Abstract
Gametophytic self-incompatibility (GSI), which prevents growth of a pollen tube through the style, provides a means of preventing self-pollination. Seen in most eudicot plant families, GSI in the genus Petunia was described by Darwin in the 19th century. By the time the first edition of this monograph was published in 1984, nearly a century later, most of the readily observable phenomena associated with self-incompatibility in the genus Petunia had been described and, as in a number of other plant systems, it had been demonstrated to depend on the actions of genes encoded at a single highly polymorphic S-locus. Molecular research of the past two decades has provided a depth of understanding into the mechanisms underlying the earlier observations, particularly in the identification of a number of SI-associated genes, their sites of action, and to some extent the mechanisms involved. This chapter summarizes what has been learned, with a focus on the molecular biology underlying GSI in Petunia, and highlights the major questions that remain unanswered.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
....protected flowers with their own pollen placed on the stigma never yielded nearly a full complement of seed; whilst those left uncovered produced fine capsules, showing that pollen from other plants must have been brought to them, probably by moths. Plants growing vigorously and flowering in pots in the green-house, never yielded a single capsule; and this may be attributed, at least in chief part, to the exclusion of moths (Darwin 1891).
Self-incompatibility (SI), the phenomenon by which plants can recognize “self ” pollen and therefore prevent inbreeding, while accepting “non-self” pollen, has been the subject of study ever since Darwin first described his observations of self- and cross-fertilization in Petunia in his book The Effects of Self and Cross Fertilization in the Vegetable Kingdom (Darwin 1891). At the time that the first Petunia monograph was published (Sink 1984), much of the essential phenomenology of gametophytic self-incompatibility (GSI) had been described. Researchers such as Mather (1943), Linskens (1975), and de Nettancourt (1977) had determined that GSI in Petunia was governed by a single, multiallelic S-locus, and that recognition and rejection of self pollen was controlled gametophytically by alleles expressed in pollen. Mutations unilaterally inactivating self-incompatibility in pollen (pollen-part mutations) had been identified and associated with centric chromosomal fragments (Brewbaker and Natarajan 1960). Tetraploid plants with diploid heteroallelic pollen had been shown to be self-compatible due to “competitive interaction” in pollen. Shivanna and Rangaswamy (1969) had demonstrated that pollination of immature styles could be used to overcome self-incompatibility (a phenomenon now understood to result from high-level expression of the S-RNase late in the development of the style). Ascher (1984) had demonstrated quantitative variation in the strength of the self-incompatibility reaction, which he termed pseudo-self-compatibility. In the ensuing years, research in this area has resulted in a far better understanding of the molecular biology underlying many of the observations just described.
A variety of experimental approaches resulted in identification of the S-ribonuclease (S-RNase) as the style-recognition component of gametophytic SI, acting together with both the previously elusive “pollen-S” gene and a number of other genes that play critical, supporting, or yet unclear roles in this response. Despite the enormous progress that has been made in understanding the molecular basis of pollen recognition and rejection, many of the fundamental aspects of gametophytic self-incompatibility remain to be fully deciphered. The most widely accepted model for S-RNase-based incompatibility proposes that self and non-self S-RNase proteins are imported into growing pollen tubes. In a compatible pollination, non-self S-RNases are inhibited from acting whereas in incompatible pollinations, self S-RNases act to degrade pollen RNA and inhibit growth. Recognition of S-RNases as self or non-self is determined via the action of a pollen-expressed S-locus-encoded F-box protein, SLF. What remains elusive, however, is how haplotype recognition (determined by the S-RNase and by the SLF protein) is integrated with either the release or continued inhibition of S-RNase activity.
1.1 Genetics, Physiology, and Distribution of GSI
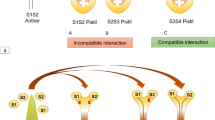
Gametophytic self-incompatibility has been estimated to occur in up to three-quarters of eudicot families (Igic and Kohn 2001; Steinbachs and Holsinger 2002). The most widespread form of GSI, as found in Petunia hybrida, is based on the interaction of style- and pollen-expressed allelic proteins encoded by a single, multiallelic S-locus. The style and pollen components together form a recognition “haplotype” (two or more tightly linked allelic variants). Recognition and rejection of pollen depend on whether there is a match of haplotypes between the growing pollen tube and the style. If the haplotype expressed in the pollen (“pollen-S”) matches one of the two S-haplotypes expressed in the style (an incompatible cross), growth of the pollen tube is inhibited in the transmitting tract, and fertilization rarely occurs. Conversely, if there is no match between the haplotype expressed in the pollen and that expressed in the style (compatible cross), pollen tube growth continues to the ovary, resulting in fertilization and seed set. In most cases, compatible versus incompatible crosses can be distinguished on the basis of seed capsule formation. In compatible crosses, large seed capsules that may contain up to a few hundred seeds are formed. In a fully incompatible cross, no seed capsules are formed, and no seed is produced. An alternative method of assaying pollination success is the use of fluorescence microscopy, traditionally using aniline blue as a fluorochrome, to monitor the extent of growth of pollen tubes. Aniline blue stains callose, a β–1, 3 glucan found in pollen tubes, and fluoresces with UV illumination. During incompatible pollinations, the majority of pollen tubes terminate growth in the upper third of the style, whereas in compatible pollinations pollen tubes reach the ovules.
As will be described more completely below, it is now understood that a self-incompatibility haplotype is defined by the presence of a specific S-RNase allele and a matching SLF allele linked at the same S-locus and inherited as a single unit due to a local suppression of recombination. A number of other proteins, including HT-B, the 120 kDa protein, SBP1, and SSK1, have been shown or hypothesized to play critical roles in the expression of the self-incompatibility response. The known and/or hypothesized roles of these genes and proteins are described in the sections below.
Gametophytic self-incompatibility has been well studied not only in Petunia, but also in other members of the Solanaceae, as well as in the Plantaginaceae and Rosaceae (Igic and Kohn 2001; Olmstead et al. 2001). Although this chapter focuses on the study of GSI in Petunia, much of our current understanding of this response comes from experiments involving other genera of the Solanaceae, such as Nicotiana, and Solanum, as well as from Antirrhinum (Plantaginaceae) and Prunus (Rosaceae). Experimental results from these systems will be discussed as appropriate. Where similar experiments have been performed in both Petunia and other plant systems, in most cases only the Petunia experiments are mentioned.
1.2 Early History of SI in Petunia
Although Darwin (1891) described the essential features of self-sterility in Petunia, it was not until later that Harland and Atteck (1933) established that a gametophytic mechanism controls the specificity of the pollen. Mather (1943) questioned the identity of the Petunia material used in some of these early studies and considered that it was probably P. hybrida rather than P. violacea (synonymous with P. integrifolia; see Chapter 1 and references therein). The current consensus is that P. hybrida has arisen from hybridization between the purple-flowered SI species P. integrifolia and the white-flowered self-compatible (SC) species P. axillaris. These interspecific crosses exhibit unilateral incompatibility (UI) and are successful only with P. axillaris as the female parent (Mather 1943). This is typical of UI that has been described in other genera of the Solanaceae. A further important discovery in these early studies was the phenomenon of breakdown in SI that occurs when a diploid undergoes colchicine-induced tetraploidy (Stout and Chandler 1941).
Several authors have commented on the suitability of P. hybrida and its wild relatives for SI research (Linskens 1975; Ascher 1984). These features include a generally strong SI reaction leading to the absence of seed set, although this can conveniently be overcome by bud pollination. The latter technique allows homozygous stocks to be established and maintained by either bud pollination or vegetative propagation. The large floral organs facilitate controlled pollinations but are also advantageous for biochemical or physiological studies. This has allowed detailed studies of pollen tube growth rates in both compatible and incompatible pollinations (Herrero and Dickinson 1980, 1981). These indicated that the difference in growth rate occurs in the stylar transmitting tissue, consistent with what is now known about the distribution of S-RNase. For a detailed discussion of these and other cytological studies the reader should refer to Ascher (1984).
1.3 Pseudo-Self-Compatibility, Partial Breakdown of SI
In early studies using Petunia hybrida, Ascher and coworkers revealed considerable variation in the responses of different GSI plants (Ascher 1984). Detailed genetic studies have shown that self-compatibility can arise because either the maternal or the paternal aspects became nonfunctional. Ascher coined the term “non-discriminating styles” and “pollen-mediated pseudo-self-compatibility” to describe each of these conditions (Flaschenreim and Ascher 1979a, b). However, there is also a state that is intermediate between the two extremes, in which some seed set occurs. Described by Ascher as “Pseudo-self compatibility” (PSC), the phenomenon has been widely studied in a number of systems. In one such study in Nemesia, part of the system that causes this self-compatibility could not be described as being attributed to either the pollen or the style (Robacker and Ascher 1982). A similar intermediate level of response has also been described in Senecio (Hiscock 2000).
Ascher defined PSC as the ability of an otherwise self-incompatible plant to set seed when self-fertilized or crossed to other individuals bearing the same S-allele. This definition distinguished the partial breakdown of self-incompatibility found in PSC plants from the compatibility seen in plants lacking an SI system. Ascher further defined a quantitative measure of PSC, percent PSC, by taking the ratio of the number of seeds produced in an incompatible cross to that produced in a fully compatible cross using the same individual. By expressing SI behavior as % PSC, Ascher could distinguish the partial breakdown of SI from generalized effects on fertility. Because all of Ascher’s studies took place prior to the identification and cloning of specific genes that govern self-incompatibility interactions, the molecular basis of PSC behavior has not been fully described. It is likely that the different levels of stylar-based PSC described (Flaschenreim and Ascher 1979b; Dana and Ascher 1986b) resulted from reduced expression or activity of the S-RNase protein in styles. The molecular basis of pollen tube–expressed PSC (Flaschenreim and Ascher 1979a; Dana and Ascher 1986a) is yet to be elucidated, and will be an interesting area of investigation now that the pollen-recognition component of GSI has been identified.
In a study that attempted to determine whether PSC in Petunia hybrida resulted from the hybrid origin of cultivated Petunia, Dana and Ascher (1985) selected for the presence of PSC in Petunia integrifolia plants grown from seeds collected in the wild. They found that individual self-pollinated P. integrifolia plants were capable of expressing greater than 20% PSC (20% of the number of seeds produced in a fully compatible cross). Thus, whatever the molecular basis of the partial breakdown of self-incompatibility, it does not appear to have arisen as an artifact of the hybrid cross(es) between P. integrifolia and P. axillaris that resulted in P. hybrida. PSC or full SC is widespread in cultivated P. hybrida and in some cases appears to be associated with a particular S-allele, S O (Ai, Kron, and Kao 1991; Robbins, Harbord, Sonneveld, and Clarke 2000).
1.4 Early Biochemical Studies of SI
The early characterization of S-proteins in the pistils of Nicotiana alata plants of defined S-genotypes paved the way for the subsequent isolation of the first cDNAs encoding S-proteins in the Solanaceae (Anderson et al. 1986). In Petunia hybrida, similar studies by Kamboj and Jackson (1986) identified electrophoretic variants of abundant pistil proteins that correlated with different S-genotypes. These proteins were basic (pI 8.3–8.7) but no N-terminal sequence was reported. In a subsequent study using S-alleles obtained from H. Linskens at the University of Nijmegen, a similar range of pI values (8.7–9.3) was reported (Broothaerts et al. 1989). S-proteins were identified that cosegregated with three S-alleles (S 1–S 3) and were shown to accumulate in the stigma and style during flower development, peaking at anthesis. Protein purification allowed for the recovery of N-terminal sequences of all three alleles, providing clear evidence of amino acid differences (Table 5.1). These S-proteins were subsequently shown to be glycosylated, and the apparent differences in MW (28–32 kD) could be accounted for by variations in the number and length of the carbohydrate side chains (Broothaerts, Vanvinckenroye, Decock, Van Damme, and Vendrig 1991). Similar patterns of glycosylation have been established for the S-proteins of Nicotiana alata (Woodward, Bacic, Jahnen, and Clarke 1989).
2 S-RNase: The Style-Recognition Component
The first cDNA sequences to be reported for S-proteins in Petunia were for P. hybrida (Clark et al. 1990) and P. inflata (Ai et al. 1990). These sequences revealed a similarity with the T2-type of fungal ribonucleases initially observed in Nicotiana alata in a work that led to the term “S-RNase.” The S-RNase gene is expressed at high levels late during the development of the pistil (Clark, Okuley, Collins, and Sims 1990), and encodes a secreted protein that accumulates to high levels in the transmitting tract of the style (Anderson et al. 1989; Ai et al. 1990). Comparative sequence analysis of S-RNase genes isolated from a number of species (Anderson et al. 1989; Ai et al. 1990; Clark et al. 1990; Ioerger, Gohlke, Xu, and Kao 1991; Xue, Carpenter, Dickinson, and Coen 1996; Ishimizu, Shinkawa, Sakiyama, and Norioka 1998) demonstrated that S-RNase proteins show a regular pattern of interspersion of highly conserved amino acid sequence with more variable sequence. Conserved domains C2 and C3 contain two histidine residues, His32 and His91, that along with Lys90 make up the catalytic site of the ribonuclease (Ida et al. 2001). S-RNase proteins in the Solanaceae and Plantaginaceae contain two highly variable sequence domains, HVa and HVb (Ioerger et al. 1991; Xue et al. 1996). Gain-of-function experiments (Lee, Huang, and Kao 1994) in which a S3-RNase of Petunia inflata was transferred to a plant of the S 1 S 2 genotype showed that transgenic plants expressing the S3 protein at levels comparable to endogenous S-RNase had acquired the ability to reject S 3 pollen. Lee et al. (1994) also used an antisense approach to downregulate the S3-RNase in a S 2 S 3 background. Plants with reduced levels of S3-RNase were no longer capable of inhibiting S 3 -pollen. In a subsequent experiment, McCubbin, Chung, and Kao (1997) introduced a RNase- (H93R) variant of the S3 S-RNase of Petunia inflata into an S 2 S 3 background. The resulting transgenic S 2 S 3 (S3H93R) plant demonstrated a dominant-negative phenotype that affected only the S3 allele; when self-pollinated, the transgenic plant was self-compatible. Crosses using pollen from other testers indicated that the dominant-negative transgenic plant had lost the ability to reject S 3 pollen but was unaffected in its ability to reject S 2 pollen. This result suggests that the S 3H93R allele somehow blocks an interaction or prevents the normal function of the S 3 allele. In all of these experiments, only the style recognition was altered. Pollen-recognition specificity was not affected, confirming that a separate gene product from the S-RNase encoded the “pollen-S” component.
The ribonuclease activity of the S-RNase is correlated with pollen rejection. McClure, Gray, Anderson, and Clarke (1990) labeled pollen RNA in vivo by watering plants with a solution containing 32P-orthophosphate, and showed that incompatible pollinations were associated with degradation of pollen-tube RNA, whereas pollen tube RNA was not degraded following compatible pollination. Both transgenic experiments (Huang, Lee, Karunanandaa, and Kao 1994) and analysis of spontaneous mutants (Royo et al. 1994) demonstrated that eliminating the catalytic ribonuclease activity of the S-RNase (e.g., by mutation of the active site histidine to asparagine), also eliminated the ability to reject pollen.
S-RNase proteins are glycoproteins, and show variability in the number, type, and fine structure of glycan chains associated with S-RNases (Woodward et al. 1989). The carbohydrate group does not appear to be essential for self-incompatibility, as elimination of the glycosylation site has no effect on the ability of a S-RNase to reject self pollen (Karunanandaa, Huang, and Kao 1994).
2.1 Basis of Recognition Specificity of the S-RNase
Experiments investigating the basis for allelic specificity in the S-RNase protein have generally focused on the role of the hypervariable regions. An experiment in Solanum chacoense appeared to provide strong evidence that the hypervariable regions were both necessary and sufficient for the specificity (Matton et al. 1997). The S 11 and S 13 S-RNase alleles of S. chacoense differ by only 10 amino acids across the entire protein, three of which are found in HVa and one in HVb. Matton et al. (1997) used in vitro mutagenesis to change the four S11 residues in the HVa and HVb regions to those found in S13, then expressed the altered allele transgenically in an S 12 S 14 background. Pollination with S 11 and S 13 pollen demonstrated that changing only these residues changed the recognition specificity of the transferred S-RNase from S 11 to S 13 . In an extension of this experiment (Matton et al. 1999), changing only two residues in HVa plus the HVb residue, resulted in a “dual-specificity” S-RNase that retained the ability to reject S 11 pollen while acquiring the ability to also reject S 13 pollen. Other experiments, however, have suggested that this may not be the outcome in all cases. Zurek, Mou, Beecher, and McClure (1997) made constructs exchanging the complete HVa and HVb domains of the S A2 and SC10 S-RNase alleles of Nicotiana alata. When expressed transgenically, the resulting protein had lost the ability to reject S A2 pollen, while not acquiring the ability to reject S C10 pollen, suggesting that protein regions outside of the hypervariable domains play a role in recognition.
The protein crystal structure has been determined for the SF11 S-RNase of Nicotiana alata (Ida et al. 2001) and provides support for the involvement of the hypervariable domains in allelic recognition and interaction. In Nicotiana the two hypervariable regions are separated in the primary amino acid sequence but located next to each other on the surface of the tertiary structure (Ida et al. 2001). The HVa region is further characterized by a cluster of positively charged side chains, whereas the HVb region has a cluster of negative charges. Comparative sequence analysis showed that the most highly variable amino acids in the hypervariable regions are located on the surface of the SF11 S-RNase and readily accessible to solvent (Ida et al. 2001). These include all four of the residues equivalent to those targeted in the mutagenesis experiments of Matton et al. (1997, 1999).
2.2 Allelic Diversity of S-RNases
Allelic diversity at the S-locus in Petunia hybrida is much more limited than that of the wild species from which it is derived: P. axillaris and P. integrifolia (see Chapter 1). The number of S-alleles found in the cultivated forms of P. hybrida is probably fewer than 10 (Robbins et al. 2000), compared to that of the natural populations in South America, which may have 40 or more distinct S-alleles (Tsukamoto et al. 2003). This reflects the bottleneck of plant breeding in which relatively few individuals were used in the initial interspecific hybridizations. Moreover, the low allelic diversity suggests that subsequent hybridizations and introgressions from wild relatives have been relatively infrequent during the 150 years or so of P. hybrida cultivation.
The estimate of the minimum number of S-alleles in P. hybrida is most easily based on the number of S-RNase sequences rather than on phenotypic assays based on pollinations (Robbins et al. 2000). Broothaerts and coworkers reported the N-terminal sequences for purified S-RNases derived from Linsken’s original three S-alleles, S 1 , S 2 , and S 3 (Broothaerts et al. 1989). They also reported an S-allele derived from a commercial variety which was named S b (Broothaerts et al. 1991). Sims and coworkers were the first to report cDNA sequences for three S-RNases from P. hybrida, derived from stocks originally described by Ascher (Clark et al. 1990). Rather confusingly, the Ascher S-alleles were also named S 1 –S 3 and yet sequence comparisons indicated that these were distinct from the Linskens alleles. Robbins et al. (2000) proposed that the Linskens S-alleles be given the suffix “L” to avoid future confusion. It is also important to distinguish these S-alleles from the S 1 –S 3 alleles of the species P. integrifolia ssp. inflata.
To this initial set of seven P. hybrida S-alleles (S 1 –S 3 , S 1L –S 3L , S b ) should also be added a functional allele Sx, derived from crosses between P. hybrida and P. inflata (Ai, Tsai, and Kao 1992). The S x allele was derived from the P. hybrida parent and was conditionally functional depending on background modifiers that segregated from the P. hybrida parent (Ai et al. 1991). One additional functional allele was identified from the self-incompatible stock V13 maintained by the Free University of Amsterdam (Harbord, Napoli, and Robbins 2000). Surprisingly, this stock had been maintained as an inbred line (perhaps by inadvertent early bud pollination), yet it was homozygous for a novel S-RNase sequence (Robbins et al. 2000). This allele has also been identified in an independent cultivar by Entani et al. (1999). Table 5.1 presents the N-terminal regions for all nine reported P. hybrida S-alleles and of S 1–S 3 of P. inflata for comparison. The cDNA sequences are available for all alleles except S 1L and S 2L, and these alleles may no longer be available. It is possible that they are equivalent to extant alleles, as the N-terminal sequences of S 1L and S x and of S 2L and Sv are identical. It follows that the number of distinct functional S-alleles identified in P. hybrida may be as few as seven.
The number of S-alleles identified in natural populations of Petunia has not been rigorously determined at the level of population genetics. One study of a single population of 100 individuals of P. inflata from Argentina identified 19 different S-haplotypes (Wang, Hughes, Tsukamoto, Ando, and Kao 2001). The cDNA sequence was obtained for 15 haplotypes maintained as bud self-homozygotes. Surprisingly, one of these was identical to the S 1 allele that has been extensively studied in transgenic plants (Ai et al. 1990). Phylogenetic analysis of the P. inflata S-RNase sequences revealed a trans-specific pattern of similarity with S-RNase sequences from other members of the Solanaceae, as demonstrated previously for a small sample of alleles (Ioerger, Clark, and Kao 1990). Natural populations of P. axillaris have been found to contain as many as 40 different haplotypes, although the situation is complicated by self-compatibility that can occur as a result of loss of style or pollen function (Tsukamoto et al. 2003).
2.3 RNase Mapping Studies
The S-locus has been mapped near the centromere of chromosome I in two closely related members of the Solanaceae, Lycopersicon/Solanum esculentum (n=12) (Tanksley and Loaiza-Figueroa 1985) and Solanum tuberosum (Gebhardt et al. 1991). In Petunia hybrida (n=7) an indirect approach was taken, using fluorescence in situ hybridization (FISH) localization of T-DNA inserts (ten Hoopen, Harbord, Maes, Nanninga, and Robbins 1998) that were known to be linked to the S-locus (Harbord et al. 2000). This approach physically mapped the S-locus in the V26 cultivar to a sub-centromeric region of chromosome III. In a more direct approach, Entani et al. (1999) also employed FISH to localize the SB1-RNase gene of P. hybrida to a sub-centromeric region of chromosome III in a SI line “PB”.
These two results are contradicted by an independent RFLP study by Strommer, Gerats, Sanago, and Molnar (2000) who mapped the S-RNase gene to chromosome IV using a VR hybrid mapping strategy. The situation is further complicated by the RFLP mapping using a potato marker CP100 reported by ten Hoopen et al. (1998). This heterologous RFLP marker has shown consistent cosegregation with the S-locus of P. hybrida (Harbord et al. 2000). When mapped with the same VR mapping population used by Strommer et al. (2000), the localization of CP100 was to chromosome III rather than chromosome IV. This makes it difficult to explain the different localizations due to cultivar-specific differences in genome organization, although such variability is known to be a recurring feature of P. hybrida cytological studies (Montijn, ten Hoopen, Fransz, Oud, and Nanninga 1998; see also Chapter 10).
A common feature of these cytological studies in P. hybrida (ten Hoopen et al. 1998; Entani et al. 1999) is a localization for the S-locus at or near the centromere. Several authors have noted that a centromeric location of the S-locus in the Solanaceae may provide tight linkage between the S-RNase and pollen-S, as this region is characterized by suppressed recombination (Round, Flowers, and Richards 1997; Copenhaver, Browne, and Preuss 1998). However, this does not appear to be a general feature, because in Antirrhinum the S-locus is located toward the telomere (Yang, Zhang, Li, Cheng, and Xue 2007).
Physical mapping of S-locus genes in Petunia has also been achieved by genomic cloning strategies. McCubbin, Wang, and Kao (2000a) isolated pollen cDNAs linked to the S-locus, and subsequently McCubbin, Zuniga, and Kao (2000b) and Wang and colleagues (Wang, Wang, McCubbin, and Kao 2003; Wang et al. 2004) used these cDNAs, along with the S-RNase, to screen BAC libraries for S-locus contig clones. This work suggested that the S-locus of Petunia inflata may span >4.4 Mb of chromosomal DNA. Sequencing of a 328 kb region containing the S2-RNase revealed the presence of approximately 50 genes, one of which was a pollen-expressed polymorphic F-box gene termed PiSLF 2 . PiSLF 2 was subsequently shown by a transgenic approach to be a pollen-S gene (Sijacic et al. 2004).
3 Non-S-Locus Stylar Factors
Although the S-RNase plays a key recognition (and cytotoxic) role in the style, other factors are required for expression of self-incompatibility. For example, using different species of Nicotiana for transgenic experiments, Murfett et al. (1996) showed that expression of the S-RNase gene in transgenic SC Nicotiana plumbaginifolia was insufficient for S-allele-specific pollen rejection, whereas expression of the S-RNase in N. plumbaginifolia X SC N. alata hybrid plants did result in S-allele-specific pollen rejection. The clear implication was that some factor(s) must be expressed in Nicotiana alata that is/are not expressed in N. plumbaginifolia, and that the factor(s) is/are required for pollen rejection. Using a differential screen based on the above observation, McClure, Mou, Canevascini, and Bernatzky (1999) cloned a small (101 amino acid) asparagine-rich protein from SC N. alata that they named HT. This protein was predicted to be secreted and processed to a mature form of 86 kDa. Antisense experiments in transgenic plants showed that downregulation of HT in styles resulted in the inability to reject pollen, even though the S-RNase was expressed at normal levels (McClure et al. 1999).
O’Brien et al. (2002), working in Solanum chacoense, extended this work to demonstrate two different isoforms of HT, which they named HT-A and HT-B. Antisense downregulation of HT-B duplicated the results of McClure et al. (1999) in that incompatible plants were converted to self-compatible plants. Downregulation of HT-A had no effect on the self-incompatibility response. The requirement of HT-B for pollen rejection was further demonstrated by analyzing different SC and SI species of tomato as well as cultivated tomato. Kondo et al. (2002a, b) showed that SC tomato species had various defects in expression of both S-RNase and HT-B, ranging from deletion of the genes, to low expression, to mutations that prevented production of normal protein. The role of HT and S-RNase expression in SC in P. hybrida is unclear at present. However, there is clear evidence for SC factors that are unlinked to the S-locus (Ai et al. 1991; Harbord et al. 2000), and the mapping of the HT genes in Petunia will be informative.
The style expresses several proteins at high levels in addition to the S-RNase. Among these are TTS (Cheung, May, Kawata, Ou, and Wu 1993), PELPIII (Goldman, Pezzotti, Seurinck, and Mariana 1992; de Graaf, Knuiman, Derksen, and Mariani 2003), and a 120 kDa protein (Lind, Bacic, Clarke, and Anderson 1994). In affinity-gel binding assays, Cruz-Garcia, Hancock, Kim, and McClure (2005) showed that all three of these stylar glycoproteins formed high molecular weight complexes with the S-RNase. These authors hypothesized that S-RNase may be taken up into pollen tubes in the form of a complex that includes one or more of these proteins. Indeed, in a recent work, Goldraij et al. (2006) showed that S-RNase taken up into pollen tubes is sequestered in a vacuolar-like compartment that is bounded by the 120 kDa protein. The 120 kDa protein, like HT-B, is also required for S-allele-specific pollen rejection. Downregulating the 120 kDa protein using RNA interference in Nicotiana plumbaginifolia X N. alata hybrids eliminated the ability to reject S-allele specific pollen. The same RNAi plants could, however, continue to reject pollen from N. plumbaginifolia, whereas plants downregulated for HT-B failed to reject either S-allele-specific pollen or N. plumbaginifolia pollen (McClure et al. 1999; Hancock, Kent, and McClure 2005).
4 SLF: the Pollen-Recognition Component
‘The first cDNA encoding an S-RNase protein was reported in 1986 (Anderson et al. 1986), but it would be sixteen years before the first published cloning of a gene that would turn out to be pollen-S (Lai et al. 2002). Despite numerous attempts over the ensuing period to identify pollen-S, it was only the improvement in techniques for cloning large-insert DNA libraries along with the ability to sequence long stretches of DNA the made it possible to finally identify and clone the S-locus F-box gene subsequently shown to be pollen-S. Prior research led to several predictions for the expected properties of the pollen-S component, which formed the basis for attempts to clone this gene. First, mutants defective in style or pollen expression of GSI are often fully functional for GSI in the complementary organ, indicating that the style (S-RNase) component and the pollen component (pollen-S) are encoded by separate genes. This observation was reinforced by transgenic experiments (discussed above) in which gain-of-function or loss-of-function experiments that altered S-RNase specificity had no effect on recognition specificity in the pollen. Second, because recombination between the S-RNase and pollen-S is rarely observed, it has been assumed that the two genes are physically linked, or may be located in chromosomal regions suppressed for recombination. Third, the pollen-S gene must be expressed in pollen, most likely in a pollen-specific fashion. Fourth, models of GSI recognition strongly suggested that pollen-S and the S-RNase must physically interact. Fifth, because S-RNase alleles are highly polymorphic (primarily in the hypervariable regions) and recombination is suppressed at the S-locus, pollen-S alleles were expected to be similarly polymorphic. As will be elaborated below, the first four of these predictions hold true, while the expectations of high levels of polymorphism for the pollen-S component are only partially supported. Finally, the observation that GSI breaks down in tetraploid plants, and in plants where the S-locus is partially or fully duplicated such that heteroallelic pollen is produced, provided a clear test for the behavior of a gene putatively identified as pollen-S.
4.1 The Inhibitor Model for Pollen-S
Most current evidence supports a cytotoxic model for pollen-tube rejection wherein the S-RNase acts to degrade pollen RNA, thereby inhibiting protein synthesis and further elongation of pollen tubes (Sims 2005). Cross-compatibility, therefore, must result from the absence of S-RNase activity in pollen tubes, either by preventing the initial import of S-RNase or by inhibiting the action of S-RNases inside pollen tubes. Several different experimental approaches have now demonstrated that the model of S-RNase inhibition is correct, although the precise nature of that inhibition has not yet been conclusively demonstrated.
Direct observations of pollen tubes using electron microscopy and immunogold labeling of S-RNases demonstrated that both compatible and incompatible S-RNases were imported into pollen tubes (Luu, Qin, Morse, and Cappadocia 2000). These results are inconsistent with a receptor model for pollen-S, in which it acts as a “gatekeeper” to exclude non-self S-RNases. Rather, these results support a model in which the S-RNase is imported into all pollen tubes regardless of genotype, but is specifically prevented from acting in non-self (compatible) pollen tubes. More recently, Goldraij et al. (2006) showed that S-RNases imported into Nicotiana pollen tubes are apparently sequestered in a vacuolar-like compartment in compatible pollen tubes and in the early stages of self-incompatible pollinations.
4.2 Breakdown of Incompatibility and Competitive Interactions
Prior to direct observations of S-RNase import into pollen tubes, most of the evidence supporting the hypothesis of a pollen-expressed inhibitor came from investigations of “competitive interaction” in diploid heteroallelic pollen from tetraploid plants. It is a well-established observation (Crane and Lewis 1942; Lewis and Modlibowska 1942; Brewbaker and Natarajan 1960; de Nettancourt 1977) that gametophytic self-incompatibility breaks down in tetraploid plants, provided that the diploid parent is heterozygous at the S-locus. Under these conditions, however, the breakdown of self-incompatibility is only on the pollen side. That is, tetraploid heterozygous styles remain capable of rejecting haploid pollen with a matching S-allele, but diploid, heteroallelic pollen is self-compatible on either diploid or tetraploid styles. This phenomenon of breakdown of self-incompatibility in the pollen has been termed “competitive interaction.”
Competitive interaction does not require complete duplication of genomes. Brewbaker and Natarajan (1960) showed that self-compatible mutants of Petunia inflata had centric chromosome fragments that presumably duplicated the S-locus. In a similar mutational analysis of the pollen component in Nicotiana alata, Golz and coworkers (Golz, Su, Clarke, and Newbigin 1999; Golz, Oh, Su, Kusaba, and Newbigin 2001) induced pollen-part mutations by gamma irradiation, and then characterized the genetic behavior and molecular basis of the induced mutations. All of the recovered pollen-part mutants were self-compatible and demonstrated competitive interaction in pollen with other S-alleles. All of the pollen-part mutants resulted from duplications of all or part of the S-locus, and no pollen-part mutations were associated with chromosomal deletions. Thus, pollen-part mutations induced by radiation phenocopy the tetraploid condition via duplication of the S-locus, most likely by duplication of pollen-S. These results are consistent with a model in which pollen-S is an inhibitor of self-S-RNases. According to this model, in tetraploid plants (or in plants with radiation-induced duplicated S-loci), heteroallelic pollen would possess two inhibitors, each capable of inhibiting all S-RNases except their cognate inhibitor, and therefore all S-RNases would be inhibited. According to this model, deletions in pollen-S would not be recoverable, since any pollen tube having a deleted pollen-S would be unable to inhibit the S-RNase, and would be rejected.
4.3 Evidence that the S-Locus F-Box Protein Is Pollen-S
Transgenic experiments that relied on the phenomenon of competitive interaction in pollen (described above) provided definitive proof that the S-locus F-box genes are pollen-S. Sijacic et al. (2004) transformed S 1 S 1 Petunia inflata with a PiSLF 2 gene construct. Transgenic plants expressing both the PiSLF 2 transgene and the endogenous F-box gene PiSLF 1 were self-compatible, as would be predicted from the phenomenon of competitive interaction. The breakdown in self-incompatibility occurred only in the pollen and did not affect stylar expression of self-incompatibility. Pollen from the transgenic S 1 S 1 /PiSLF 2 plants was compatible on non-transgenic S 1 S 1 plants, while S 1 pollen from the non-transgenic S 1 S 1 plants was rejected by styles of the transgenic S 1 S 1 /PiSLF 2 plants. Progeny resulting from the compatible pollinations carried the PiSLF 2 transgene, and all were self-compatible. In a further experiment, PiSLF 2 was used to transform S 2 S 3 Petunia inflata. Transgenic S 2 S 3 /PiSLF 2 plants produced a mixture of pollen genotypes: haploid S 2 and S 3 pollen (rejected via the standard self-incompatibility response), heteroallelic S 3 /PiSLF 2 pollen, and homoallelic S 2 /PiSLF 2 pollen. The transgenic S 2 S 3 /PiSLF 2 plants were all self-compatible. Analysis of the progeny showed that the resulting plants were all S 2 S 3 or S 3 S 3 . Failure to recover S 2 S 2 plants in the progeny indicates that only S 3 /PiSLF 2 was functional, as predicted, because competitive interaction should not occur in S 2 /PiSLF 2 pollen.
In similar experiments, Qiao et al. (2004) transformed S 3 S 3 Petunia hybrida with the AhSLF-S 2 pollen F-box gene together with the S 2-RNase gene from Antirrhinum hispanicum. Transgenic plants carrying intact genes for AhSLF-S 2 and the S 2-RNase and expressing both at normal levels were self-compatible, and all produced seed when used as the pollen donor to non-transformed S 3 S 3 Petunia. This is the expected result from competitive interaction between different pollen-S genes expressed in the same pollen grain. In the reciprocal cross, these plants rejected non-transgenic S 3 pollen. Taken together with the results reported by Sijacic et al. (2004), these experiments demonstrate that the F-box gene encodes the pollen-recognition factor of gametophytic self-incompatibility, pollen-S. It is striking in the work reported by Qiao et al. (2004) that the transferred AhSLF-S 2 demonstrated functional conservation in the ability to induce competitive interaction in the pollen, despite sharing only 30% amino acid sequence identity with endogenous Petunia SLF genes.
4.4 Pollen-Part Mutants in the Rosaceae
Several Prunus species, in the Rosaceae, have been shown to carry SLF (also called SFB) genes linked to the S-locus (Ushijima et al. 2003; Entani et al. 2003; Yamane, Ikeda, Ushijima, Sassa, and Tao 2003). Transgenic assays to demonstrate that the SFB/SLF genes encode functional pollen components of GSI are not yet possible in Prunus. Several self-compatible mutants have, however, been identified in different species of Prunus, and all show various defects in SFB. Sequence analysis of S-locus F-box genes showed that the F-box motif was located at the N-terminus of the protein. The Prunus SFB/SLF proteins also have two hypervariable regions, HVa and HVb, located near the C-terminus of the protein. Ushijima et al. (2004) reported the characterization of two self-compatible mutants of Prunus avium and P. mume. DNA sequence analysis predicted that the HVa and HVb domains should be missing in the two mutant SFB proteins. SFB 4' has a frame shift mutation that produces an altered amino acid sequence in the HVa region and a stop codon just upstream of the HVb region. SFB f has a 6.8 kb insertion sequence in the region encoding the C-terminal portion of the protein. The insertion would code for 37 amino acids before a stop codon is reached; the mutant protein would lack the C-terminal 195 amino acids found in normal SFB proteins, and therefore lack both HVa and HVb regions. Sonneveld, Tobutt, Vaughan, and Robbins (2005) characterized two pollen-part mutants of Prunus avium. One of these was the same SFB 4' mutant described by the previous group. A second mutant, S 3', had a deletion that removed the entire SFB gene. The S 13' self-compatible mutant in Prunus cerasus also shows alterations in the SFB gene (Tsukamoto, Hauck, Tao, Jiang, and Iezzoni 2006). In this allele, a 1 bp guanine-to-thymine substitution at position +733 produces a UAA stop codon that truncates the SFB protein and eliminates the HVa and HVb regions. Two additional SFB mutations have been reported in self-compatible peach, Prunus persica (Tao et al. 2007). SFB 1 contains a 155 bp insertion that results in a truncated SFB protein, while SFB 2 has a 5 bp insertion that produces a stop codon in the middle of the protein, truncating the protein upstream of the HVa and HVb regions (Tao et al. 2007). Together, the identification of different pollen-part mutants in these species provides strong support for the identification of SFB/SLF as the pollen component of GSI in the Rosaceae. It is noteworthy that pollen-part mutants resulting from a deletion or mutation of SLF have not been reported in the Solanaceae.
5 The Role of Ubiquitination in GSI
The identification of F-box proteins as pollen-S, along with the identification of a RING-HC protein PhSBP1 (see below), suggested a role for the ubiquitin-proteosome system in self-incompatibility recognition. F-box proteins are the recognition components of multiprotein SCF-type E3 ubiquitin ligases. These complexes target proteins for ubiquitination and degradation via the 26S proteosome. The prototypical SCF complex consists of the F-box protein, SKP1, a Cullin protein and a RING domain protein RBX1 (Cardozo and Pagano 2004; Schwechheimer and Villalobos 2004).
A potential role for ubiquitination in gametophytic self-incompatibility was first suggested by the isolation of a gene encoding the RING-HC-containing protein PhSBP1 from Petunia hybrida (Sims and Ordanic 2001). In an attempt to identify pollen-expressed proteins interacting with the S-RNase, Sims and Ordanic (2001) screened a yeast two-hybrid library from mature pollen of P. hybrida with a bait construct for the N-terminal half of the P. hybrida S1-RNase. This screen identified a gene, named PhSBP1 (for P. hybrida S-RNase binding protein), that bound to N-terminal but not C-terminal regions of the S-RNase. Sequence characterization of PhSBP1 indicated that it contained a C-terminal RING-HC (or C3HC4) protein domain. Such domains have been shown to be characteristic of E3 ubiquitin ligases, the components of the ubiquitin-proteosome system that interact with specific substrates targeted for ubiquitination and protein turnover (Freemont 2000). O’Brien, Major, Chantha, and Matton (2004) similarly screened a pollen two-hybrid library from Solanum chacoense with a bait consisting of the HVa and HVb domains of the S. chacoense S 11 allele. This screen resulted in the isolation of a Solanum PhSBP1 ortholog, ScSBP1. In a confirmation of the apparently key role of SBP1, Hua and Kao (2006) carried out a yeast two-hybrid screen using three separate bait constructs of the PiSLF 2 F-box gene of Petunia inflata. All three of these baits bound to a protein 98% identical to PhSBP1, which was named PiSBP1. Further protein interaction assays showed that PiSBP1 also bound to PiSLF1 of P. inflata, as well as to an unrelated F-box protein PiFBP2411 (Hua and Kao 2006).
These results suggested an attractive model for how SLF/SFB, and possibly SBP1, function in recognition of self/non-self pollen and inhibition of the growth of self pollen (Kao 2004; Sims 2005). This model can be stated as follows: during pollination, pollen grains (either self or non-self) are deposited on the stigmatic surface, germinate and produce pollen tubes that begin to grow through the transmitting tract of the style, where they encounter secreted S-RNases. Both self and non-self S-RNases are imported into pollen tubes. S-RNases recognized as non-self are recognized by a SCFSLF (SCF-S-locus F-box)-E3 ubiquitin ligase complex, and targeted for ubiquitination and degradation via the 26S proteosome. S-RNases recognized as self are not ubiquitinated, retain ribonuclease activity, and act to degrade pollen tube RNA, thereby inhibiting protein synthesis and pollen tube growth. This model makes at least two predictions: first, that S-RNase should be polyubiquitinated and degraded in compatible pollinations, and second, that downregulation of SLF/SFB should result in complete self-incompatibility. Available evidence, however, provides only modest support for these two predictions. Experiments designed to determine if S-RNases were degraded in pollen tubes (Qiao et al. 2004) suggested a possible reduction in S-RNase levels. The observed reduction was far from dramatic however, and it was difficult to determine from the data presented whether the observed reduction was statistically significant at all time points. Goldraij et al. (2006) also assayed the level of S-RNase protein in both compatible and incompatible pollinations, and found little evidence to support large-scale degradation of S-RNase in compatible pollen. Hua et al. (Hua, Fields, and Kao 2008) have argued, however, that the failure to observe significant differences in S-RNase levels between compatible and incompatible pollen tubes cannot be used as evidence against the protein degradation model. To date there have been no reports regarding the effect of downregulation of SLF in transgenic plants. As discussed above, however, mutants of different Prunus species in which truncated SFB/SLF proteins are produced or the gene is completely deleted are self-compatible rather than universally incompatible. The study of downregulation or mutation of the SLF gene in Solanaceae will be an important area for future comparative studies of the S-RNase-based mechanism in these two families.
5.1 Evidence for a SCFSLF-Like E3 Ubiquitin Ligase Complex
Although the precise role of ubiquitination and/or protein degradation in GSI remains unclear, there is strong evidence for the involvement of a SCF-type complex in self-incompatibility. It appears, however, that the proposed SCFSLF complex differs somewhat from the prototypical SCF complex. The canonical SCF complex contains the core subunit SKP1; however, Hua and Kao (2006) showed that SKP1 proteins did not bind to products of PiSLF alleles in protein interaction assays. Similarly, Huang, Zhao, Yang, and Xue (2006) could not detect binding between SKP1 proteins in Antirrhinum and AhSLF. Huang et al. (2006) identified a SKP1-like protein, AhSSK1, by using a bait construct of the Antirrhinum F-box protein AhSLF-S2 to screen a yeast two-hybrid library. In addition to its interaction with AhSLF, AhSSK1 interacted with another scaffold component of SCG complexes, CUL1. Huang et al. (2006) proposed that AhSSK1 may act as a bridge between AhSLF and CUL1. The prototypical SCF complex contains a small RING protein RBX1. In the experiments of Hua and Kao, no interaction could be detected between RBX1 from Petunia inflata and either CUL1 or SLF. Lastly, both Sims (unpublished) and Hua and Kao (2006) found that SBP1 binds to the E2 ubiquitin conjugation protein PhUBC1. Together, these results suggest that a SCFSLF-like E3 ubiquitin ligase complex may function in gametophytic self-incompatibility. Components of this complex would include SLF, SBP1 (replacing RBX1), CUL1, and SSK1 (replacing SKP1). This complex could play a role in recognizing either all S-RNases or in specific recognition and inhibition of non-self S-RNases.
6 Vacuolar Sequestration of S-RNase in Compatible Pollen Tubes
Two style-expressed proteins, HT-B and the 120 kDa glycoprotein, have been shown to be required for expression of self-incompatibility (McClure et al. 1999; Hancock et al. 2005). Downregulation of either renders otherwise self-incompatible plants incapable of rejecting pollen. In an examination of the role that they play in compatible and incompatible pollination, Goldraij et al. (2006) investigated the subcellular localization of these proteins, the S-RNase, and compartment-marker proteins in growing pollen tubes. S-RNase, HT-B and the 120 kDa protein were all imported into both compatible and incompatible pollen tubes. In compatible pollen tubes, and in pollen tubes at early stages of incompatible pollinations, the S-RNase appeared to be sequestered in a vacuolar compartment bounded by the 120 kDa glycoprotein. HT-B appeared to be degraded in compatible pollen tubes. In incompatible pollen tubes, late in pollination, this compartment appeared to break down. S-RNase levels persisted, as did HT-B, but the 120 kDa protein was no longer evident. In antisense HT-B plants, which were completely self-compatible, S-RNase remained sequestered (Goldraij et al. 2006).
7 Conclusion: Models for Pollen Recognition and Rejection
Taken together, current results support two alternative models for how S-RNase activity is inhibited in compatible pollen tubes, but released in incompatible pollinations. According to the SCFSLF complex model, an E3 ubiquitin ligase complex preferentially recognizes non-self S-RNases and ubiquitinates them, most likely leading to degradation. In the sequestration model, compatibility occurs due to the continued sequestration of S-RNase in a vacuolar compartment, which is coupled to the degradation of HT-B. In an incompatible pollination, according to this model, HT-B remains intact and the vacuolar compartment breaks down, releasing the S-RNase. The challenge for researchers in this field is in how to distinguish and/or resolve these models. Particularly in the sequestration model, it is unclear what the role of SLF or the proposed SCF complex might be, and also how a vacuolar-sequestered S-RNase can interact in any fashion with pollen-S, which is cytoplasmic. It is possible that the role of ubiquitination in this response is not to target proteins for degradation, but instead to target a specific protein substrate to the endomembrane system. Indeed, mono-ubiquitination (as opposed to poly-ubiquitination) has been shown to target proteins for endocytosis rather than degradation (D’Azzo, Bongiovanni, and Nastasi 2005). Given the significant contributions of Petunia inflata and P. hybrida as model systems for GSI research to date, Petunia will undoubtedly continue to provide insights into the mechanisms of this agronomically significant trait.
References
Ai, Y., Singh, A., Coleman, C.E., Ioerger, T.R., Kheyr-Pour, A. and Kao T.-H. (1990) Self-incompatibility in Petunia inflata: Isolation and characterization of complementary DNA encoding three S-allele-associated proteins. Sex. Plant Reprod. 3, 130–138.
Ai, Y., Kron, E. and Kao, T.-H. (1991) S-alleles are retained and expressed in a self-compatible cultivar of Petunia hybrida. Mol. Gen. Genet. 230, 353–358.
Ai, Y., Tsai, D.S. and Kao, T.-H. (1992) Cloning and sequencing of cDNAs encoding two S proteins of a self-compatible cultivar of Petunia hybrida. Plant Mol. Biol. 19, 523–528.
Anderson, M.A., Cornish, E.C., Mau, S.-L., Williams, E.G., Hoggart, R., Atkinson, A., Bonig, I., Grego, B., Simpson, B., Roche, P.J., Haley, J.D., Penschow, J.D., Niall, H.D., Tregear, G.W., Coghlan, J.P., Crawford, R.J. and Clarke, A.E. (1986) Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature 321, 38–44.
Anderson, M.A., McFadden, G.I., Bernatzky, R., Atkinson, A., Orpin, T., Dedman, H., Tregear, G., Fernley, R. and Clarke, A.E. (1989) Sequence variability of three alleles of the self-incompatibility gene of Nicotiana alata. Plant Cell 1, 483–491.
Ascher, P.D. (1984) Self-incompatibility. In: K.C. Sink (Ed.), Petunia:Monographs on Theoretical and Applied Genetics 9. Springer-Verlag, Berlin, pp. 22–109.
Brewbaker, J.L. and Natarajan, A.T. (1960) Centric fragments and pollen-part mutation of incompatibility alleles in Petunia. Genetics 45, 699–704.
Broothaerts, W.J., van Laere, A., Witters, R., Préaux, G., Decock, B., van Damme, J. and Vendrig, J.C. (1989) Purification and N-terminal sequencing of style glycoproteins associated with self-incompatibility in Petunia hybrida.Plant Molec. Biol. 14, 93–102.
Broothaerts, W., Vanvinckenroye, P., Decock, B., Van Damme, J. and Vendrig, J.C. (1991) Petunia hybrida S-proteins: Ribonuclease Activity and the Role of their Glycan Side Chains in Self-Incompatibility. Sex. Plant Reprod. 4, 258–266.
Cardozo, T. and Pagano, M. (2004) The SCF ubiquitin ligase: Insights into a molecular machine. Nature Rev. Molec. Cell Biol. 5, 739–751.
Cheung, A.Y., May, B., Kawata, E.E., Ou, Q. and Wu, H. (1993) Characterization of cDNAs for stylar transmitting tissue-specific proline-rich proteins in tobacco. Plant J. 3, 151–160.
Clark, K.R., Okuley, J., Collins, P.D. and Sims, T.L. (1990) Sequence variability and developmental expression of S-alleles in self-incompatible and pseudo-self-compatible Petunia. Plant Cell 2, 815–826.
Copenhaver, G.P., Browne, W.E. and Preuss, D. (1998) Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl. Acad. Sci, USA 95, 247–252.
Crane, M.B. and Lewis, D. (1942) Genetical studies in pears III. Incompatibility and sterility. J. Genet. 43, 31–43.
Cruz-Garcia, F., Hancock, C.N., Kim, D. and McClure, B. (2005) Stylar glycoproteins bind to S-RNase in vitro. Plant J. 42, 295–305.
D’Azzo, A., Bongiovanni, A. and Nastasi, T. (2005) E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6, 429–441.
Dana, M.N. and Ascher, P.D. (1985) Pseudo-self compatibility (PSC) in Petunia integrifolia J. Hered. 76, 468–470.
Dana, M.N. and Ascher, P.D. (1986a) Sexually localized expression of pseudo-self compatibility in Petunia-hybrida 1. Pollen inactivation. Theor. Appl. Genet. 71, 573–577.
Dana, M.N. and Ascher, P.D. (1986b) Sexually localized expression of pseudo-self compatibility in Petunia-hybrida 2. Stylar inactivation. Theor. Appl. Genet. 71, 578–584.
Darwin, C. (1891) The Effects of Cross and Self Fertilisation in the Vegetable Kingdom, 3rd Ed. John Murray, London, pp. 188–189.
de Graaf, B.H.J., Knuiman, B.A., Derksen, J. and Mariani, C. (2003) Characterization and localization of the transmitting tissue-specific PELPIII proteins of Nicotiana tabacum. J. Exp. Bot. 54, 55–63.
de Nettancourt, D. (1977) Incompatibility in Angiosperms: Monographs on Theoretical and Applied Genetics 3. Springer-Verlag, NY.
Entani, T., Iwano, M., Shiba, H., Takayama, S., Fukui, K. and Isogai, A. (1999) Centromeric localization of an S-RNase gene in Petunia hybrida Vilm. Theor. Appl. Genet. 99, 391–397.
Entani, T., Iwano, M., Shiba, H., Che, F.-S., Isogai, A. and Takayama, S. (2003) Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: Identification of a pollen-expressed F-box gene with allelic diversity. Genes to Cells 8, 203–213.
Flaschenreim, D.R. and Ascher, P.D. (1979a) Pollen tube expression of pseudo-self-compatibility (PSC) in Petunia hybrida. Theor. Appl. Genet. 54, 97–101.
Flaschenreim, D.R. and Ascher, P.D. (1979b) S allele discrimination in styles of Petunia hybrida bearing stylar-conditioned pseudo-self-compatibility. Theor. Appl. Genet. 55, 23–28.
Freemont, P.S. (2000) Ubiquitination: RING for destruction? Curr. Biol. 10, 84–87.
Gebhardt, C., Ritter, E., Barone, A., Debener, T., Walkemeier, B., Schachtschabel, U. et al. (1991) RFLP maps of potato and their alignment with the homoeologous tomato genome. Theor. Appl. Genet. 83, 49–57.
Goldman, M.H.D.S., Pezzotti, M., Seurinck, J. and Mariana, C. (1992) Developmental expression of tobacco pistil-specific genes encoding novel extension-like proteins. Plant Cell 4, 1041–1051.
Goldraij, A., Kondo, K., Lee, C.B., Hancock, C.N., Sivaguru, M., Vazquez-Santana, S. et al. (2006) Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439, 805–810.
Golz, J.F., Su, V., Clarke, A.E. and Newbigin, E. (1999) A molecular description of mutations affecting the pollen component of the Nicotiana alata S locus. Genetics 152, 1123–1135.
Golz, J.F., Oh, H.-Y., Su, V., Kusaba, M. and Newbigin, E. (2001) Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S locus. Proc. Natl. Acad. Sci., USA 98, 15372–15376.
Hancock, C.N., Kent, L. and McClure, B. (2005) The 120 kDa glycoprotein is required for S-specific pollen rejection in Nicotiana. Plant J. 43, 716–723.
Harbord, R.M., Napoli, C.A. and Robbins, T.P. (2000) Segregation distortion of T-DNA markers linked to the self-incompatibility (S) locus in Petunia hybrida. Genetics 154, 1323–1333.
Harland, S.C. and Atteck, O.S. (1933) The inheritance of self-sterility in Petunia violacea Genetica 15, 89–102.
Herrero, M. and Dickinson, H.G. (1980) Pollen tube growth following compatible and incompatible intraspecific pollinations in Petunia hybrida. Planta 148, 217–221.
Herrero, M. and Dickinson, H.G. (1981) Pollen tube development in Petunia hybrida following compatible and incompatible intraspecific matings. J. Cell Sci. 47, 365–385.
Hiscock, S.J. (2000) Genetic control of self-incompatibility in Senecio squalius L. (Asteraceae): A successful colonizing species. Heredity 85, 10–19.
Hua, Z. and Kao, T.-H. (2006) Identification and characterization of components of a putative Petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell 18, 2531–2553.
Hua, Z.H., Fields, A. and Kao, T.-H. (2008) Biochemical Models for S-RNase-Based Self-Incompatibility. Molecular Plant 1, 575–585.
Huang, S., Lee, H.-S., Karunanandaa, B. and Kao, T.-H. (1994) Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell 6, 1021–1028.
Huang, J., Zhao, L., Yang, Q. and Xue, Y. (2006) AhSSK1, a novel SKP1-like protein that interacts with the S-locus F-box protein SLF. Plant J. 46, 780–793.
Ida, K., Norioka, S., Yamamoto, M., Kumasaka, T., Yamashita, E., Newbigin, E., Clarek, A.E., Sakiyama, F. and Sato, M. (2001) The 1.55 Å resolution structure of Nicotiana alata SF11 –RNase associated with gametophytic self-incompatibility. J. Molec. Biol. 314, 103–112.
Igic, B. and Kohn, J.R. (2001) Evolutionary relationships among self-incompatibility S-RNases. Proc. Natl. Acad. Sci., USA 98, 13167–13171.
Ioerger, T.R., Clark, A.G. and Kao, T.-H. (1990) Polymorphism at the self-incompatibility locus in the Solanaceae predates speciation. Proc. Natl. Acad. Sci., USA 87, 9732–9735.
Ioerger, T.R., Gohlke, J.R., Xu, B. and Kao, T.-H. (1991) Primary structural features of the self-incompatibility protein in Solanaceae. Sex. Plant Reprod. 4, 81–87.
Ishimizu, T., Shinkawa, T., Sakiyama, F. and Norioka, S. (1998) Primary structural features of rosaceous S-RNases associated with gametophytic self-incompatibilty. Plant Molec. Biol. 37, 931–941.
Kamboj, R.K. and Jackson, J.F. (1986) Self-incompatibility alleles control a low molecular weight, basic protein in pistils of Petunia hybrida. Theor. Appl. Genet. 71, 815–819.
Kao, T.-H. and Tsukamoto, T. (2004) The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16, S72–S83.
Karunanandaa, B. Huang, S. and Kao, T.-H. (1994) Carbohydrate moiety of the Petunia inflata S3 protein is not required for self-incompatibility interactions between pollen and pistil. Plant Cell 6, 1933–1940.
Kondo, K., Yamamoto, M., Itahashi, R., Sato, T., Egashira, H., Hattori, T. and Kowyama, Y. (2002a) Insights into the evolution of self-incompatibility from a study of stylar factors. Plant J. 30, 143–153.
Kondo, K., Yamamoto, M., Matton, D.P., Sato, T., Hirai, M., Norioka, S., Hattori, T. and Kowyama, Y. (2002b) Cultivated tomato has defects in both S-RNase and HT genes required for stylar function of self-incompatibility. Plant J. 29, 627–636.
Lai, Z., Ma, W., Han, B., Liang, L., Zhang, Y., Hong, G. and Xue, Y. (2002) An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Molec. Biol. 50, 29–41.
Lee, H.S., Huang, S. and Kao, T.H. (1994) S proteins control rejection of incompatible pollen in Petunia inflate. Nature 367, 560–563.
Lewis, D. and Modlibowska, I. (1942) Genetical studies in pears IV. Pollen tube growth and incompatibility. J. Genet. 43, 211–222.
Lind, J.L., Bacic, A., Clarke, A.E. and Anderson, M.A. (1994) A style-specific hydroxyproline-rich glycoprotein with properties of both extensions and arabinogalactan proteins. Plant J. 6, 491–502.
Linskens, H.F. (1975) Incompatibility in Petunia. Proc. Royal Soc. London, Series B, Biolog. Sci. 188, 299–311.
Luu, D.T., Qin, K.K., Morse, D. and Cappadocia, M. (2000) S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407, 649–651.
Mather, K. (1943) Specific differences in Petunia I. J. Genet. 45, 215–235.
Matton, D.P., Maes, O., Laublin, G., Xike, Q., Bertrand, C., Morse, D. and Cappadocia, M. (1997) Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition. Plant Cell 9, 1757–1766.
Matton, D.P., Luu, D.T., Xike, Q., Laublin, G., O’Brien, M., Maes, O., et al. (1999) Production of an S RNase with dual specificity suggests a novel hypothesis for the generation of new S alleles. Plant Cell 11, 2087–2098.
McClure, B.A., Gray, J.E., Anderson, M.A. and Clarke, A.E. (1990) Self incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 347, 757–760.
McClure, B.A., Mou, B., Canevascini, S. and Bernatzky, R. (1999) A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proc. Natl. Acad. Sci., USA 96, 13548–13553.
McCubbin, A.G., Chung, Y.-Y. and Kao, T.-H. (1997) A mutant S-RNase of Petunia inflata lacking RNase activity has an allele-specific dominant negative effect on self-incompatibility interactions. Plant Cell 9, 85–95.
McCubbin, A.G., Wang, X. and Kao, T.-H. (2000a) Identification of self-incompatibility (S-) locus linked pollen cDNA markers in Petunia inflata. Genome 43, 619–627.
McCubbin, A.G., Zuniga, C. and Kao, T.-H. (2000b) Construction of a binary bacterial artificial chromosome library of Petunia inflata and the isolation of large genomic fragments linked to the self-incompatibility (S-) locus. Genome 43, 820–826.
Montijn, M.B., ten Hoopen, R., Fransz, P.F., Oud, J.L. and Nanninga, N. (1998) Characterisation of the nucleolar organising regions during the cell cycle in two varieties of Petunia hybrida as visualised by fluorescence in situ hybridisation and silver staining. Chromosoma 107, 80–86.
Murfett, J., Strabala, T.J., Zurek, D.M., Mou, B., Beecher, B. and McClure, B. (1996) S-RNase and interspecific pollen rejection in the genus Nicotiana: Multiple pollen-rejection pathways contribute to unilateral incompatibility between self-incompatible and self-compatible species. Plant Cell 8, 943–958.
O’Brien, M., Kapfer, C., Major, G., Laurin, M., Bertrand, C., Kondo, K., Kowyama, Y. and Matton, D.P. (2002) Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J. 32, 985–996.
O’Brien, M., Major, G., Chantha, S.C. and Matton, D.P. (2004) Isolation of S-RNase binding proteins from Solanum chacoense: Identification of an SBP1 (RING finger protein) orthologue. Sex. Plant Reprod. 17, 81–87.
Olmstead, R.G., DePamphilis, C.W., Wolfe, A.D., Young, N.D., Eilisons, W.J. and Reeves, P.A. (2001) Disintegration of the Scrophulariaceae. Amer. J. Bot. 88, 348–361.
Qiao, H., Wang, F., Zhao, L., Zhou, J.L., Lai, Z., Zhang, Y.S., Robbins, T.P. and Xue, Y.B. (2004) The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 16, 2307–2322.
Robacker, C.D. and Ascher, P.D. (1982) Discriminating styles (DS) and pollen-mediated pseudo-self-compatibility (PMPSC) in Nemesia strumosa benth. Theor. Appl. Genet. 61, 289–296.
Robbins, T.P., Harbord, R.M., Sonneveld, T. and Clarke, K. (2000) The molecular genetics of self-incompatibility in Petunia hybrida. Ann. Bot. 85 (Suppl. A), 105–112.
Round, E.K., Flowers, S.K. and Richards, E.J. (1997) Arabidopsis thaliana centromere regions: Genetic map positions and repetitive DNA structure. Genome Res. 7, 104–1053.
Royo, J., Kunz, C., Kowyama, Y., Anderson, M., Clarke, A.E. and Newbigin, E. (1994) Loss of a histidine residue at the active site of S-locus ribonuclease is associated with self-compatibility in Lycopersicon peruvianum. Proc. Natl. Acad. Sci., USA 91, 6511–6514.
Schwechheimer, C. and Villalobos, L.I.A.C. (2004) Cullin-containing E3 ubiquitin ligases in plant development. Curr. Opin. Plant Biol. 7, 677–686.
Shivanna, K.R. and Rangaswamy, N.S. (1969) Overcoming self-incompatibility in Petunia axillaris. I. Delayed pollination, pollination with stored pollen, and bud pollination. Phytomorph. 19, 372–380.
Sijacic, P., Wang, X., Skirpan, A.L., Wang, Y., Dowd, P.E., McCubbin, A.G., Huang, S. and Kao, T.-H. (2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429, 302–305.
Sims, T.L. and Ordanic, M. (2001) Identification of a S-ribonuclease binding protein in Petunia hybrida. Plant Molec. Biol. 47, 771–783.
Sims, T.L. (2005) Pollen recognition and rejection in different self-incompatibility systems. Rec. Res. Devel. Plant Molec. Biol. 2, 31–62.
Sink, K. (1984) Petunia: Monographs on Theoretical and Applied Genetics 9. Springer-Verlag, Berlin.
Sonneveld, T., Tobutt, K.R., Vaughan, S.P. and Robbins, T.P. (2005) Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17, 37–51.
Steinbachs, J.E. and Holsinger, K.E. (2002) S-RNase-mediated gametophytic self-incompatibility is ancestral in eudicots. Molec. Biol. Evol. 19, 825–829.
Stout, A.B. and Chandler, C. (1941) Change from self-incompatibility to self compatibility accompanying change from diploidy to tetraploidy. Science 94, 118.
Strommer, J., Gerats, A.G.M., Sanago, M. and Molnar, S.J. (2000) A gene-based RFLP map of petunia. Theor. Appl. Genet. 100, 899–905.
Tanksley, S.D. and Loaiza-Figueroa, F. (1985) Gametophytic self-incompatibility is controlled by a single major locus on chromosome 1 in Lycopersicon peruvianum. Proc. Natl. Acad. Sci., USA 82, 5093–5096.
Tao, R., Watari, A., Hanada, T., Habu, T., Yaegaki, H., Yamaguchi, M. and Yamane, H. (2007) Self-compatible peach (Prunus persica) has mutant versions of the S haplotypes found in self-incompatible Prunus species. Plant Molec. Biol. 63, 109–123.
ten Hoopen, R., Harbord, R.M., Maes, T., Nanninga, N. and Robbins, T.P. (1998) The self-incompatibility (S) locus in Petunia hybrida is located on chromosome III in a region syntenic for the Solanaceae. Plant J. 16, 729–734.
Tsukamoto, T., Ando, T., Takahashi, K., Omori, T., Watanabe, H., Kokubun, H., Marchesi, E. and Kao, T.H. (2003) Breakdown of self-incompatibility in a natural population of Petunia axillaris caused by loss of pollen function. Plant Physiol. 131, 1903–1912.
Tsukamoto, T., Hauck, N.R., Tao, R., Jiang, N. and Iezzoni, A.F. (2006) Molecular characterization of three non-functional S-haplotypes in sour cherry (Prunus cerasus). Plant Molec. Biol. 62, 371–383.
Ushijima, K., Sassa, H., Dandekar, A.M., Gradziel, T.M., Tao, R. and Hirano, H. (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond: Identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15, 771–781.
Ushijima, K., Yamane, H., Watari, A., Kakehi, E., Ikeda, K., Hauck, N.R. et al. (2004) The S haplotype-specific F-box protein gene SFB is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J. 39, 573–586.
Wang, X., Hughes, A.L., Tsukamoto, T., Ando, T. and Kao, T.-H. (2001) Evidence that intragenic recombination contributes to allelic diversity of the S-RNase gene at the self-incompatibility (S) locus in Petunia inflata. Plant Physiol. 125, 1012–1022.
Wang, Y., Wang, X., McCubbin, A.G. and Kao, T.-H. (2003) Genetic mapping and molecular characterization of the self-incompatibility (S) locus in Petunia inflate. Plant Molec. Biol. 53, 565–580.
Wang, Y., Tsukamoto, T., Yi, K.-W., Wang, X., Huang, A., McCubbin, A.G. and Kao, T.-H. (2004) Chromosome walking in the Petunia inflata self-incompatibility (S-) locus and gene identification in an 881 kb contig containing S2-RNase. Plant Molec. Biol. 54, 727–742.
Woodward, J.R., Bacic, A., Jahnen, W. and Clarke, A.E. (1989) N-linked glycan chains on S-allele-associated glycoproteins from Nicotiana alata. Plant Cell 1, 511–514.
Xue, Y., Carpenter, R., Dickinson, H.G. and Coen, E.S. (1996) Origin of allelic diversity in Antirrhinum S locus RNases. Plant Cell 8, 805–814.
Yamane, H., Ikeda, K., Ushijima, K., Sassa, H. and Tao, R. (2003) A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol. 44, 764–769.
Yang, Q., Zhang, D., Li, Q., Cheng, Z. and Xue, Y. (2007) Heterochromatic and genetic features are consistent with recombination suppression of the self-incompatibility locus in Antirrhinum. Plant J. 51, 140–151.
Zurek, D.M., Mou, B., Beecher, B. and McClure, B. (1997) Exchanging sequence domains between S-RNases from Nicotiana alata disrupts pollen recognition. Plant J. 11, 797–808.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2009 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Sims, T.L., Robbins, T.P. (2009). Gametophytic Self-Incompatibility in Petunia. In: Gerats, T., Strommer, J. (eds) Petunia. Springer, New York, NY. https://doi.org/10.1007/978-0-387-84796-2_5
Download citation
DOI: https://doi.org/10.1007/978-0-387-84796-2_5
Publisher Name: Springer, New York, NY
Print ISBN: 978-0-387-84795-5
Online ISBN: 978-0-387-84796-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)