Abstract
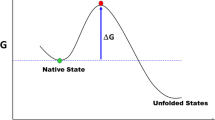
The analysis of protein folding reactions by monitoring the kinetic effects of specifically designed single-point mutations, the so-termed phi-value analysis, has been a favorite technique to experimentally probe the mechanisms of protein folding. The idea behind phi-value analysis is that the effects that mutations have on the folding and unfolding rate constants report on the energetic/structural features of the folding transition state ensemble (TSE), which is the highest point in the free energy surface connecting the native and unfolded states, and thus the rate limiting step that ultimately defines the folding mechanism. For single-domain, two-state folding proteins, the general procedure to perform the phi-value analysis of protein folding is relatively simple to implement in the lab. Once the mutations have been produced and purified, the researcher needs to follow a few specific guidelines to perform the experiments and to analyze the data so produced. In this chapter, a step-by-step description of how to measure and interpret the effects induced by site-directed mutations on the folding and unfolding rate constants of a protein of interest is provided. Some possible solutions to the most typical problems that arise when performing phi-value analysis in the lab are also provided.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Matouschek A, Kellis JT Jr, Serrano L, Fersht AR (1989) Mapping the transition state and pathway of protein folding by protein engineering. Nature 340(6229):122–126
Onuchic JN, Wolynes PG (2004) Theory of protein folding. Curr Opin Struct Biol 14(1):70–75
Campos LA, Sadqi M, Liu J, Wang X, English DS, Muñoz V (2013) Gradual disordering of the native state on a slow two-state folding protein monitored by single-molecule fluorescence spectroscopy and NMR. J Phys Chem B 117(42):13120–13131
Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG (1995) Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins 21(3):167–195
Muñoz V, Campos LA, Sadqi M (2016) Limited cooperativity in protein folding. Curr Opin Struct Biol 36:58–66
Matthews CR (1987) Effect of point mutations on the folding of globular proteins. Methods Enzymol 154:498–511
Jackson SE, Fersht AR (1991) Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry 30(43):10428–10435
Naganathan AN, Muñoz V (2010) Insights into protein folding mechanisms from large scale analysis of mutational effects. Proc Natl Acad Sci U S A 107(19):8611–8616
Goldenberg DP, Frieden RW, Haack JA, Morrison TB (1989) Mutational analysis of a protein-folding pathway. Nature 338(6211):127–132
Serrano L, Matouschek A, Fersht AR (1992) The folding of an enzyme. III. Structure of the transition state for unfolding of barnase analysed by a protein engineering procedure. J Mol Biol 224(3):805–818
Akmal A, Muñoz V (2004) The nature of the free energy barriers to two-state folding. Proteins 57(1):142–152
Sanchez IE, Kiefhaber T (2003) Origin of unusual phi-values in protein folding: evidence against specific nucleation sites. J Mol Biol 334(5):1077–1085
Li L, Mirny LA, Shakhnovich EI (2000) Kinetics, thermodynamics and evolution of non-native interactions in a protein folding nucleus. Nat Struct Biol 7(4):336–342
Viguera AR, Vega C, Serrano L (2002) Unspecific hydrophobic stabilization of folding transition states. Proc Natl Acad Sci U S A 99(8):5349–5354
Ventura S, Vega MC, Lacroix E, Angrand I, Spagnolo L, Serrano L (2002) Conformational strain in the hydrophobic core and its implications for protein folding and design. Nat Struct Biol 9(6):485–493
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18(15):2714–2723
DeLano WL (2002) Pymol: an open-source molecular graphics tool. CCP4 Newslet Protein Crystallograph 40:82–92
Raleigh DP, Plaxco KW (2005) The protein folding transition state: what are Phi-values really telling us? Protein Pept Lett 12(2):117–122
Itzhaki LS, Otzen DE, Fersht AR (1995) The structure of the transition state for folding of chymotrypsin inhibitor 2 analysed by protein engineering methods: evidence for a nucleation-condensation mechanism for protein folding. J Mol Biol 254(2):260–288
Burton RE, Huang GS, Daugherty MA, Calderone TL, Oas TG (1997) The energy landscape of a fast-folding protein mapped by Ala-->Gly substitutions. Nat Struct Biol 4(4):305–310
Villegas V, Martinez JC, Aviles FX, Serrano L (1998) Structure of the transition state in the folding process of human procarboxypeptidase A2 activation domain. J Mol Biol 283(5):1027–1036
Chiti F, Taddei N, White PM, Bucciantini M, Magherini F, Stefani M, Dobson CM (1999) Mutational analysis of acylphosphatase suggests the importance of topology and contact order in protein folding. Nat Struct Biol 6(11):1005–1009
Fulton KF, Main ER, Daggett V, Jackson SE (1999) Mapping the interactions present in the transition state for unfolding/folding of FKBP12. J Mol Biol 291(2):445–461
Kragelund BB, Osmark P, Neergaard TB, Schiodt J, Kristiansen K, Knudsen J, Poulsen FM (1999) The formation of a native-like structure containing eight conserved hydrophobic residues is rate limiting in two-state protein folding of ACBP. Nat Struct Biol 6(6):594–601
Martinez JC, Serrano L (1999) The folding transition state between SH3 domains is conformationally restricted and evolutionarily conserved. Nat Struct Biol 6(11):1010–1016
Riddle DS, Grantcharova VP, Santiago JV, Alm E, Ruczinski I, Baker D (1999) Experiment and theory highlight role of native state topology in SH3 folding. Nat Struct Biol 6(11):1016–1024
Ternstrom T, Mayor U, Akke M, Oliveberg M (1999) From snapshot to movie: phi analysis of protein folding transition states taken one step further. Proc Natl Acad Sci U S A 96(26):14854–14859
Choe SE, Li L, Matsudaira PT, Wagner G, Shakhnovich EI (2000) Differential stabilization of two hydrophobic cores in the transition state of the villin 14T folding reaction. J Mol Biol 304(1):99–115
Hamill SJ, Steward A, Clarke J (2000) The folding of an immunoglobulin-like Greek key protein is defined by a common-core nucleus and regions constrained by topology. J Mol Biol 297(1):165–178
Guerois R, Serrano L (2000) The SH3-fold family: experimental evidence and prediction of variations in the folding pathways. J Mol Biol 304(5):967–982
Kim DE, Fisher C, Baker D (2000) A breakdown of symmetry in the folding transition state of protein L. J Mol Biol 298(5):971–984
McCallister EL, Alm E, Baker D (2000) Critical role of beta-hairpin formation in protein G folding. Nat Struct Biol 7(8):669–673
Rodriguez HM, Vu DM, Gregoret LM (2000) Role of a solvent-exposed aromatic cluster in the folding of Escherichia coli CspA. Protein Sci 9(10):1993–2000
Chu R, Pei W, Takei J, Bai Y (2002) Relationship between the native-state hydrogen exchange and folding pathways of a four-helix bundle protein. Biochemistry 41(25):7998–8003
Northey JG, Di Nardo AA, Davidson AR (2002) Hydrophobic core packing in the SH3 domain folding transition state. Nat Struct Biol 9(2):126–130
Otzen DE, Oliveberg M (2002) Conformational plasticity in folding of the split beta-alpha-beta protein S6: evidence for burst-phase disruption of the native state. J Mol Biol 317(4):613–627
Friel CT, Capaldi AP, Radford SE (2003) Structural analysis of the rate-limiting transition states in the folding of Im7 and Im9: similarities and differences in the folding of homologous proteins. J Mol Biol 326(1):293–305
Gianni S, Guydosh NR, Khan F, Caldas TD, Mayor U, White GW, DeMarco ML, Daggett V, Fersht AR (2003) Unifying features in protein-folding mechanisms. Proc Natl Acad Sci U S A 100(23):13286–13291
Garcia-Mira MM, Boehringer D, Schmid FX (2004) The folding transition state of the cold shock protein is strongly polarized. J Mol Biol 339(3):555–569
Hedberg L, Oliveberg M (2004) Scattered Hammond plots reveal second level of site-specific information in protein folding: phi' (beta++). Proc Natl Acad Sci U S A 101(20):7606–7611
Anil B, Sato S, Cho JH, Raleigh DP (2005) Fine structure analysis of a protein folding transition state; distinguishing between hydrophobic stabilization and specific packing. J Mol Biol 354(3):693–705
Went HM, Jackson SE (2005) Ubiquitin folds through a highly polarized transition state. Protein Eng Des Sel 18(5):229–237
Wilson CJ, Wittung-Stafshede P (2005) Role of structural determinants in folding of the sandwich-like protein Pseudomonas aeruginosa azurin. Proc Natl Acad Sci U S A 102(11):3984–3987
Teilum K, Thormann T, Caterer NR, Poulsen HI, Jensen PH, Knudsen J, Kragelund BB, Poulsen FM (2005) Different secondary structure elements as scaffolds for protein folding transition states of two homologous four-helix bundles. Proteins 59(1):80–90
Bueno M, Ayuso-Tejedor S, Sancho J (2006) Do proteins with similar folds have similar transition state structures? A diffuse transition state of the 169 residue apoflavodoxin. J Mol Biol 359(3):813–824
Petrovich M, Jonsson AL, Ferguson N, Daggett V, Fersht AR (2006) Phi-analysis at the experimental limits: mechanism of beta-hairpin formation. J Mol Biol 360(4):865–881
Ferguson N, Sharpe TD, Johnson CM, Fersht AR (2006) The transition state for folding of a peripheral subunit-binding domain contains robust and ionic-strength dependent characteristics. J Mol Biol 356(5):1237–1247
Li Y, Gupta R, Cho JH, Raleigh DP (2007) Mutational analysis of the folding transition state of the C-terminal domain of ribosomal protein L9: a protein with an unusual beta-sheet topology. Biochemistry 46(4):1013–1021
Sato S, Fersht AR (2007) Searching for multiple folding pathways of a nearly symmetrical protein: temperature dependent phi-value analysis of the B domain of protein A. J Mol Biol 372(1):254–267
Campbell-Valois FX, Michnick SW (2007) The transition state of the ras binding domain of Raf is structurally polarized based on Phi-values but is energetically diffuse. J Mol Biol 365(5):1559–1577
Sharpe TD, Ferguson N, Johnson CM, Fersht AR (2008) Conservation of transition state structure in fast folding peripheral subunit-binding domains. J Mol Biol 383(1):224–237
Campos LA, Bueno M, Lopez-Llano J, Jimenez MA, Sancho J (2004) Structure of stable protein folding intermediates by equilibrium phi-analysis: the apoflavodoxin thermal intermediate. J Mol Biol 344(1):239–255
Krantz BA, Sosnick TR (2001) Engineered metal binding sites map the heterogeneous folding landscape of a coiled coil. Nat Struct Biol 8(12):1042–1047
Sosnick TR, Krantz BA, Dothager RS, Baxa M (2006) Characterizing the protein folding transition state using psi analysis. Chem Rev 106(5):1862–1876
Fersht AR, Sato S (2004) Phi-value analysis and the nature of protein-folding transition states. Proc Natl Acad Sci U S A 101(21):7976–7981
Nolting B, Andert K (2000) Mechanism of protein folding. Proteins 41(3):288–298
de los Rios MA, Muralidhara BK, Wildes D, Sosnick TR, Marqusee S, Wittung-Stafshede P, Plaxco KW, Ruczinski I (2006) On the precision of experimentally determined protein folding rates and phi-values. Protein Sci 15(3):553–563
Mok YK, Elisseeva EL, Davidson AR, Forman-Kay JD (2001) Dramatic stabilization of an SH3 domain by a single substitution: roles of the folded and unfolded states. J Mol Biol 307(3):913–928
Northey JG, Maxwell KL, Davidson AR (2002) Protein folding kinetics beyond the phi value: using multiple amino acid substitutions to investigate the structure of the SH3 domain folding transition state. J Mol Biol 320(2):389–402
Lawrence C, Kuge J, Ahmad K, Plaxco KW (2010) Investigation of an anomalously accelerating substitution in the folding of a prototypical two-state protein. J Mol Biol 403(3):446–458
Muñoz V, Cerminara M (2016) When fast is better: protein folding fundamentals and mechanisms from ultrafast approaches. Biochem J 473(17):2545–2559
Jackson SE, Fersht AR (1991) Folding of chymotrypsin inhibitor 2. 2. Influence of proline isomerization on the folding kinetics and thermodynamic characterization of the transition state of folding. Biochemistry 30(43):10436–10443
Sanchez IE, Kiefhaber T (2003) Evidence for sequential barriers and obligatory intermediates in apparent two-state protein folding. J Mol Biol 325(2):367–376
Kaya H, Chan HS (2003) Origins of chevron rollovers in non-two-state protein folding kinetics. Phys Rev Lett 90(25 Pt 1):258104
de Los Rios MA, Plaxco KW (2005) Apparent Debye-Huckel electrostatic effects in the folding of a simple, single domain protein. Biochemistry 44(4):1243–1250
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Campos, L.A. (2022). Mutational Analysis of Protein Folding Transition States: Phi Values. In: Muñoz, V. (eds) Protein Folding. Methods in Molecular Biology, vol 2376. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1716-8_1
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1716-8_1
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1715-1
Online ISBN: 978-1-0716-1716-8
eBook Packages: Springer Protocols