Abstract
Drought, cold, high-salinity and heat are major abiotic stresses that severely reduce the yield of food crops worldwide. Traditional plant breeding approaches to improve abiotic stress tolerance of crops had limited success due to multigenic nature of stress tolerance. In the last decade, molecular techniques have been used to understand the mechanisms by which plants perceive environmental signals and further their transmission to cellular machinery to activate adaptive responses. This knowledge is critical for the development of rational breeding and transgenic strategies to impart stress tolerance in crops. Studies on physiological and molecular mechanisms of abiotic stress tolerance have led to characterisation of a number of genes associated with stress adaptation. Techniques like microarrays have proven to be invaluable in generating a list of stress-related genes. Some of these genes are specific for a particular stress while others are shared between various stresses. Interestingly, a number of genes are shared in abiotic and biotic stress responses. This highlights the complexity of stress response and adaptation in plants. There is a whole cascade of genes involved in abiotic stress tolerance; starting from stress perception to transcriptional activation of downstream genes leading to stress adaptation and tolerance. A number of these genes have been discovered but we still do not have the complete list with all interactions. There is also significant number of genes with unknown functions found to be regulated by abiotic stresses. Understanding the function of these genes and their interaction with other known genes to effect stress adaptation is required.
The recent discovery that microRNAs regulate gene expression adds another layer of complexity to our understanding of abiotic stress tolerance. Significant amount of work will be needed to identify microRNAs associated with abiotic stress response, and understand their interaction with each other and their mechanism of regulating abiotic stress response. The promising side is the development of next-generation sequencing techniques that has allowed deep sequencing of mRNAs and microRNAs associated with abiotic stress response. A complete understanding on physiological and molecular mechanisms especially signalling cascades in response to abiotic stresses in tolerant plants will help to manipulate susceptible crop plants and increase agricultural productivity in the near future.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Abiotic stress
- Antioxidants
- Ion homeostasis
- MicroRNA
- Osmotic adjustments
- Signal transduction
- Transgenic approaches
1 Introduction
The major abiotic stresses (drought, high salinity, cold, and heat) negatively influence the survival, biomass production and yields of staple food crops up to 70% (Vorasoot et al. 2003; Kaur et al. 2008; Thakur et al. 2010) hence, threaten the food security worldwide. Dehydration stress imparted by drought, salinity and temperature severity is the most prevalent abiotic stress that limits plant growth and productivity (Vorasoot et al. 2003; Jaleel et al. 2009; Thakur et al. 2010). Since tolerance to this stress is multigenic and quantitative in nature (Collins et al. 2008), a massive challenge exists to understand the key molecular mechanisms for advanced selective breeding purposes. Traditional plant-breeding approaches have been marginally successful in improving the tolerances to these stresses (Flowers and Yeo 1995; Flowers et al. 2000). The molecular mechanisms underlying abiotic stress tolerances in plants are being unravelled with various high throughput sequencing and functional genomics tools in particular to advance the understanding of stress signal perception and transduction of the associated molecular regulatory networks (Heidarvand and Amiri 2010; Ray et al. 2010; Sanchez et al. 2011). Understanding the mechanisms by which plants perceive environmental signals and further their transmission to cellular machinery to activate adaptive responses is of critical importance for the development of rational breeding and transgenic strategies to impart stress tolerance in crops. Ultimately, marrying the physiological, biochemical and gene regulatory network knowledge will be essential to develop or select for stress-tolerant and high-yielding food crop cultivars.
2 Physiological and Molecular Mechanisms of Abiotic Stress Tolerance
2.1 Ion Transport and Homeostasis
The effect of salinity on plant growth limitation is proposed to be due to the osmotic effect from the ion imbalance in the earlier phase and a direct effect of the ions themselves in the latter phase of low to moderate stress (Munns and Tester 2008). At high salinity levels, salt-sensitive species lack the ability to control Na+ transport, where ionic effects dominate the osmotic effect. For normal metabolic reactions, plant cells need to maintain high K+ (100–200 mM) and low Na+ (less than 10–20 mM) levels (Flowers and Dalmond 1992; Carden et al. 2003). Therefore, tolerance to salinity stress must involve maintaining or quickly re-establishing both osmotic and ionic homeostasis (Munns and Tester 2008).
In general, plants employ one or both of the following to survive high salinity environments to ensure internal osmotic and ionic homeostasis; (1) avoidance – to keep sensitive plant tissues away from regions of concentrated salt ions and (2) tolerance – to exclude ions from roots or compartmentalise ions away from the cytoplasm of physiologically active cells (Silva et al. 2010). Indeed, efficient exclusion of excess Na+ ions from the cytoplasm and accumulation of Na+ ions within vacuoles are the main adaptive tolerance mechanisms to salinity stress (Munns and Tester 2008). Exclusion is typically carried out by transmembrane transport proteins that exclude Na+ from the cytosol in exchange for H+, a secondary transport process which is energy-dependent and driven by the proton motive force generated by the plasma membrane H+-ATPase. Likewise, compartmentalisation is generally carried out by vacuolar membrane H+-ATPase and H+-pyrophosphatase proteins (Rodríguez-Rosales et al. 2009; Ye et al. 2009; Leidi et al. 2010; Pasapula et al. 2011). By increasing the cellular levels of proteins (such as vacuolar antiporter proteins), number of abiotic stress-tolerant transgenic plants have been produced to control the transport functions. AtSOS from Arabidopsis has been shown to encode plasma membrane Na+/H+ antiporter (NHX) with significant sequence similarity to the respective antiporter from bacteria and fungi (Shi et al. 2000). Overexpression of the vacuolar Na+/H+ antiporter (NHX1) or the Arabidopsis thaliana vacuolar H + -translocating pyrophosphatase (AVP1) gene energized the pumping of Na+ into the vacuole and increased both accumulation and tolerance to Na+ (Gaxiola et al. 2001; Pasapula et al. 2011). More efficient sequestration of these ions to the vacuole may improve tissue tolerance to salinity by reducing the cytosolic Na+ concentrations. The importance of Na+ sequestration in salt tolerance has been further demonstrated in transgenic plants overexpressing AtNHX1 (Leidi et al. 2010; Silva et al. 2010).
2.2 Osmotic Adjustments and Controlling Factors
Intracellular water lost from the cell due to salt, drought and cold, leads to cellular dehydration. To prevent this and protect the cellular proteins, plants accumulate many organic compounds such as amino acids (proline), quaternary and other amines (glycine betaine and polyamines), a variety of sugars (mainly fructose and sucrose), sugar alcohols, complex sugars (like trehalose and fructans) and organic acids (oxalate, malate) (Valliyodan and Nguyen 2006). These metabolites with osmolytic function are also known as compatible solutes or osmoprotectants and may accumulate to high levels without disturbing the intracellular biochemistry (Ford 1984). By reducing the water potential within the cell, water loss is prevented and osmotic adjustment is facilitated (Delauney and Verma 1993).
Transgenic studies have been carried out for developing tolerant genotypes through manipulation of enzymes that synthesize specific osmolytes (Chen and Murata 2008; Szabados and Savoure 2010). The success of these studies on imparting stress tolerance has varied since the function of the targeted osmolytes is not restricted to osmotic adjustment, but also confers osmoprotection (Krishnan et al. 2008). In several studies, accumulation of osmolytes provided protection through scavenging of reactive oxygen species (ROS) and chaperone-like activities in maintaining protein structures and functions (Bohnert and Shen 1999; Krishnan et al. 2008). Pleiotropic effects such as necrosis and growth retardation were also observed due to disturbances in endogenous pathways of primary metabolisms. Patade et al. (in press) reported differential osmotic adjustment in sugarcane where salt-stressed plants appeared to use salt as an osmoticum and PEG stressed plants relied on accumulation of sugars.
A substantial number of transgenic studies have been performed to overexpress genes encoding osmoprotectants such as glycine-betaine (Bensen et al. 2008; Chen and Murata 2008) and proline (Delauney and Verma 1993; Verdoy et al. 2006; Szabados and Savoure 2010). Also, a number of sugars and sugar alcohols (mannitol, trehalose, myo-inositol and sorbitol) have been targeted for the engineering of compatible-solute overproduction, thereby protecting the membrane and protein complexes during stress (Gao et al. 2000; Suprasanna et al. 2005). In particular, there is a growing body of research on trehalose metabolism as a means of engineering stress tolerance in crops (Suprasanna 2003). In transgenic tomato, trehalose overproduction using the yeast trehalose-6-phosphate synthase gene led to significant tolerance to salinity, drought and oxidative stress (Cortina and Culianez-Macia 2005). Similarly, transgenic plants engineered for the overexpression of polyamines exhibited increased tolerance to multiple abiotic stresses such as heavy metal, salinity, drought, low and high temperature and fungal disease resistance (Capell et al. 2004; Prabhavathi and Rajam 2007).
Aside from osmotolerance, osmolyte accumulation also plays a vital role in the maintenance of cellular activities. For example, proline accumulation through overexpression of the P5CS gene in Medicago truncatula resulted in enhanced osmotolerance and also aided in maintaining nitrogen-fixing activity under osmotic stress (Verdoy et al. 2006). In order to minimize possible negative pleiotropic effects such as those previously mentioned, engineering of pathways for overproduction of compatible solutes should be through stress-inducible and/or tissue specific regulation (Su and Wu 2004).
2.3 Cold Acclimation
Plants survive freezing temperatures either through avoidance, primarily by super cooling of tissue water, or through freezing tolerance. Several plant species have the ability to increase freezing tolerance (FT) in response to low non-freezing temperatures (below 10°C) within a short photoperiod, a phenomenon known as cold acclimation (Thomashow 2010). The level of FT obtained through cold acclimation is not static but can vary seasonally and is rapidly lost upon return to a warm non-acclimating temperature. FT can be induced by osmotic stresses (Li et al. 2002) as well as treatment with abscisic acid (Li et al. 2003). Programmed dehydration is characteristic of overwintering tissues and at least partly contributes to FT (Welling et al. 2004). Further, cellular changes related to accumulation of storage proteins, sugars and starch are triggered by alteration of source–sink relationships after growth cessation in response to low temperature exposure (Zhu and Coleman 2001). The essential accumulation of sugars for cold acclimation was demonstrated by the inability of an Arabidopsis sucrose synthase mutant to cold acclimate (Uemura et al. 2003). The high abundance of sugars in cold acclimated plants suggests a role in osmoregulation, whereas less abundant sugars might have a role in cryoprotection or as signalling molecules (Stitt and Hurry 2002).
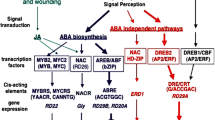
Recent progress has been made in elucidating the physiological and molecular mechanisms underpinning freezing tolerance. FT is a genetically complex trait, reflected by large number of genes that are affected by low temperature, thus estimated to be up to 25% of the entire transcriptome (Krebs et al. 2002). Altered expression of specific cold responsive-COR genes results in various physiological and biochemical changes during the process of cold acclimation, and the combined effect of the gene products is manifested in the level of FT obtained (Chinnusamy et al. 2006; Novillo et al. 2007; Thomashow 2010). The activation of COR genes is controlled by a set of signalling pathways triggered by exposure to the LT stimulus (Chinnusamy et al. 2006). The A. thaliana CBF (C-repeat binding factor) cold response pathway is most likely the best understood regulatory pathway involved in cold acclimation. This occurs through a rapid cold induction of CBF transcription factors, followed by expression of the regulon genes, which imparts freezing tolerance (Thomashow 2010). Specifically, in Arabidopsis, three CBF genes, CBF1 (DREB1b), CBF2 (DREB1c) and CBF3 (DREB1a), were induced within 15 min of low temperature exposure (Gilmour et al. 1998; Liu et al. 1998). These CBFs encodes closely related members of the AP2/ERF (Apetala2/Ethylene-responsive element binding factor), a family of transcription factors (Riechmann et al. 2000) that binds to CRT/DRE (C-repeat/dehydration responsive element) DNA regulatory elements found in the promoters of CBF-targeted genes (Stockinger et al. 1997; Liu et al. 1998). The CBF proteins induce the expression of many CBF regulon genes (Maruyama et al. 2004; Vogel et al. 2005). This leads to an increase in freezing tolerance (Jaglo-Ottosen et al. 1998; Liu et al. 1998) through the accumulation of low molecular weight cryoprotective metabolites, such as raffinose, sucrose and proline (Cook et al. 2004; Kaplan et al. 2004), along with the production of cryoprotective polypeptides, such as COR15a (Steponkus et al. 1998).
Cytoskeletal reorganization serves as a link between membrane rigidification and Ca2+ influx in the early stages of cold acclimation, and is needed for the development of maximum FT (Orvar et al. 2000; Sangwan et al. 2002). Low-temperature-induced changes in cytosolic calcium correlate with the expression of cold-responsive genes and the development of FT. In Arabidopsis, the increase in cytosolic calcium comes from rapid cold-induced release of calcium from both extracellular and vacuolar stores (Knight et al. 1996). Following the cold stimulus, the Ca2+ homeostasis in cells is restored to resting levels by active Ca2+ transporters. A connection between the calcium spikes and cold-regulated gene expression has been demonstrated to involve induction of DREB genes (Shinozaki and Yamaguchi-Shinozaki 1996). Overexpression of CBF1, a DREB1A homolog, enhanced freezing-stress tolerance and increased the expression of cold regulated genes (cor15a, cor6.6, and cor47) (Jaglo-Ottosen et al. 1998). Overexpression of DREB1A also enhanced drought and salt tolerance in transgenic plants, demonstrating the cross-stress protective function of this gene family (Kasuga et al. 1999).
In Arabidopsis, the rapid influx of calcium into the cytosol is required for normal cold induction of the CBF target genes KIN1 and KIN2 (Knight et al. 1996; Tahtiharju et al. 1997). Accumulation of dehydrins, proteins which accumulate in vegetative tissues during dehydration stresses, was linked to the development of FT both in herbaceous and woody plants (Peng et al. 2008; Xu et al. 2008). Recently, a close link between the up-regulation of low temperature-associated proteins and vernalization fulfilment in wheat (Triticum aestivum) was reported (Sarhadi et al. 2010).
2.4 Antioxidant Defence for Abiotic Stress Tolerance
Reactive Oxygen Species (ROS) such as singlet oxygen, hydrogen peroxide molecules, superoxide and hydroxyl radicals are constantly produced in chloroplasts, mitochondria and peroxisomes by aerobic processes (Apel and Hirt 2004). Thought to be integral to downstream defense/tolerance responses, the elevated levels of ROS are often associated with exposure to biotic (e.g. pathogens or pests) and abiotic (e.g. high light, UV radiation, temperature extremes, heavy metals, air pollutants, drought stress, salt stress, mechanical/physical stress) factors (Neill et al. 2002; Imlay 2003; Einset et al. 2007). Overproduction of ROS leads to oxidative damage such as lipid peroxidation of cell membranes (Imlay 2003) or even cell death (Jones 2000). In order to control ROS levels and protect cells from oxidative injury, plants possess both enzymes and non-enzymatic metabolites that may play a significant role in ROS signalling in plants (Vranova et al. 2002).
The harmful effects of ROS are prevented by the presence of lipid soluble antioxidants (α-tocopherol and carotenoids), water-soluble reductants (glutathione and ascorbate) and antioxidant enzymes such as catalase (CAT, EC 1.11.1.6), ascorbate peroxidase (APX, EC 1.11.1.11) and superoxide dismutase (SOD, EC 1.15.1.1) present in plant cells (Desikan et al. 2004). In response to stress, some of the osmolytes accumulate in plant cells, besides a role in scavenging of free radicals and protecting enzymes (Krishnan et al. 2008). The ability to activate protective mechanisms, such as an increase in the activity of scavenging enzymes, is vital for oxidative stress tolerance. Transgenic improvements for abiotic stress tolerances have been achieved through detoxification strategies by overexpressing the enzymes involved in oxidative protection. For example, salt or thermal stress treatment inhibited the growth of wild tobacco and caused increased lipid peroxidation, while overexpression of tobacco glutathione-S-transferase (GST) and glutathione peroxidase (GPX) reduced oxidative damage in the stressed transgenic seedlings (Roxas et al. 2000). Furthermore, overexpression of CuZn superoxide dismutase (SOD) and ascorbate peroxidase (APX) in transgenic sweet potato enhanced tolerance and recovery from drought stress. This was due to a considerable increase in expression of antioxidant enzymes that reduced the levels of malondialdehyde and electrolyte leakage (Lu et al. 2010). Likewise, Arabidopsis transformed with antisense barley 2-cystein peroxiredoxin sequence resulted in high expression of APX and monodehydroascorbate reductase (MDHAR; Baier et al. 2000).
Overexpression of an alternative oxidase (AOX) gene reduced oxidative stress in transgenic Arabidopsis under cold exposure (Sugie et al. 2006). Vitamin E was shown to be another participant in the protective mechanism against oxidative stress since Vitamin-E deficient Arabidopsis mutants were chilling sensitive. This was proposed to be because of a defective export of photoassimilate (Zhu et al. 2007).
2.5 Signal Transduction in Response to Abiotic Stresses: Specificity and Cross-Talk
Abiotic stresses are complex stimuli (ionic imbalance and osmotic stress), perceived by multiple primary sensors that cause alteration in the expression of many genes. The cascade of molecular responses ranges from stress perception, to signal transduction to cytoplasm and nucleus, to gene expression and finally metabolic changes leading to stress tolerance. Rapid increase in cytosolic Ca2+ levels in response to the various environmental stress stimuli are controlled by four major families of calcium-binding proteins; calmodulins, calmodulin-like proteins, calcineurin B-like proteins and calcium-dependent protein kinases (CDPKs) (Snedden and Fromm 2001; Luan et al. 2002; Sanders et al. 2002). Following the Ca2+ influx, signals are proposed to be mediated by combinations of phosphorylation/dephosphorylation cascades and is thought to be controlled by members of the Ca2+-dependent protein kinase (CDPK) gene family (Zhang et al. 2005). Members of the CDPK family are also reported to activate ABA/stress responsive gene expression. Altered expression of Oryza sativa CDPK (OsCDPK) has been correlated with tolerance to cold, salt and drought stress (Saijo et al. 2000).
Plants demonstrate both, stress-specific as well as shared responses that protect them from several environmental stresses (Mantri et al. 2010b). Plants respond to stress by regulating gene expression leading to both common and distinctive changes in transcript levels of stress responsive genes (Shinozaki and Yamaguchi-Shinozaki 2000). Indeed, overlap has been reported in gene expression induced by different stresses (Chen et al. 2002; Mantri et al. 2007; Seki et al. 2009). Plants universally appear to suffer from osmotic and oxidative stress under salt, drought and cold stress (Beck et al. 2007; Munns and Tester 2008). However, prevention of the osmotic stress is performed by stress-specific and general tolerance mechanisms. For example, in salt stress, osmotic adjustment maintains osmotic homeostasis while endurance through the period of freezing-induced osmotic stress relies on avoidance or interruption of ice nuclei formation (Pearce 2001).
Chen et al. (2002) identified groups of transcription factors regulated either singly i.e., abiotic stress (class I) or by both, biotic and abiotic stresses (class II) in Arabidopsis. Among the class I group, ∼20 genes were preferentially induced by abiotic stresses such as salinity, osmotic, cold and jasmonic acid treatments. These transcription factors include DRE/CRT binding factors activated by cold stress, CCA1 and Athb-8 (regulated by hormones, Baima et al. 2001), Myb proteins as well as bZIP/HD-ZIPs and AP2/EREBP domain proteins (Kizis et al. 2001). Further, Seki et al. (2002) employed a full-length cDNA microarray, containing 7,000 independent Arabidopsis cDNAs to identify cold, drought and salinity-induced target genes and stress-related transcription factor family members such as DREB, ERF, WRKY, MYB, bZIP, helix-loop-helix and NAC. ABA is not only involved in drought-specific responses but also there is a cross-talk in cold and salinity stress responses (Seki et al. 2002).
2.5.1 Cross Talk Between Biotic and Abiotic Stress Signalling
Plants have developed various methods to deal with biotic and abiotic stresses. Traditionally, the molecular mechanisms associated with tolerance to each stress have been studied independently. Therefore, the knowledge of signalling pathways that are shared during biotic and abiotic stress responses remain rudimentary. In a recent study in chickpea, plant responses to fungal infection (Ascochyta blight) were found to be more similar to high-salinity stress than drought and cold stresses (Mantri et al. 2010a). Supporting this, abscisic acid-induced myb1 (SlAIM1) gene from tomato (Solanum lycopersicum) encoding an R2R3MYB transcription factor was induced by pathogens, plant hormones, salinity and oxidative stress (Abuqamar et al. 2009). Further, silencing the SlAIM1 by RNA interference led to an increased susceptibility to the necrotrophic fungus Botrytis cinerea, and increased sensitivity to salt and oxidative stress. Also an ectopic expression of SlAIM1 led to high salinity and oxidative stress tolerance (Abuqamar et al. 2009). This suggested that SlAIM1 regulates a transmembrane ion flux, an indication of an early response to abiotic stress and pathogen infection, perhaps preceding hypersensitive cell death and necrosis.
Misregulation of ion fluxes can result in impaired plant tolerance to necrotrophic infection or abiotic stress (Abuqamar et al. 2009). Emerging evidence suggests that hormone signalling pathways like those controlled by jasmonic acid, abscisic acid, ethylene, and salicylic acid are central to the crosstalk between abiotic and biotic stress responses (Fujita et al. 2006). Recent studies have indicated several transcription factors and kinases are important candidates leading to crosstalk in stress signalling pathways. Mitogen-activated protein kinases (MAPKs) have been shown to be involved in developmental, hormonal, abiotic, and biotic stress signalling (Colcombet and Hirt 2008; Rodriguez et al. 2010). The activation of components of MAPK cascades by more than one type of stress, suggests that MAPK cascades serve as crossroads for numerous abiotic and biotic stress signalling pathways. Furthermore, as the Arabidopsis genome is reported to have around 20 MAPKs, 10 MAPKKs and 60 MAPKKKs, the signals recognized by the 60 MAPKKKs have to be transferred via 10 MAPKKs to 20 MAPKs, offering great chances for crosstalk between different stress signals.
Spatial and temporal expression patterns based on cell biological analysis combined with biochemical characterization of the signalling components, mainly identification of signalling complexes, is necessary to establish specificity or crosstalk of the signalling pathways (Chinnusamy et al. 2004). In the coming years, with the further development and incorporation of “omics” tools and computational approaches, deeper understanding of the signalling pathways, specificity and cross talk should be targeted. Currently, only a limited number of pathways and their components have been unravelled. In nature, however, plants face and respond to an overabundance of stimuli (including biotic as well as abiotic) simultaneously.
3 Involvement of Other Novel Genes Like MicroRNA in Plant Stress Tolerance
Discovery and functional association of microRNAs (miRNAs) have led to a large new research area in the previously unsuspected world of non-coding RNAs (Lee et al. 1993; Reinhart et al. 2002). The miRNAs are endogenous, small 21–24 nucleotide, single stranded, non-protein coding RNAs that have recently emerged as important regulators of gene expression (Bartel 2004). These regulate target gene expression by catalyzing posttranscriptional gene silencing (Palatnik et al. 2003) or translation repression (Chen 2004). Targets of miRNA comprises transcription factors or other regulatory proteins that function in plant development or signal transduction. Recently, research on micro-RNAs (miRNAs) have suggested an association between miRNAs and plant stress responses (Patade and Suprasanna 2010). However, the relationship between micro-RNAs and stress response is just beginning to be explored. Several miRNAs are either up- or down-regulated by abiotic stresses, suggesting to be involved in stress-responsive gene expression and stress adaptation affecting a variety of cellular and physiological processes (Sunkar and Zhu 2004; Shukla et al. 2008).
Sunkar and Zhu (2004) identified novel and abiotic stress-regulated miRNAs and reported differential expression of some of the identified miRNAs in Arabidopsis seedlings exposed to dehydration, salinity, or cold stress. In order to unravel function of microRNA, Zhao et al. (2007) studied transcript expression profiles of miRNAs in rice (O. sativa) under drought stress. The drought-induced expression of miR-169g and miR393 was validated by microarray expression profiling and confirmed greater expression of miR-169g in roots rather than shoots. Sequence analysis revealed occurrence of two proximate DREs (dehydration-responsive element) in the upstream of the MiR-169g, suggesting transcript expression regulation of miR-169g by CBF/DREBs.
Sunkar et al. (2006) provided evidence on involvement of miRNA in oxidative stress responses by targeting cytosolic and chloroplastic superoxide dismutases that detoxify superoxide radicals. Transcript expression of miR398 in response to oxidative stress was down-regulated, leading to posttranscriptional accumulation of the SOD mRNA and thus oxidative stress tolerance. Moreover, transgenic Arabidopsis plants overexpressing a miR398-resistant form of SOD accumulated more mRNA than plants overexpressing a regular form and were consequently much more tolerant to high light, heavy metals and other oxidative stresses. Arabidopsis have been shown to trigger the accumulation of miR159 in response to ABA, drought stress, and gibberellic acid (GA) treatment and the miRNA was predicted to target four MYB transcription factors (Reyes and Chua 2007). Recently, Patade and Suprasanna (2010) characterized transcript expression of mature miR159 in response to short- and long-term salt and PEG-induced osmotic stress in sugarcane. A change in mature transcript levels of miR159 was not detected in response to long-term (15 days) NaCl or iso-osmotic (−0.7 MPa) PEG stress. However, short-term (up to 24 h) salt or PEG stresses increased transcript level of the mature miRNA as compared to the control. The early induction of the gene under the short treatments supports its involvement in the regulation of genes involved in stress perception and/or signalling.
Zhou et al. (2008) developed a computational transcriptome-based approach to annotate stress-inducible miRNAs in plants. Interestingly, the promoter analysis of the miRNA genes revealed the presence of many known stress-responsive cis-regulatory elements. Continued efforts are needed to identify the complete set of miRNAs and other small RNAs that are fundamental to the stress regulation pathways. The identification and functional validation of stress-regulated small RNAs including miRNAs will help in designing new strategies for improving stress tolerance (Sunkar et al. 2006; Katiyar-Agarwal et al. 2007).
4 Strategies for Improving Abiotic Stress Tolerance
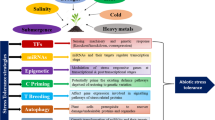
Many strategies undertaken for improving abiotic stress tolerance in a particular genetic background have included screening of diverse genetic resources, wide crossing and subsequent recurrent backcrossing; identification and selection of the major conditioning genes through linkage mapping and quantitative trait loci (QTL) analysis; the production and screening of mutant populations and the transgenic introduction of novel genes (Fig. 1.1). Although some success has been achieved in introducing tolerance traits into crop varieties from wild relatives (i.e. barley; Forster et al. 2000 and tomato; Foolad et al. 2001), in general there has been very little success reported in achieving high abiotic tolerance into elite germplasm (Flowers 2004).
As previously mentioned, breeding for, or induction of, abiotic stress tolerance traits is almost always limited by the genetic complexity of the underpinning mechanisms as well as potential interaction among genetic determinants. Also, differential selection of a particular stress may be affected by additional environmental factors, plant development stage, poor or irreproducible selection techniques, and the logistical constraints of physiological screening of large breeding populations on a field scale (Flowers et al. 2000). In this regard, the identification of discrete chromosomal regions that have a major effect on the specific tolerance trait through quantitative trait loci (QTL) mapping and marker-assisted selection remains a valuable option for many breeding programs (Cushman 2009; Cuartero et al. 2010). This is particularly so when whole genome knowledge is lacking and no candidate tolerance genes are known.
For accurate selection of the related phenotype, reliable and realistic screening techniques are required. However, uniformity and reliability of field-based screening may suffer from heterogeneity in the stress across the site (i.e. boron or salinity level) as well as the potential compounding environmental factors (i.e. disease, rainfall, temperature). Also, when the starting material is genetically wide, heterogeneity among the genetic backgrounds may also impact on the ability to accurately select the most superior or different tolerances. As an alternative, cellular-based mutant induction and subsequent selection initially under controlled in vitro conditions offers a method to quickly screen large populations with homogeneous backgrounds for novel fortuitous changes related to tolerance. Subsequent field screening then ensures adequate performance of the tolerance trait under the external potentially mitigating factors previously mentioned. Unsurprisingly, this method has generated great interest in selecting for abiotic stress tolerances in several crop species (Suprasanna et al. 2008).
4.1 Transgenic Approaches for Engineering Tolerance
Many genes linked to different pathways and processes such as stress perception and signalling, contributing to molecular, biochemical, cellular, physiological and morphological adaptations are differentially regulated in response to plant stress (Munns and Tester 2008). Stress responsive genes include those that alleviate the effect of the stress and lead to adjustment of the cellular environment and plant tolerance. The gene products are classified into three major groups: those encoding products that directly protect plant cells against stress, those that are involved in signalling cascades and in transcriptional control and those that are involved in water and ion uptake and transport.
Engineering metabolic and stress-signalling pathways to produce stress-tolerant crops is one of the major interests of agricultural research. Genetic transformation with stress-inducible genes has been employed to gain an understanding of their functional role in the tolerance response and ultimately to improve the tolerance trait in the target genotype (Zhang et al. 2004, Cuartero et al. 2010). To date, by far, majority of these studies have been limited to single-gene transfers within known multigenic pathways and mostly those involved in signalling and regulatory pathways, or effector genes that code for enzymes catalysing the synthesis of structural and functional defendants (Wang et al. 2003; Chinnusamy et al. 2005; Jewell et al. 2010). When selecting for success of the transformation experiment, a common prime consideration is whether the transgenic plants express a higher level of the transgene (i.e. an osmoprotectant or a protein) only under the stress conditions (Zhu 2001). In general, specific inducible promoters are used rather than constitutive promoters since the tolerance/stress-induced mechanisms may be energy and nucleic acid greedy and divert essential resources away from normal growth processes (Su et al. 1998).
As examples, transgenic rice plants developed with choline oxidase (codA), d-pyrroline-5-corboxylate synthase (P5CS), LEA protein group 3 (HVA1), alcohol dehydrogenase (ADH) and pyruvate decarboxylase (PDC) genes exhibited drought tolerance (Datta 2002; Soren et al. 2010). Potato and rice (Yeo et al. 2000 and Garg et al. 2002, respectively) transformed with trehalose synthesis genes displayed tolerance to drought (in case of potato), and salt, drought, and low-temperature stress (in case of rice). Tobacco plants transformed with ectoine biosynthesis genes from the halophilic bacterium Halomonas elongate showed enhanced salt tolerance. Also transformation with genes for sorbitol (Sheveleva et al. 1997) or mannitol (Shen et al. 1997) resulted in an increased accumulation of these osmolytes and tolerance to high salinity (Table 1.1). Overexpression of genes encoding the enzymes pyrroline-5-carboxylate (P5C) synthetase (P5CS) and P5C reductase (P5CR) resulted in proline overproduction and enhanced abiotic stress tolerance (Szabados and Savoure 2010). P5CS overexpression in transgenic tobacco dramatically elevated free proline (Kishor et al. 1995) with improved germination and growth of seedlings under salt stress. Transgenic petunia plants transformed with Arabidopsis P5CS gene showed resistance to drought conditions for longer duration than control plants (Yamada et al. 2005).
The enhancement of glycine betaine (GB) synthesis in transgenic plants using genes that encode for enzymes (choline monooxygenase, betaine aldehyde dehydrogenase and choline oxidase) in GB biosynthesis is another strategy to achieve enhanced tolerance to drought, salt and chilling stress (Rontein et al. 2002; Chen and Murata 2008). Transgenic rice plants expressing the codA (choline oxidase) gene recovered from an initial growth inhibition under salt and low-temperature stress, and grew normally than the wild type (Sakamoto et al. 1998). Several other plants that have been genetically engineered for obtaining salt, drought, freezing and heat tolerance through GBS accumulation include; A. thaliana, Brassica napus, Brassica juncea, Gossypium hirsutum, Lycopersicon esculentum, Nicotiana tabacum, Solanum tuberosum and Zea mays (Chen and Murata 2008).
Trehalose is a non-reducing disaccharide and an effective osmoprotectant (Goddijn and van Dunn 1999). Transgenic plants overexpressing trehalose biosynthetic genes showed increased tolerance to different abiotic stress conditions (Penna 2003; Almeida et al. 2007). A stress-inducible promoter has been utilised to overexpress Escherichia coli trehalose biosynthesis genes (otsA and otsB) as a fusion gene (TPSP) in rice, to confer tolerance to different abiotic stresses (Garg et al. 2002). The TPSP fusion gene is dually advantageous as both the genes can be simultaneously introduced into the rice genome leading to increased catalytic efficiency for trehalose synthesis (Jang et al. 2003; Almeida et al. 2007).
Research on genetic engineering efforts with other osmolytes such as mannitol, fructans, ononitol, proline, glycinebetaine and ectoine have also shown promise for generating tolerant genotypes (Suprasanna et al. 2005). To avoid overproduction of compatible solutes burdening the plant’s metabolic machinery and potentially diminishing pleiotropic effects, engineering for overproduction should be done under stress-inducible and/or tissue specific regulation. In addition, production of the osmolytes should be targeted to the chloroplast by placing a signal sequence in front of the engineered enzymes (Shen et al. 1997).
As previously stated, abiotic stress generates an increase in reactive oxygen species that may be deleterious to normal cellular functions. Therefore, several oxidative-stress-related genes have been employed in developing transgenic plants tolerant to various stresses (Hussain et al. 2008). For example, transgenic tobacco plants overexpressing chloroplastic Cu/Zn-SOD showed increased resistance to oxidative stress caused by salt exposure (Tanaka et al. 1999; Bartel 2001). Transgenic alfalfa (Medicago sativa) plants expressing Mn-SOD had reduced injury from water-deficit stress, as determined by chlorophyll fluorescence, electrolyte leakage and regrowth (McKersie et al. 1996). Simultaneous expression of genes encoding three antioxidant enzymes: copper zinc superoxide dismutase, ascorbate peroxidase and dehydroascorbate reductase in the chloroplasts of tobacco plants conferred enhanced tolerance to oxidative stresses caused by paraquat and salt (Lee et al. 2007). Similarly, overexpression of AtNDPK2 efficiently modulated oxidative stress caused by various environmental stresses in sweet potato through enhanced antioxidant enzyme activities such as peroxidase, ascorbate peroxidase and catalase (Kim et al. 2010). Thus it seems promising to target detoxification pathways as an approach for obtaining plants with multiple stress-tolerance traits.
Transgenic manipulation of detoxification pathways through overexpressing genes involved in oxidative protection, such as glutathione peroxidase, superoxide dismutase, ascorbate peroxidases and glutathione reductases is an area of current interest. Constitutive expression of the Nicotiana PK1 gene (regulatory protein NPK1) enhanced freezing, heat and salinity tolerance in transgenic maize plants (Shou et al. 2004b). In a further study, Shou et al. (2004a) expressed a tobacco MAPKKK (NPK1) constitutively in maize resulting in enhanced drought tolerance. The transgenic maize plants maintained significantly higher photosynthesis rates, suggesting, NPK1 induced a mechanism that protected photosynthesis machinery from dehydration damage.
Under salt stress, tolerant plant cells must maintain high K+ (100–200 mM) and lower Na+ (less than 1 mM) levels for normal metabolic function. An important strategy for achieving greater tolerance to salinity stress is to help plants to re-establish homeostasis under stressful environments, restoring both ionic and osmotic homeostasis. This strategy continues to be a major approach to improve salt tolerance in plants through genetic engineering, where the target is to achieve Na+ excretion, or vacuolar storage. A number of abiotic stress-tolerant transgenic plants have been produced by increasing the cellular levels of proteins (such as vacuolar antiporter proteins) that control the transport functions. For example, AtSOS from Arabidopsis has been shown to encode a plasma membrane Na+/H+ antiporter (NHX) with significant sequence similarity to the respective antiporter from bacteria and fungi (Shi et al. 2000). Constitutive expression of vacuolar Na+/H+ antiporter (NHX1) or AVP1 (A. thaliana vacuolar H+-translocating pyrophosphatase) gene energized the pumping of Na+ into the vacuole, and increased both accumulation and Na+ tolerance in Arabidopsis (Gaxiola et al. 2001). Thus more efficient sequestration of these ions to the vacuole could improve tissue tolerance to salinity by reducing the cytosolic Na+ concentrations. The importance of Na+ sequestration in salt tolerance has been further demonstrated in transgenic tomato plants overexpressing the AtNHX1 gene (Zhang and Blumwald 2001). Also, a vacuolar chloride channel gene, AtCLCd, involved in cation detoxification has been cloned as well as overexpressed in Arabidopsis and shown to confer salt tolerance. Up-regulation of the Salt Overly Sensitive 1 (SOS1) gene in Arabidopsis resulted in a greater proton motive force necessary for elevated Na+/H+ antiporter activities (Shi et al. 2000).
Apart from the single gene approach, tolerance towards multiple stresses may be achieved by targeting a stress inducible signal transduction molecule and/or transcription factor (Chinnusamy et al. 2005). The transcription factors play an important role in the acquisition of stress tolerance, which ultimately contribute to agricultural and environmental practices (Century et al. 2008). A large number of transcription factors are involved in the plant response to abiotic stress (Vincour and Altman 2005). Most of these falls into several large transcription factor families, such as AP2/ERF, bZIP, NAC, MYB, MYC, Cys2His2 zinc-finger and WRKY. Accordingly, overexpression of the functionally conserved At-DBF2 gene resulted in wide and high levels of multiple stress tolerances in Arabidopsis (Lee et al. 1999). Salt stress-tolerant tobacco plants were produced by overexpressing the calcineurin, a Ca2+/calmodulin-dependent protein phosphatase gene, formally identified as being involved in salt-stress signal transduction in yeast (Pardo et al. 1998; Grover et al. 1999).
Some stress responsive genes may share the same transcription factors, as indicated by the significant overlap of the gene expression profiles that are induced in response to drought and cold stress (Seki et al. 2001; Chen and Murata 2002; Mantri et al. 2007). The activation of stress-induced genes has been possible in transgenic plants by overexpressing one or more transcription factors that recognize regulatory elements of these genes. In Arabidopsis, the transcription factor DREB1A specifically interacts with the DRE and induces expression of stress tolerance genes (Shinozaki and Yamaguchi-Shinozaki 1997). CaMV 35S promoter-driven overexpression of DREB1A cDNA in transgenic Arabidopsis plants provided tolerance to salt, freezing and drought stress through strong constitutive expression of the stress inducible genes (Liu et al. 1998).
The transcription factors involved in the ABA-dependent (such as NAC, AREB/ABF, and MYB) and –independent (AP2/ERF gene) stress response pathways regulate cascade of downstream genes and events that enhance tolerance to drought stress. Transforming crops with such transcription factor genes should be more meaningful in the development of drought tolerance (Zhang et al. 2004; Ashraf 2010). Overexpressing Arabidopsis CBF1 (CRT/DRE) cDNA in tomato improved tolerance to salt, chilling and drought stress; however, the plants exhibited a dwarf phenotype as well as reduced fruit set and seed number (Hsieh et al. 2002). Overexpression of Alfin1 (transcriptional regulator) in alfalfa plants exhibited salinity tolerance through regulated endogenous MsPRP2 (NaCl-inducible gene) mRNA levels (Winicov and Bastola 1999).
4.2 The Future of Transgenic Approaches
The current plant genetic engineering approach for developing salt stress-tolerant transgenic plants includes altering the expression levels of native genes or incorporating alien genes for osmolytes, ion transporters, transcription factors and other signalling molecules. The advent of global transcription profiling has demonstrated that large numbers of other genes are also up- and down-regulated simultaneously in response to salt stress. This second category of genes encode proteins related to the regulation of transcriptional and translational machineries with distinct roles in mediating the salt stress response. Particularly, coordinated induction and action of the transcript of several RNA binding proteins, ribosomal genes, helicases, cyclophilins, F-box proteins, dynamin-like proteins, translation initiation and elongation factors seems to be important in salt stress tolerance. The functionality of these genes at the cellular level should also be investigated to assess aptness for targeted transgenic approaches (Sahi et al. 2006).
The evaluation of genetically engineered salt-tolerant transgenic lines needs critical, careful, and thorough experimentation (Flowers 2004). The fourth or fifth generation genotypes should be evaluated along with parental (wild-type) lines under controlled saline and non-saline treatment conditions. Validation should not stop at the laboratory or green house level, since quantitative measures of growth are required throughout the plant life cycle in field conditions.
5 Conclusions and Future Perspective
In the last decade, significant progress has been made in our understanding of the complex mechanisms governing abiotic stress tolerance in crop plants. However, we are still far from pinning the exact battery of gene activation responsible for tolerance to a particular abiotic stress condition. This situation is complicated when one considers plants have to simultaneously cope with numerous biotic stresses along with various abiotic stresses. Our struggle to understand these complex mechanisms is ongoing and recent development of new tools for high-throughput genotyping and phenotyping gives us a new ray of hope. A complete understanding on physiological and molecular mechanisms especially signalling cascades in response to abiotic stresses in tolerant plants will help to manipulate susceptible crop plants and increase agricultural productivity in the near future.
References
Abuqamar S, Luo H, Laluk K, Mickelbart MV, Mengiste T (2009) Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J 58(2):347–360
Almeida A, Cardoso L, Santos D, Torné J, Fevereiro P (2007) Trehalose and its applications in plant biotechnology. In Vitro Cell Dev Biol Plant 43(3):167–177
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Ashraf M (2010) Inducing drought tolerance in plants: recent advances. Biotechnol Adv 28:169–183
Baier M, Noctor G, Foyer CH, Dietz KJ (2000) Antisense suppression of 2-Cysteine peroxiredoxin in Arabidopsis specifically enhances the activities and expression of enzymes associated with ascorbate metabolism but not glutathione metabolism. Plant Physiol 124(2): 823–832
Baima S, Possenti M, Matteucci A, Wisman E, Altamura M, Ruberti I, Morelli G (2001) The Arabidopsis ATHB-8 HD-Zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 126(2):643–655
Bartel D (2001) Targeting detoxification pathways: an efficient approach to obtain plants with multiple stress tolerance. Trends Plant Sci 6:284–286
Bartel D (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Beck E, Fettig S, Knake C, Hartig K, Bhattarai T (2007) Specific and unspecific responses of plants to cold and drought stress. J Biosci 32(3):501–510
Bensen R, Castiglioni P, Korte J, Bell E, Hinchey B, Loida P, Ahrens J (2008) Transgenic plants with increased glycine-betaine. United States Patent 7410800. Application Number:10/839092
Bohnert HJ, Shen B (1999) Transformation and compatible solutes. Sci Hortic 78:237–260
Capell T, Bassie L, Christou P (2004) Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc Natl Acad Sci USA 101:9909–9914
Carden DE, Walker DJ, Flowers TJ, Miller AJ (2003) Single-cell measurements of the contributions of cytosolic Na+ and K+ to salt tolerance. Plant Physiol 131:676–683
Century K, Reuber TL, Ratcliffe OJ (2008) Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol 147:20–29
Chen X (2004) A microRNA as translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022–2025
Chen TH, Murata N (2002) Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr Opin Plant Biol 5:250–257
Chen THH, Murata N (2008) Glycinebetaine: an effective protectant against abiotic stress in plants. Trends Plant Sci 13(9):499–505
Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, Budworth PR, Toa Y, Xie Z, Chen X, Lam S, Kreps JA, Harper JF, Si-Ammour A, Mauch-Mani B, Heinlein M, Kobayashi K, Hohn T, Dangl JL, Wang X, Zhu T (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14:559–574
Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. J Exp Bot 55(395): 225–236
Chinnusamy V, Jagendorf A, Zhu JK (2005) Understanding and improving salt tolerance in plants. Crop Sci 45:437–448
Chinnusamy V, Zhu J, Zhu JK (2006) Gene regulation during cold acclimation in plants. Physiol Plant 126:52–61
Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413(2):217–226
Collins NC, Tardieu F, Tuberosa R (2008) Quantitative trait loci and crop performance under abiotic stress: where do we stand? Plant Physiol 147:469–486
Cook D, Fowler S, Fiehn O, Thomashow MF (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101: 15243–15248
Cortina C, Culianez-Macia FA (2005) Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Sci 169:75–82
Cuartero J, Bolarin MC, Moreno V, Pineda B (2010) Molecular tools for enhancing salinity tolerance in plants. In: Jain SM, Brar DS (eds) Molecular techniques in crop improvement. Springer, New York, pp 373–405
Cushman JC (2009) Functional genomics of plant abiotic stress tolerance. In: Genomics of plants and fungi (Book Chapter). Marcel Dekker, Inc., New York
Datta SK (2002) Recent developments in transgenics for abiotic stress tolerance in rice. JIRCAS Work Rep: 43–53
Delauney AJ, Verma DP (1993) Proline biosynthesis and osmoregulation in plants. Plant J 4:215–223
Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ (2004) ABA, hydrogen peroxide and nitric oxide signaling in stomatal guard cells. J Exp Bot 55:205–212
Einset J, Winge P, Bones A (2007) ROS signaling pathwaysin chilling stress. Plant Signal Behav 2(5):365–367
Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55:307–319
Flowers TJ, Dalmond D (1992) Protein synthesis in halophytes: the influence of potassium, sodium and magnesium in vitro. Plant Soil 146:153–161
Flowers TJ, Yeo AR (1995) Breeding for salinity tolerance in crop plants: where next? Aust J Plant Physiol 22:875–884
Flowers TJ, Koyama ML, Flowers SA, Sudhakar C, Singh KP, Yeo AR (2000) QTL: their place in engineering tolerance of rice to salinity. J Exp Bot 51:99–106
Foolad MR, Zhang LP, Lin GY (2001) Identification and validation of QTLs for salt tolerance during vegetative growth in tomato by selective genotyping. Genome 44:444–454
Ford CW (1984) Accumulation of low molecular weight solutes in water stress tropical legumes. Phytochemistry 22:875–884
Forster BP, Ellis RP, Thomas WT, Newton AC, Tuberosa R, This D, el-Enein RA, Bahri MH, Ben Salem M (2000) The development and application of molecular markers for abiotic stress tolerance in barley. J Exp Bot 51:19–27
Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9:436–442
Gao M, Sakamoto A, Miura K, Murata N, Sugiura A, Tao R (2000) Transformation of Japanese persimmon (Diospyros kaki Thunb.) with a bacterial gene for choline oxidase. Mol Breed 6:501–510
Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YC, Kochian LV, Wu RJ (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99: 15898–15903
Gaxiola RA, Li J, Unurraga S, Dang LM, Allen GJ, Alper SL, Fink GR (2001) Drought- and salt-tolerant plants result from over expression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98:11444–11449
Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16:433–442
Goddijn OJM, van Dunn K (1999) Trehalose metabolism in plants. Trends Plant Sci 4:315–319
Grover A, Sahi C, Sanan N, Grover A (1999) Taming abiotic stresses in plants through genetic engineering: current strategies and perspective. Plant Sci 143:101–111
Heidarvand L, Amiri RM (2010) What happens in plant molecular responses to cold stress? Acta Physiol Plant 32:419–431
Holmström KO, Somersalo S, Mandal A, Palva TE, Welin B (2000) Improved tolerance to salinity and low temperature in transgenic tobacco producing glycine betaine. Journal of Experimental Botany 51(343): 177–185
Hsieh TH, Lee JT, Charng YY, Chan MT (2002) Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol 130:618–626
Huang J, Hirji R, Adam L, Rozwadowski KL, Hammerlindl JK, Keller WA, Selvaraj G (2000) Genetic Engineering of Glycinebetaine Production toward Enhancing Stress Tolerance in Plants: Metabolic Limitations. Plant Physiology 122:747–756
Hussain TM, Thummala C, Mahamed H, Zafar S, Saleh BK, Rama GG (2008) Recent advances in salt stress biology – a review. Biotechnol Mol Biol Rev 3(1):008–013
Imlay JA (2003) Pathways of oxidative damage. Annu Rev Microbiol 57:395–418
Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 over expression induces COR genes and enhances freezing tolerance. Science 280:104–106
Jaleel CA, Manivannan P, Wahid A, Farooq M, Somasundaram R, Panneerselvam R (2009) Drought stress in plants: a review on morphological characteristics and pigments composition. Int J Agric Biol 11:100–105
Jang IC, Oh SJ, Seo JS, Choi WB, Song SI, Kim CH, Kim YS, Seo HS, Choi YD, Nahm BH, Kim JK (2003) Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate. Phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth. Plant Physiol 131:516–524
Jewell MC, Campbell BC, Ian D (2010) Transgenic plants for abiotic stress resistance. In: Kole C et al (eds) Transgenic crop plants. Springer, Berlin, pp 67–132
Jones A (2000) Does the plant mitochondrion integrate cellular stress and regulate programmed cell death? Trends Plant Sci 5:273–278
Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature stress metabolome of Arabidopsis. Plant Physiol 136:4159–4168
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291
Katiyar-Agarwal S, Gao A, Vivian-Smith J (2007) A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev 21:3123–3134
Kaur G, Kumar S, Nayyar H, Upadhyaya HD (2008) Cold stress injury during the pod-filling phase in chickpea (Cicer arietinum L.): effects on quantitative and qualitative components of seeds. J Agron Crop Sci 194(6):457–464
Kim Y-H, Kim MD, Choi YI, Park S-C, Yun D-J, Noh EW, Lee H-S, Kwak S-S (2010) Transgenic poplar expressing Arabidopsis NDPK2 enhances growth as well as oxidative stress tolerance. Plant Biotechnol J 9:334–347
Kishor PB, Hong Z, Miao G-H, Hu C-AA, Verma DP (1995) Overexpression of delta-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol 108: 1387–1394
Kizis D, Lumbreras V, Pagès M (2001) Role of AP2/EREBP transcription factors in gene regulation during abiotic stress. FEBS Lett 498(2–3):187–189
Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8:489–503
Krebs JA, Wu Y, Chang H-S, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130:2129–2141
Krishnan N, Dickman MB, Becker DF (2008) Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic Biol Med 44(4):671–681
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Lee JH, van Montagu M, Verbruggen N (1999) A highly conserved kinase is an essential component for stress tolerance in yeast and plant cells. Proc Natl Acad Sci USA 96:5873–5877
Lee YP, Kim SH, Bang JW (2007) Enhanced tolerance to oxidative stress in transgenic tobacco plants expressing three antioxidant enzymes in chloroplasts. Plant Cell Rep 26:591–598
Leidi EO, Barragán V, Rubio L, El-Hamdaoui A, Ruiz MT, Cubero B, Fernández JA, Bressan RA, Hasegawa PM, Quintero FJ, Pardo JM (2010) The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J 61(3):495–506
Li C, Puhakainen T, Welling A, Viherä-Aarnio A, Ernstsen A, Junttila O, Heino P, Palva ET (2002) Cold acclimation in silver birch (Betula pendula). Development of freezing tolerance in different tissues and climatic ecotypes. Physiol Plant 116:478–488
Li C, Junttila O, Heino P, Palva ET (2003) Different responses of northern and southern ecotypes of Betula pendula to exogenous ABA application. Tree Physiol 23:481–487
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Lu YY, Deng XP, Kwak SS (2010) Over expression of CuZn superoxide dismutase (CuZn SOD) and ascorbate peroxidase (APX) in transgenic sweet potato enhances tolerance and recovery from drought stress. Afr J Biotechnol 9(49):8378–8391
Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14(Suppl): S389–S400
Mantri NL, Ford R, Coram TE, Pang ECK (2007) Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought. BMC Genomics 8:303
Mantri NL, Ford R, Coram TE, Pang ECK (2010a) Evidence of unique and shared responses to major biotic and abiotic stresses in chickpea. Environ Exp Bot 69(3):286–292
Mantri NL, Pang ECK, Ford R (2010b) Molecular biology for stress management. In: Yadav SS, McNeil DN, Weeden N, Patil SS (eds) Climate change and management of cool season grain legume crops. Springer, Heidelberg, pp 377–408. ISBN 978-90-481-3708-4
Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38:982–993
McKersie BD, Bowley SR, Harjanto E, Leprince O (1996) Water-deficit tolerance and field performance of transgenic alfalfa overexpressing superoxide dismutase. Plant Physiol 111(4):1177–1181
McNeil SD, Nuccio ML, Ziemak MJ, Hanson AD (2001) Enhanced synthesis of choline and glycine betaine in transgenic tobacco plants that overexpress phosphoethanolamine N-methyltransferase. PNAS 98: 10001–10005
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Nakayama H, Yoshida K, Ono H, Murooka Y, Shinmyo A (2000) Ectoine, the Compatible Solute of Halomonas elongata, Confers Hyperosmotic Tolerance in Cultured Tobacco Cells. Plant Physiology 122:1239–1248
Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (1999) Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Letters 461:205–210
Neill S, Desikan R, Hancock J (2002) Hydrogen peroxide signaling. Curr Opin Plant Biol 5:388–395
Novillo F, Medina J, Salinas J (2007) Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc Natl Acad Sci USA 104:21002–21007
Nuccio ML, Russell BL, Nolte KD, Rathinasabapathi B, Gage DA, Hanson AD (1998) The endogenous choline supply limits glycine betaine synthesis in transgenic tobacco expressing choline monooxygenase. The Plant Journal 16(4):487–496
Nuccio ML, McNeil SD, Ziemak MJ, Hanson AD, Jain RK, Selvaraj G (2000) Choline Import into Chloroplasts Limits Glycine Betaine Synthesis in Tobacco: Analysis of Plants Engineered with a Chloroplastic or a Cytosolic Pathway. Metabolic Engineering 2:300–311
Orvar BL, Sangwan V, Omann F, Dhindsa RS (2000) Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant J 23:785–794
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Pardo JM, Reddy MP, Yang S (1998) Stress signaling through Ca2+/Calmodulin dependent protein phosphatase calcineurin mediates salt adaptation in plants. Proc Natl Acad Sci USA 95:9681–9683
Park EJ, Jeknic Z, Sakamoto A, DeNoma J, Yuwansiri R, Murata N, Chen TH (2004) Genetic engineering of glycinebetaine synthesis in tomato protects seeds, plants, and flowers from chilling damage. The Plant Journal 40(4):474–487
Pasapula V, Shen G, Kuppu S, Paez-Valencia J, Mendoza M, Hou P, Chen J, Qiu X, Zhu L, Zhang X, Auld D, Blumwald E, Zhang H, Gaxiola R, Payton P (2011) Expression of an Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) in cotton improves drought- and salt tolerance and increases fibre yield in the field conditions. Plant Biotechnol J 9(1):88–99
Patade VY, Suprasanna P (2010) Short-term salt and PEG stresses regulate expression of MicroRNA, miR159 in sugarcane leaves. J Crop Sci Biotechnol 13(3): 177–182
Patade VY, Bhargava S, Suprasanna P. Salt and drought tolerance of sugarcane under iso-osmotic salt and water stress: growth, osmolytes accumulation and antioxidant defense. J Plant Interact, in press
Pearce R (2001) Plant freezing and damage. Ann Bot 87:417–424
Peng Y, Reyes JL, Wei H, Yang Y, Karlson D, Covarrubias AA, Krebs SL, Fessehaie A, Arora R (2008) RcDhn5, a cold acclimation-responsive dehydrin from Rhododendron catawbiense rescues enzyme activity from dehydration effects in vitro and enhances freezing tolerance in RcDhn5-overexpressing Arabidopsis plants. Physiol Plant 134(4):583–597
Penna S (2003) Building stress tolerance through over-producing trehalose in transgenic plants. Trends Plant Sci 8(8):355–357
Prabhavathi VR, Rajam MV (2007) Polyamine accumulation in transgenic eggplant enhances tolerance to multiple abiotic stresses and fungal resistance. Plant Biotechnol 24:273–282
Ray S, Dansana PK, Bhaskar A, Giri J, Kapoor S, Khurana JP, Tyagi AK (2010) Emerging trends in functional genomics for stress tolerance in crop plants. In: Hirt H (ed) Plant stress biology: from genomics to systems biology. Wiley-VCH Verlag GmbH and Co. KGaA, Weinheim, Germany
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) microRNAs in plants. Genes Dev 16: 1616–1626
Reyes JL, Chua NH (2007) ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49:592–606
Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu GL (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110
Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61:621–649
Rodríguez-Rosales MP, Gálvez FJ, Huertas R, Aranda MN, Baghour M, Cagnac O, Venema K (2009) Plant NHX cation/proton antiporters. Plant Signal Behav 4(4):265–276
Rontein D, Basset G, Hanson AD (2002) Metabolic engineering of osmoprotectant accumulation in plants. Metab Eng 4:49–56
Roxas VP, Lodhi SA, Garrett DK, Mahan JR, Allen RD (2000) Stress tolerance in transgenic tobacco seedlings that overexpress Glutathione S-Transferase/Glutathione Peroxidase. Plant Cell Physiol 41(11): 1229–1234
Sahi S, Singh A, Blumwald E, Grover A (2006) Beyond osmolytes and transporters: novel plant stress tolerance related genes from transcriptional profiling data. Physiol Plant 127:1–9
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Sakamoto A, Murata A, Murata N (1998) Metabolic engineering of rice leading to biosynthesis of glycinebetaine and tolerance to salt and cold. Plant Mol Biol 38:1011–1019
Sanchez DH, Pieckenstain FL, Szymanski J, Erban A, Bromke M, Hannah MA, Kraemer U, Kopka J, Udvardi MK (2011) Comparative functional genomics of salt stress in related model and cultivated plants identifies and overcomes limitations to translational genomics. PLoS One 6(2):e17094
Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signalling. Plant Cell 14(Suppl):S401–S417
Sangwan V, Orvar BL, Beyerly J, Hirt H, Dhindsa RS (2002) Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J 31:629–638
Sarhadi E, Mahfoozi S, Hosseini SA, Salekdeh GH (2010) Cold acclimation proteome analysis reveals close link between the up-regulation of low-temperature associated proteins and vernalization fulfillment. J Proteome Res 9(11):5658–5667
Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13:61–72
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K (2002) Monitoring expression profile of 7000 Arabidopsis genes under drought, cold-and high-salinity stresses using a full-length cDNA microarray. Plant J 31:279–292
Seki M, Okamoto M, Matsui A, Kim JM, Kurihara Y, Ishida J, Morosawa T, Kawashima M, Kim T, Shinozaki K (2009) Microarray analysis for studying the abiotic stress responses in plants. Mol Tech Crop Improve 3:333–355
Shen B, Jensen RG, Bohnert HJ (1997) Increased resistance to oxidative stress in transgenic plants by targeting mannitol biosynthesis to chloroplasts. Plant Physiol 113:1177–1183
Sheveleva E, Chmara W, Bohnert HJ, Jensen RG (1997) Increased salt and drought tolerance by d-ononitol production in transgenic Nicotiana tabacum L. Plant Physiol 115:1211–1219
Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS I encodes a putative Na−/H+ antiporter. Proc Natl Acad Sci USA 97:6896–6901
Shinozaki K, Yamaguchi-Shinozaki K (1996) Molecular responses to drought and cold stress. Curr Opin Biotechnol 7:161–167
Shinozaki K, Yamaguchi-Shinozaki K (1997) Gene expression and signal transduction in water–stress response. Plant Physiol 115:327–334
Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular response to dehydration and low temperature: differences and cross-talk between two stress signalling pathways. Curr Opin Plant Biol 3:217–223
Shou H, Bordallo P, Fan JB, Yeakley JM, Bibikova M, Sheen J, Wang K (2004a) Expression of an active tobacco mitogen-activated protein kinase kinase kinase enhances freezing tolerance in transgenic maize. Proc Natl Acad Sci USA 101:3298–3303
Shou H, Bordallo P, Wang K (2004b) Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize. J Exp Bot 55: 1013–1019
Shukla LI, Chinnusamy V, Sunkar R (2008) The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim Biophys Acta 1779:743–748
Silva P, Facanha AR, Tavares RM, Geros H (2010) Role of tonoplast proton pumps and Na+/H+ antiport system in salt tolerance of Populus euphratica oliv. J Plant Growth Regul 29:23–34
Snedden WA, Fromm H (2001) Calmodulin as a versatile calcium signal transducer in plants. New Phytol 151:35–66
Soren KR, Ali K, Tyagi A, Tyagi V (2010) Recent developments in transgenics for abiotic stress tolerance in rice. Indian J Biotechnol 9:233–251
Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF (1998) Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA 95:14570–14575
Stitt M, Hurry V (2002) A plant for all seasons: alterations in photosynthetic carbon metabolism during cold acclimation in Arabidopsis. Curr Opin Plant Biol 5:199–206
Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94:1035–1040
Su J, Wu R (2004) Stress-inducible synthesis of proline in transgenic rice confers faster growth under stress conditions than that with constitutive synthesis. Plant Sci 166(4):941–948
Su J, Shen Q, Ho T-H, Wu R (1998) Dehydration-stress-regulated transgene expression in stably transformed rice plants. Plant Physiol 117:913–922
Sugie A, Naydenov N, Mizuno N, Nakamura C, Takumi S (2006) Overexpression of wheat alternative oxidase gene Waox1a alters respiration capacity and response to reactive oxygen species under low temperature in transgenic Arabidopsis. Genes Genet Syst 81: 349–354
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNA from Arabidopsis. Plant Cell 16:2001–2019
Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by down regulation of miR398 and important for oxidative stress tolerance. Plant Cell 18:2051–2065
Suprasanna P (2003) Building stress tolerance through over-producing trehalose in transgenic plants. Trends Plant Sci 8(8):355–357
Suprasanna P, Teixeira da Silva JA, Bapat VA (2005) Plant abiotic stress, sugars and transgenics: a perspective. In: Teixeira da Silva JA (ed) Floriculture, ornamental and plant biotechnology: advances and topical issues, 1st edn. Global Science Publishers, London, UK, pp 86–93
Suprasanna P, Sidha M, Bapat VA (2008) Integrated approaches of mutagenesis and in vitro selection for crop improvement. In: Kumar A, Shekhawat NS (eds) Plant tissue culture, molecular markers and their role in crop productivity. IK International Publishers, New Delhi, pp 73–92
Szabados L, Savoure A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15(2):89–97
Tahtiharju S, Sangwan V, Monroy AF, Dhindsa RS, Borg M (1997) The induction of kin genes in cold-acclimating Arabidopsis thaliana: evidence of a role for calcium. Planta 203:442–447
Tanaka Y, Hibino T, Hayashi Y, Tanaka A, Kishitani S, Takabe T, Yokota S, Takabe T (1999) Salt tolerance of transgenic rice overexpressing yeast mitochondrial Mn-SOD in chloroplasts. Plant Sci 148:131–138
Thakur P, Kumar S, Malik JA, Berger JD, Nayyar H (2010) Cold stress effects on reproductive development in grain crops: an overview. Environ Exp Bot 67(3):429–443
Thomashow MF (2010) Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol 154:571–577
Uemura M, Warren G, Steponkus PL (2003) Freezing sensitivity in the sfr4 mutant of Arabidopsis is due to low sugar content and is manifested by loss of osmotic responsiveness. Plant Physiol 131:1800–1807
Valliyodan B, Nguyen HT (2006) Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol 9:1–7
Verdoy D, Coba de la peña T, Redondo FJ, Lucas MM, Pueyo JJ (2006) Transgenic Medicago truncatula plants that accumulate proline display nitrogen-fixing activity with enhanced tolerance to osmotic stress. Plant Cell Environ 29:1913–1923
Vincour B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Plant Biol 16:123–132
Vogel JT, Zarka DG, van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41:195–211
Vorasoot N, Songsri P, Akkasaeng C, Jogloy S, PatanothaiA (2003) Effect of water stress on yield and agronomiccharacters of peanut. Songklanakarin J Sci Technol25(3):283–288
Vranova E, Inze D, Breusegem F (2002) Signal-transduction during oxidative stress. J Exp Bot 53: 1227–1236
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218: 1–14
Welling A, Rinne P, Vihera-Aarnio A, Kontunen-Soppela S, Heino P, Palva ET (2004) Photoperiod and temperature differentially regulate the expression of two dehydrin genes during overwintering of birch (Betula pubescens Ehrh.). J Exp Bot 55:507–516
Winicov I, Bastola DR (1999) Transgenic over expression of the transcription factor Alfin1 enhances expression of the endogenous MsPRP2 gene in alfalfa and improves salinity tolerance of the plants. Plant Physiol 120:473–480
Xu J, Zhang Y, Guan Z, Wei W, Han L, Chai T (2008) Expression and function of two dehydrins under environmental stresses in Brassica juncea L. Mol Breed 21(4):431–438
Yamada M, Morishita H, Urano K, Shiozaki N, Yamaguchi-Shinozaki K, Shinozaki K, Yoshiba Y (2005) Effects of free proline accumulation in petunias under drought stress. J Exp Bot 56:1975–1981
Ye CY, Zhang HC, Chen JH, Xia XL, Yin WL (2009) Molecular characterization of putative vacuolar NHX-type Na(+)/H(+) exchanger genes from the salt-resistant tree Populus euphratica. Physiol Plant 137(2): 166–174
Yeo ET, Kwon HB, Han SE, Lee JT, Ryu JC, Byu MO (2000) Genetic engineering of drought resistant potato plants by introduction of the trehalose-6-phosphate synthase (TPS1) gene from Saccharomyces cerevisiae. Mol Cells 10:263–268
Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19:765–768
Zhang JZ, Creelman RA, Zhu JK (2004) From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol 135:615–621
Zhang T, Wang Q, Chen X, Tian C, Wang X, Xing T, Li Y, Wang Y (2005) Cloning and biochemical properties of CDPK gene OsCDPK14 from rice. J Plant Physiol 162(10):1149–1159
Zhao BT, Liang RQ, Ge LF, Li W, Xiao HS, Lin HX, Ruan KC, Jin YX (2007) Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun 354:585–590
Zhou X, Wang G, Sutoh K, Zhu JK, Zhang W (2008) Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim Biophys Acta 1779:780–788
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Zhu B, Coleman GD (2001) Phytochrome-mediated photoperiod perception, shoot growth, glutamine, calcium, and protein phosphorylation influence the activity of the poplar bark storage protein gene promoter (bspA). Plant Physiol 126:342–351
Zhu J, Dong CH, Zhu JK (2007) Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr Opin Plant Biol 10:290–295
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Mantri, N., Patade, V., Penna, S., Ford, R., Pang, E. (2012). Abiotic Stress Responses in Plants: Present and Future. In: Ahmad, P., Prasad, M. (eds) Abiotic Stress Responses in Plants. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-0634-1_1
Download citation
DOI: https://doi.org/10.1007/978-1-4614-0634-1_1
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-0633-4
Online ISBN: 978-1-4614-0634-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)