein(s) form a tunnel through which the phage DNA passes to the host cytoplasm (Chang et al. 2010b).
However, in addition to shared common features with the T7 core, the cores of Epsilon15, P-SSP7, and N4 also display dissimilarities to the T7 core. These differences include size differences, the Epsilon15 core is slightly smaller than that of T7 (200 Å high and about 180 Å) (Jiang et al. 2006), while the N4 core is much smaller (110 Å in height and ∼80 Å in width) (Choi et al. 2008). The Epsilon15 core potentially lacks symmetry (Jiang et al. 2006), and no symmetry could be identified for the P-SSP7 core (Liu et al. 2010), although both these results could be a result of the difficulties of resolving the components of such small structures. The N4 core structure is apparently different from that of T7, and is composed of at least two components, an upper disk-like structure above a lower, longer cylindrical density that sits above a density attributed to the vRNAP (see above) (Choi et al. 2008).
Better comparisons of the cores of these phages with those of T7, and other phages, will be enhanced when the core proteins, and their copy numbers and functions are known. Currently, no protein in any of these three phages has been structurally allocated to the core, although candidates for core proteins exist in Epsilon15 and P-SSP7 based on other considerations. For instance, Epsilon15 gp17 has been named a core protein (Chang et al. 2010b), probably based on it being identified as a component of the phage particle by mass spectrometry (MS) (Jiang et al. 2006), and in an appropriate copy number (7.5 copies) (Kropinski et al. 2007). Epsilon15 gp15 is also a good candidate for a core protein, being identified by MS (Jiang et al. 2006) and present in the virion in about 20 copies (Kropinski et al. 2007). Epsilon gp14 is another potential core candidate having homology to the T7 core protein gp14 using Hidden Markov Modeling (2.5 × 10−12) (our observation). Proteins identified by MS in P-SSP7 (gp15, gp16, and possibly gp14, equating to proteins 33, 34, and 35, respectively) are also good candidates for core components in this phage (Liu et al. 2010). It is also of note that the genes encoding the main candidates for inner core proteins in Epsilon15 and P-SSP7, like T7, are located within the same genome locale, a characteristic of phage virion genes with related functions (such as head genes). Such clustering of genes with related functions enables their transcription, and subsequent translation, to occur at a time appropriate for morphogenesis. Gene clustering may also protect essential structural protein interactions from being lost by recombinational exchanges with related coinfecting bacteriophages genomes. Currently, there is less indication of candidates for the components of the N4 core; however, it also has proteins identified as a component of the virion by MS whose functions are yet to be determined (e.g., gps 51, 52, 54, and 67) (Choi et al. 2008). Similarly, it will need to be determined whether the N4 vRNAP, which sits below the core, is technically a part of the core or not.
Of the proteins known to be components of a core structure in a mature capsid, there is no set of proteins, or even a single protein, that is strongly conserved at the sequence level (such as at the level of conservation seen between the major head proteins or terminase proteins of many tailed phages) between the different types of podoviruses with cores discussed above. This diversity of core proteins suggests that their current functions are also likely to be highly diversified. However, it is possible that some core proteins could share a distant ancestor, but have diverged greatly from that original state. This is reminiscent of scaffold proteins, found within the procapsid but not the mature capsid, between which there is typically little or no indication of a common ancestor at the sequence level in phages of different types. However, the fact that all share a conservation of structure (highly alpha helical) and function, argues otherwise (Steven et al. 2005). Considering the T7 precedent, where each of the core proteins has function(s) individualized to enable/enhance infection of its host, diversity in core proteins could potentially be akin to the diversity seen in phage tail fiber proteins, and in the T-even internal proteins. These proteins are under pressure constantly to adapt to changes in the host to preserve the ability of that phage to infect that host. Whether the core proteins in different phages have different impacts on the packaging, structure, and release of DNA, or whether they retain certain traits or structural elements to keep changes in DNA interactions to a minimum, remains to be determined.
3.3.1 Inner Bodies Found in Giant Myoviruses
A group of Pseudomonas phages, which includes PhiKZ, EL and 201phi2-1, also contains a multiprotein inner structure within their unusually large (T = 27) capsids, based on electron microscopic observations (Krylov et al. 1984; Matsko et al. 2001) (Fig. 21.4). This structure, or inner body, is considerably larger than that of the T7 inner core, being roughly 90-nm in length and 35-nm in diameter in PhiKZ (Krylov et al. 1984). In fact, the volume occupied by this structure is potentially greater than the total capsid volume of some smaller phages. The PhiKZ inner body has a spring- or spool-like appearance and may disappear after phage particles adsorb to bacteria (Krylov et al. 1984) (Fig. 21.4c). However, whether this structure has an inner channel and/or is anchored to the portal complex, as in the T7 core, is yet to be determined. Recently, mass spectrometric studies of PhiKZ and EL (Lecoutere et al. 2009), and 201phi2-1 (Thomas et al. 2008, 2010), have supported earlier indications that these phages were structurally complex, based on the numbers of different proteins identified in their virions (>62). These phages also have an exceptionally large number of different proteins associated with their heads, even in comparison with T4 whose mature capsid is composed of 13 different proteins (Black et al. 1993). The latter observation is based on (1) the identification of 30 proteins in a tailless mutant of PhiKZ (Lecoutere et al. 2009) and (2) that nineteen 201phi2-1 (Thomas et al. 2010) and eighteen PhiKZ (Thomas and Black, unpublished) proteins are processed at a cleavage motif that ends in a glutamic acid residue comparable with the morphogenetic protease processing motif (LxE or IxE) in T4 head proteins. The latter suggests these proteins are processed by a T4-like prohead morphogenic protease, and are therefore head proteins. Overall, in these phages, there are enough different proteins to account for the capsid shell (including vertex and portal structures) as well as protein mass to account for the inner body structure.
The inner body found in the large myovirus, PhiKZ, and its relative 201phi2-1. (a) Electron micrograph of disrupted heads of the PhiKZ mutant ts759. Image reproduced from Krylov et al. (1984); (b) electron micrograph of partially disrupted PhiKZ particles. Arrow indicates the inner body. Image reproduced from Matsko et al. (2001); (c) electron micrograph of a PhiKZ particle disrupted by freeze–thawing. Arrow indicates inner body released from disrupted head. Image reproduced from Krylov et al. (1984); (d) cryo-electron micrograph of 201phi2-1. The large white arrow highlights bubbles which formed in the head apparently around the protein body. Image reproduced from Thomas et al. (2008)
The function(s) of the inner body of PhiKZ, and those of its relatives, is unknown, but potential functions include roles in DNA compaction, packaging and/or injection, or in strengthening of the large capsid (Krylov et al. 1984). Two other potential functions for the inner body are that it acts to regulate capsid size [i.e., analogous to the shape-determining T4 morphogenetic core (or to a tail tape measure protein)] and/or represents a reservoir of proteins that are injected into the host cell to aid the initial steps of infection and replication. A good candidate for a protein with the latter function is the multisubunit vRNAP conserved and packaged within each of these phages (Thomas et al. 2008). There are many unknowns regarding the inner bodies of PhiKZ and its relatives; however, based on the structural and functional complexity of the T7 core, the structure and function(s) of these large inner bodies represent fascinating subjects for further studies. One particularly intriguing aspect to resolve will be if the size of these inner bodies is constant in different members of this group of phages, or whether the size, composition, and even orientation within the capsid varies from phage to phage. Thus far, it appears there are variations, for instance between PhiKZ and EL, because not all structural proteins are identifiable as homologous between these two phages (Lecoutere et al. 2009). Also, it has been observed that the capsids of these two phages have apparently very similar dimensions (Hertveldt et al. 2005), whereas their genome lengths vary by nearly 70 kb. Since the density of packaged DNA appears to be similar in these phages, a larger inner body in EL, relative to that of PhiKZ, may play a role in determining packaged genome size.
4 Conclusions
Despite the generally common features of DNA packaging, such as the packaging motor itself and the resulting conserved density of DNA packaged within capsids, there are clearly different packaged DNA structures found in different large, tailed dsDNA phages. However, the reasons for these differences are in many cases still obscure. For instance, for certain phages, such as T7, the inner core is thought to have a major impact on coaxial spooling of the DNA within the capsid. While the large portal of P22 (gp3, 12 × 82 kDa) forms a complex that apparently extends well into the capsid and may similarly impart coaxial spooling (Tang et al. 2011), the core or portal is unlikely to be the only factor to result in this structure. Thus in the case of HK97, there does not appear to be an unusually large portal subunit (gp5, 12 × 47 kDa) for this to occur, and HK97 does not have a core, although its DNA is coaxially spooled (Duda et al. 2009). Overall, while the packaged DNA structure is necessarily expected to be significantly affected by the presence of inner head proteins, either structured or unstructured, often removal of these proteins (e.g., of over 1,000 T4 internal proteins) has no significant effect on either packaging or DNA ejection. Moreover, it is unknown whether observed differences in packaged DNA structure, such as the transverse packaging in T7 versus the more nearly parallel packaging found in T4 capsids, relate to the variable speeds and mechanisms (motor or nonmotor driven) DNA ejection into host bacteria that are observed; e.g., ≥10–15 min in T7 (Molineux 2001) to less than 2 min in T4 (Kalasauskaite et al. 1983) for complete ejection. For biophysical modeling studies of DNA condensation to prove more valuable for understanding both packaging and ejection, more experimental evidence from studies of different phages should be incorporated into the models or reasons should be given for excluding such evidence (e.g., see experimental determination of the direction of T4 and lambda DNA packaging condensation above).
In spite of conserved features of the packaged DNA structure, internal/injected capsid proteins have been observed in numbers of unrelated phage groups, suggesting that they may be even more common than currently appreciated. These capsid proteins are diverse, present in different copy numbers, found in discrete structures or dispersed within the DNA, and with different modes of entry into host cells and different functions, which are often only essential functions in a narrow set of specific hosts or conditions for a given phage [e.g., T4 IPI (Rifat et al. 2008)]. The diversity of inner capsid proteins in different phages with specialized locations shows some common themes, including that these proteins are apparently often packaged within close proximity to the portal channel for injection into the host cell. As already noted, these proteins enter the DNA-free precursor prohead before the DNA is packaged. The presence of portal-localized inner proteins raises a number of interesting questions, particularly as to how DNA packaging occurs around or through them. Do any of these proteins have any specific interactions with packaged DNA? Or conversely, do some of these proteins have limited DNA interactions so as to prevent any hindering of their injection into the host cell, possibly ahead of the DNA? Other questions regarding these proteins include the mechanisms by which they are targeted into their respective capsids. In the T4-like phages, it is clear that affinity of an N-terminal CTS peptide for the major shape-determining scaffolding core protein allows virtually any protein to which the CTS is joined to be incorporated into the prohead. But what of other phages, do they still utilize targeting affinity with the core or scaffold, or with other proteins? Supporting the use of other proteins was the finding by Stiege et al. (2003) that their candidate for the SPP1 genome plug (gp7) is apparently incorporated into the capsid via its affinity for the portal protein. A related intriguing feature of injected capsid proteins to be resolved is the mechanisms by which large proteins such as Alt, Dar, N4, and PhiKZ RNAPs, and even a nonphage protein such as β-galactosidase, are apparently unfolded within the capsid, probably while the DNA is being packaged, to allow their ejection through the narrow portal and tail tube channels. Could this unfolding be a secondary but essential function of the DNA packaging motor mechanism that, through high DNA pressure within the fully packaged capsid, promotes the ejection of unfolded or partially unfolded large proteins that are then refolded upon injection to display their sometimes essential enzymatic functions in the host cell?
Ejection of highly structured inner capsid core proteins, whose structure persists within the densely packaged DNA environment (e.g., T7 and possibly PhiKZ), raises the question of how such structures are disassembled to allow exit from the capsid. The role of the T4 core in determining head size suggests the possibility that a comparable sizing mechanism may operate in even larger capsids, e.g., may be one function of the capsid inner body in PhiKZ-related phages. However, it is likely that the proteins forming such bodies have multiple functions, including incorporation and ejection of proteins that enhance infection.
References
Ackermann H-W, Tremblay D, Moineau S (2004) Long-term bacteriophage preservation. WFCC Newslett 38:35–40
Aebi U, Bijlenga RKL, Bt H et al (1976) Comparison of the structural and chemical composition of giant T-even phage heads. J Supramol Struct 5:475–495
Agirrezabala X, Martin-Benito J, Caston JR et al (2005) Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J 24:3820–3829
Arsuaga J, Vázquez M, Trigueros S et al (2002) Knotting probability of DNA molecules confined in restricted volumes: DNA knotting in phage capsids. Proc Natl Acad Sci USA 99:5373–5377
Bair CL, Rifat D, Black LW (2007) Exclusion of glucosyl-hydroxymethylcytosine DNA containing bacteriophages is overcome by the injected protein inhibitor IPI*. J Mol Biol 366:779–789
Bijlenga RKL, Aebi U, Kellenberger E (1976) Properties and structure of a gene 24-controlled T4 giant phage. J Mol Biol 103:469–498
Black LW (1989) DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol 43:267–292
Black LW, Silverman DJ (1978) Model for DNA packaging into bacteriophage T4 heads. J Virol 28:643–655
Black LW, Newcomb WW, Boring JW et al (1985) Ion etching bacteriophage T4: support for a spiral-fold model of packaged DNA. Proc Natl Acad Sci USA 82:7960–7964
Black LW, Showe MK, Steven AC (1993) Morphogenesis of the T4 head. In: Karam JD (ed) Molecular biology of bacteriophage T4. ASM, Washington, DC, pp 218–258
Cardarelli L, Lam R, Tuite A et al (2010) The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J Mol Biol 395:754–768
Cermakian N, Ikeda TM, Cedergren R et al (1996) Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res 24:648–654
Cerritelli ME, Cheng N, Rosenberg AH et al (1997) Encapsidated conformation of bacteriophage T7 DNA. Cell 91:271–280
Cerritelli ME, Conway JF, Cheng N et al (2003a) Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv Protein Chem 64:301–323
Cerritelli ME, Trus BL, Smith CS et al (2003b) A Second Symmetry Mismatch at the Portal Vertex of Bacteriophage T7: 8-fold Symmetry in the Procapsid Core. J Mol Biol 327:1–6
Chang J, Weigele P, King J et al (2006) Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14:1073–1082
Chang C-Y, Kemp P, Molineux IJ (2010a) Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology 398:176–186
Chang JT, Schmid MF, Haase-Pettingell C et al (2010b) Visualizing the structural changes of bacteriophage Epsilon15 and its Salmonella host during infection. J Mol Biol 402:731–740
Chattoraj DK, Inman RB (1974) Location of DNA ends in P2, 186, P4 and lambda bacteriophage heads. J Mol Biol 87:11–22
Chen J, Lu Z, Sakon J et al (2000) Increasing the thermostability of staphylococcal nuclease: implications for the origin of protein thermostability. J Mol Biol 303:125–130
Choi KH, McPartland J, Kaganman I et al (2008) Insight into DNA and protein transport in double-stranded DNA viruses: the structure of bacteriophage N4. J Mol Biol 378:726–736
Comolli LR, Spakowitz AJ, Siegerist CE et al (2008) Three-dimensional architecture of the bacteriophage phi29 packaged genome and elucidation of its packaging process. Virology 371:267–277
Depping R, Lohaus C, Meyer HE et al (2005) The mono-ADP-ribosyltransferases Alt and ModB of bacteriophage T4: target proteins identified. Biochem Biophys Res Commun 335:1217–1223
Duda RL, Ross PD, Cheng N et al (2009) Structure and energetics of encapsidated DNA in bacteriophage HK97 studied by scanning calorimetry and cryo-electron microscopy. J Mol Biol 391:471–483
Earnshaw WC, Casjens SR (1980) DNA packaging by the double-stranded DNA bacteriophages. Cell 21:319–331
Earnshaw WC, Harrison SC (1977) DNA arrangement in isometric phage heads. Nature 268:598–602
Earnshaw WC, King J, Harrison SC et al (1978) The structural organization of DNA packaged within the heads of T4 wild-type, isometric and giant bacteriophages. Cell 14:559–568
Ebisawa T, Yamamura A, Kameda Y et al (2010) The structure of mAG, a monomeric mutant of the green fluorescent protein Azami-Green, reveals the structural basis of its stable green emission. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:485–489
Effantin G, Boulanger P, Neumann E et al (2006) Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J Mol Biol 361:993–1002
Fang P-A, Wright ET, Weintraub ST et al (2008) Visualization of bacteriophage T3 capsids with DNA incompletely packaged in vivo. J Mol Biol 384:1384–1399
Fuller DN, Rickgauer JP, Jardine PJ et al (2007) Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29. Proc Natl Acad Sci USA 104:11245–11250
Garcia L, Molineux I (1996) Transcription-independent DNA translocation of bacteriophage T7 DNA into Escherichia coli. J Bacteriol 178:6921–6929
Gleghorn ML, Davydova EK, Rothman-Denes LB et al (2008) Structural basis for DNA-hairpin promoter recognition by the bacteriophage N4 virion RNA polymerase. Mol Cell 32:707–717
Glucksmann MA, Markiewicz P, Malone C et al (1992) Specific sequences and a hairpin structure in the template strand are required for N4 virion RNA polymerase promoter recognition. Cell 70:491–500
Gowen B, Bamford JKH, Bamford DH et al (2003) The tailless icosahedral membrane virus PRD1 localizes the proteins involved in genome packaging and injection at a unique vertex. J Virol 77:7863–7871
Hartman PS, Eisenstark A, Pauw PG (1979) Inactivation of phage T7 by near-ultraviolet radiation plus hydrogen peroxide: DNA-protein crosslinks prevent DNA injection. Proc Natl Acad Sci USA 76:3228–3232
Hertveldt K, Lavigne R, Pleteneva E et al (2005) Genome comparison of Pseudomonas aeruginosa large phages. J Mol Biol 354:536–545
Hong Y-R, Black LW (1993) Protein folding studies in vivo with a bacteriophage T4 expression-packaging-processing vector that delivers encapsidated fusion proteins into bacteria. Virology 194:481–490
Iida S, Streiff MB, Bickle TA et al (1987) Two DNA antirestriction systems of bacteriophage P1, darA, and darB: characterization of darA- phages. Virology 157:156–166
Jiang W, Chang J, Jakana J et al (2006) Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439:612–616
Johnson JE, Chiu W (2007) DNA packaging and delivery machines in tailed bacteriophages. Curr Opin Struct Biol 17:237–243
Juers DH, Jacobson RH, Wigley D et al (2000) High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation. Protein Sci 9:1685–1699
Kalasauskaite EV, Kadisaite DL, Daugelavicius RJ et al (1983) Studies on energy supply for genetic processes. Eur J Biochem 130:123–130
Karhu NJ, Ziedaite G, Bamford DH et al (2007) Efficient DNA packaging of bacteriophage PRD1 requires the unique vertex protein P6. J Virol 81:2970–2979
Kazmierczak KM, Davydova EK, Mustaev AA et al (2002) The phage N4 virion RNA polymerase catalytic domain is related to single-subunit RNA polymerases. EMBO J 21:5815–5823
Kemp P, Garcia LR, Molineux IJ (2005) Changes in bacteriophage T7 virion structure at the initiation of infection. Virology 340:307–317
Koonin EV, Senkevich TG, Dolja VV (2006) The ancient Virus World and evolution of cells. Biol Direct 1:29
Kropinski AM, Kovalyova IV, Billington SJ et al (2007) The genome of Epsilon15, a serotype-converting, Group E1 Salmonella enterica-specific bacteriophage. Virology 369:234–244
Krylov VN, Smirnova TA, Minenkova IB et al (1984) Pseudomonas bacteriophage phiKZ contains an inner body in its capsid. Can J Microbiol 30:758–762
Lander GC, Tang L, Casjens SR et al (2006) The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312:1791–1795
Lavigne R, Seto D, Mahadevan P et al (2008) Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res Microbiol 159:406–414
Lecoutere E, Ceyssens P-J, Miroshnikov K et al (2009) Identification and comparative analysis of the structural proteomes of phiKZ and EL, two giant Pseudomonas aeruginosa bacteriophages. Proteomics 9:3215–3219
Leforestier A, Livolant F (2010) The bacteriophage genome undergoes a succession of intracapsid phase transitions upon DNA ejection. J Mol Biol 396:384–395
Lepault J, Dubochet J, Baschong W et al (1987) Organization of double-stranded DNA in bacteriophages: a study by cryo-electron microscopy of vitrified samples. EMBO J 6:1507–1512
Lhuillier S, Gallopin M, Gilquin B et al (2009) Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci USA 106:8507–8512
Lipinska B, Rao AS, Bolten BM et al (1989) Cloning and identification of bacteriophage T4 gene 2 product gp2 and action of gp2 on infecting DNA in vivo. J Bacteriol 171:488–497
Liu X, Zhang Q, Murata K et al (2010) Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol 17:830–836
Marenduzzo D, Orlandini E, Stasiak A et al (2009) DNA-DNA interactions in bacteriophage capsids are responsible for the observed DNA knotting. Proc Natl Acad Sci USA 106:22269–22274
Matsko N, Klinov D, Manykin A et al (2001) Atomic force microscopy analysis of bacteriophages PhiKZ and T4. J Electron Microsc (Tokyo) 50:417–422
McAllister WT, Wu HL (1978) Regulation of transcription of the late genes of bacteriophage T7. Proc Natl Acad Sci USA 75:804–808
Mendelson EC, Newcomb WW, Brown JC (1992) Ar+ plasma-induced damage to DNA in bacteriophage lambda: implications for the arrangement of DNA in the phage head. J Virol 66:2226–2231
Moak M, Molineux IJ (2000) Role of the Gp16 lytic transglycosylase motif in bacteriophage T7 virions at the initiation of infection. Mol Microbiol 37:345–355
Moak M, Molineux I (2004) Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol Microbiol 51:1169–1183
Molineux IJ (2001) No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol Microbiol 40:1–8
Mullaney JM, Black LW (1996) Capsid targeting sequence targets foreign proteins into bacteriophage T4 and permits proteolytic processing. J Mol Biol 261:372–385
Mullaney JM, Black LW (1998) Activity of foreign proteins targeted within the bacteriophage T4 head and prohead: implications for packaged DNA structure. J Mol Biol 283:913–929
Mullaney JM, Thompson RB, Gryczynski Z et al (2000) Green fluorescent protein as a probe of rotational mobility within bacteriophage T4. J Virol Methods 88:35–40
Overman SA, Aubrey KL, Reilly KE et al (1998) Conformation and interactions of the packaged double-stranded DNA genome of bacteriophage T7. Biospectroscopy 4(5 Suppl):S47–S56
Peñalva MA, Salas M (1982) Initiation of phage phi 29 DNA replication in vitro: formation of a covalent complex between the terminal protein, p3, and 5′-dAMP. Proc Natl Acad Sci USA 79:5522–5526
Petrov AS, Boz MB, Harvey SC (2007) The conformation of double-stranded DNA inside bacteriophages depends on capsid size and shape. J Struct Biol 160:241–248
Rao V, Black L (2010) Structure and assembly of bacteriophage T4 head. Virol J 7:356
Rao VB, Feiss M (2008) The bacteriophage DNA packaging motor. Annu Rev Genet 42:647–681
Ray K, Ma JX, Oram M et al (2010) Single-molecule and FRET fluorescence correlation spectroscopy analyses of phage DNA packaging: colocalization of packaged phage T4 DNA ends within the capsid. J Mol Biol 395:1102–1113
Repoila F, Tetart F, Bouet J-Y et al (1994) Genomic polymorphism in the T-even phages. EMBO J 13:4181–4192
Rickgauer JP, Fuller DN, Grimes S et al (2008) Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage phi29. Biophys J 94:159–167
Rifat D, Wright NT, Varney KM et al (2008) Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J Mol Biol 375:720–734
Saigo K (1975) Tail-DNA connection and chromosome structure in bacteriophage T5. Virology 68:154–165
Saigo K, Uchida H (1974) Connection of the right-hand terminus of DNA to the proximal end of the tail in bacteriophage lambda. Virology 61:524–536
Salmon B, Baines JD (1998) Herpes simplex virus DNA cleavage and packaging: association of multiple forms of U (L) 15-encoded proteins with B capsids requires at least the U(L)6, U(L)17, and U(L)28 genes. J Virol 72:3045–3050
Scraba DG, Bradley RD, Leyritz-Wills M et al (1983) Bacteriophage phi W-14: the contribution of covalently bound putrescine to DNA packing in the phage head. Virology 124:152–160
Serwer P (1986) Arrangement of double-stranded DNA packaged in bacteriophage capsids: an alternative model. J Mol Biol 190:509–512
Serwer P, Wright ET, Hakala KW et al (2008) Evidence for bacteriophage T7 tail extension during DNA injection. BMC Res Notes 1:36
Sheaffer AK, Newcomb WW, Gao M et al (2001) Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J Virol 75:687–698
Smith DE, Tans SJ, Smith SB et al (2001) The bacteriophage Phi29 portal motor can package DNA against a large internal force. Nature 413:748–752
Sternberg N, Weisberg R (1975) Packaging of prophage and host DNA by coliphage lambda. Nature 256:97–103
Steven AC, Heymann JB, Cheng N et al (2005) Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol 15:227–236
Stiege AC, Isidro A, Dröge A et al (2003) Specific targeting of a DNA-binding protein to the SPP1 procapsid by interaction with the portal oligomer. Mol Microbiol 49:1201–1212
Streiff MB, Iida S, Bickle TA (1987) Expression and proteolytic processing of the darA antirestriction gene product of bacteriophage P1. Virology 157:167–171
Sun M, Serwer P (1997) The conformation of DNA packaged in bacteriophage G. Biophys J 72:958–963
Tang J, Lander Gabriel C, Olia A et al (2011) Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in P22. Structure 19(4):496–502
Thomas JO (1974) Chemical linkage of the tail to the right-hand end of bacteriophage lambda DNA. J Mol Biol 87:1–9
Thomas JA, Rolando MR, Carroll CA et al (2008) Characterization of Pseudomonas chlororaphis myovirus 201phi2-1 via genomic sequencing, mass spectrometry, and electron microscopy. Virology 376:330–338
Thomas JA, Weintraub ST, Hakala K et al (2010) Proteome of the large Pseudomonas myovirus 201φ2-1: delineation of proteolytically processed virion proteins. Mol Cell Proteomics 9:940–951
Wang GR, Vianelli A, Goldberg EB (2000) Bacteriophage T4 self-assembly: in vitro reconstitution of recombinant gp2 into infectious phage. J Bacteriol 182:672–679
Watabe K, Shin M, Ito J (1983) Protein-primed initiation of phage phi 29 DNA replication. Proc Natl Acad Sci USA 80:4248–4252
Witkiewicz H, Schweiger M (1985) A model of lambda DNA arrangement in the viral particle. J Theor Biol 116:587–605
Xiang Y, Morais MC, Battisti AJ et al (2006) Structural changes of bacteriophage phi29 upon DNA packaging and release. EMBO J 25:5229–5239
Yu TY, Schaefer J (2008) REDOR NMR characterization of DNA packaging in bacteriophage T4. J Mol Biol 382:1031–1042
Zachary A, Black LW (1991) Isolation and characterization of a portal protein-DNA complex from dsDNA bacteriophage. Intervirology 33:6–16
Zhang Z, Greene B, Thuman-Commike PA et al (2000) Visualization of the maturation transition in bacteriophage P22 by electron cryomicroscopy. J Mol Biol 297:615–626
Acknowledgments
The authors thank Julienne Mullaney, Ian Molineux, and Alasdair Steven for their reading of this manuscript and their helpful suggestions.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Black, L.W., Thomas, J.A. (2012). Condensed Genome Structure. In: Rossmann, M., Rao, V. (eds) Viral Molecular Machines. Advances in Experimental Medicine and Biology, vol 726. Springer, Boston, MA. https://doi.org/10.1007/978-1-4614-0980-9_21
Download citation
DOI: https://doi.org/10.1007/978-1-4614-0980-9_21
Published:
Publisher Name: Springer, Boston, MA
Print ISBN: 978-1-4614-0979-3
Online ISBN: 978-1-4614-0980-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)