Genomic research in Rosaceae crops is commonly directed at understanding the genetic control of important agronomic traits with the aim of improving product quality and reducing production costs. Genomic knowledge can be used for genetic improvement of cultivars through breeding or genetic engineering. Genomic knowledge can also be used for the development of new cultural practices and the tailoring of existing production practices according to genetic categories of cultivars. The translation of genomic data and fundamental discoveries into practical results with real world applications is often termed “translational genomics”. However, the term is also used to describe the transfer of genomic knowledge from model organisms, such as Arabidopsis, to crop species, with practical application sometimes only implied.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Suppression Subtractive Hybridization
- Transcript Profile
- Fire Blight
- Soluble Solid Concentration
- Fire Blight Resistance
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Use of Genomics in Rosaceae
Genomic research in Rosaceae crops is commonly directed at understanding the genetic control of important agronomic traits with the aim of improving product quality and reducing production costs. Genomic knowledge can be used for genetic improvement of cultivars through breeding or genetic engineering. Genomic knowledge can also be used for the development of new cultural practices and the tailoring of existing production practices according to genetic categories of cultivars. The translation of genomic data and fundamental discoveries into practical results with real world applications is often termed “translational genomics”. However, the term is also used to describe the transfer of genomic knowledge from model organisms, such as Arabidopsis, to crop species, with practical application sometimes only implied.
Many crop attributes are limited by the underlying genetics of the cultivars at hand. Breeders seek to raise the bar with each generation, and provide new genetic possibilities. New cultivars are designed to possess improved potential for horticultural performance, whether as incremental gains over previous cultivars, or with novel attributes that set them apart. Decisions regarding parent selection for crossing and progeny selection for advancing potential cultivars are based on knowledge, as well as educated guesses and hunches, of how controlling genes combine and are expressed in breeding populations. Genomics can shortcut or enhance the scope of genetic studies to elucidate the genetic architecture of traits by identifying, quantifying, and validating important genomic regions. It can also identify the genes that control trait variation and determine their strength of expression under varying production conditions. Armed with such knowledge, breeders can more efficiently manipulate germplasm over generations to produce optimum genetic combinations and novel genetic possibilities in the form of new cultivars that perform better for growers and produce superior products for handlers, processors, marketers, and ultimately, consumers. Breeders are therefore genetic architects, designing new products from the manipulation of genetic components.
Genomics can also impact the production of established plantings. In horticultural production, many crop attributes are readily influenced by cultural practices. However, some cultivars may respond poorly to treatments or respond differently across different environments and seasons. Knowledge of the genetics underlying the performance of each cultivar could lead to genetic “diagnostics” that allow cultural practices to be tailored to a specific functional genetic group of cultivars. Another approach is genomic-based crop “therapeutics”, or “chemical genomics”, an emerging field of research that also allows improvement of plants already under cultivation. Where specific genes are known to influence important traits, compounds can be designed that enhance or interfere with their expression to improve a crop’s performance or product quality.
New genomic technologies are also valuable for more fundamental studies. Basic biological research has traditionally avoided Rosaceae species, and numerous organismal systems are much more tractable to studying many basic biological mechanisms. However, certain biological phenomena in Rosaceae crops such as perenniality, dormancy, extended juvenility, scion-rootstock interaction, complex polyploidy, and diverse plant, flower, and fruit form are usually absent in model organisms. The genetic systems of one or more species in the Rosaceae family can offer useful platforms for uncovering the genomic networks underlying these attributes, mechanisms, and processes. In this approach, the rosaceous species is the model organism. Ultimately, fundamental genomic studies in this plant family can be turned to practical use by providing the valuable knowledge that aids in understanding and manipulating existing cultivars, and breeding the next generation.
1.1 Genetic Basis of Agronomically Significant traits
A collection of attributes sets most rosaceous crops apart from field crops and model species. These attributes include perenniality, large plant size, extended juvenility, use of rootstocks, clonal propagation and highly perishable products (strawberry being an exception to the first four attributes). Product quality, rather than yield, is critical for profitability. From the perspective of Rosaceae crop industries, key needs are to (1) improve fresh and processed product quality, shelf life and safety, including the development of novel or improved flavors, textures, aromas, and colors, for a healthier and more satisfied consumer; (2) reduce chemical pesticide use and develop stress tolerant plants for greater environmental sustainability; and (3) decrease labor and energy costs of crop production (The U.S. Rosaceae Genomics, Genetics, and Breeding Initiative White Paper, 2006). The traits associated with these needs, including (1) fruit, nut, and flower postharvest quality, (2) pest, disease and abiotic stress resistance, and (3) plant architecture and phenology, are currently designated as the highest priority targets for improvement by the U.S. genomics, genetics, and breeding community. These priorities are mirrored in the international arena.
1.1.1 Genetics or Genomics?
Improvement of Rosaceae crops in the era before formal knowledge of genetics principles, prior to the widespread acceptance of Mendel’s laws, was often based on selection and clonal propagation of superior individuals. The principles of Mendelian genetics, followed by their elaboration into quantitative genetics theory, provided a powerful framework for genetic improvement through dedicated plant breeding. The discipline of genetics holds that heritable traits are controlled by interacting alleles of individual genes, themselves interacting with a finite number of other genes in the background of the whole genome. Genomics takes a more holistic approach from the outset, considering the complexities of gene networks and gradually narrowing the focus to specific genetic elements, at which point, genetic approaches can be effectively engaged.
The success of genetics and genomics approaches to crop improvement is strongly dictated by the underlying genetic architecture of traits of interest. The main components of genetic architecture of a trait are heritability (degree of genetic as opposed to environmental control), the number of influencing loci, the genetic action and magnitude of effect of alleles at controlling loci, and genetic linkages with other traits. Important traits can be categorized as qualitative (also known as simple, Mendelian, or discrete traits) or quantitative (also known as complex or continuous traits). Qualitative traits are typically controlled by variation in one gene with high heritability, and are usually readily tackled by genetics. Quantitative traits may be influenced by many genes or by a few genes with low heritability, and can be approached by genetics or genomics, separately or together. Some quantitative horticultural traits have been previously addressed by genetics without significant success, and genomic technologies offer powerful new tools for their elucidation.
Current knowledge of the genetic architecture of important traits in Rosaceae crops can be exemplified by fruit texture attributes. Components of fruit texture, including firmness, softening rate and pattern, hardness, crispness, crunchiness, juiciness, mealiness, fibrousness, turgor, and others, cover the spectrum of genetic architecture. Various genetic and genomic approaches, integrated with physiology, molecular biology, and practical aspects of breeding, have been employed to study fruit texture, as described below for flesh softening and mealiness in peach and nectarine.
1.1.2 “Melting Flesh” in Peach and Nectarine
Fruit flesh softening is of considerable interest to the peach and nectarine industry. The market is divided into fresh, which are usually the quick-softening “melting flesh” (MF) types, and canning, which uses almost exclusively non-melting flesh types. Breeding programs are usually separated for fresh-market and canning cultivars, with crosses conducted within, but rarely between, these two categories and typically targeting greater firmness of fruit to facilitate harvest and transport. While the melting texture is most desired by consumers for fresh eating, breeders in some regions such as Florida and Spain have developed very firm non-melting flesh (NMF) peach cultivars that are suitable for the fresh market as they do not have a rubbery texture like canning peaches.
The MF/NMF attribute is qualitative, as each tree produces fruit that is either MF or NMF in most cases, and is easily determined by squeezing or biting into ripe fruit. The genetic control of this qualitative trait is stable across seasons and locations, and thus heritability is very high. The trait has long been described as under the control of a single locus, Melting flesh, and was part of the first linkage group described for peach (Bailey and French 1949). Basic genetic analysis of segregating populations easily identifies that MF is dominant over NMF (Bailey and French 1949; Peace et al. 2005b).
Various molecular genetic tools were used in several labs (Lester et al. 1994, 1996; Callahan et al. 2004; Peace et al. 2005b) to identify the controlling gene as that encoding endopolygalacturonase (endoPG), an enzyme that metabolizes pectin in the cell wall and is implicated in fruit softening and abscission processes of various crops (Hadfield and Bennett 1998). Our current understanding is that presence of the Melting flesh endoPG gene results in MF fruit, while absence of the gene results in NMF fruit. A simple PCR test is available for making this distinction (Peace et al. 2005b). With this knowledge of the gene behind an important horticultural trait, the genetic predisposition of currently grown cultivars can be better understood, and the genetic marker can be used in breeding. These applications are indeed occurring. However, this locus has other interesting aspects.
In loci where genetic polymorphism causes qualitative differences in phenotype, alleles with quantitative effects on phenotype can also be detected (Robertson 1989). Qualitative mutants are often the result of critical mutations in a gene producing a non-functional gene product or no product at all. Quantitative alleles can result from point mutations in the gene sequence, giving a less efficient product and a subtle difference in phenotype (Pflieger et al. 2001). This theory has been supported in studies of Arabidopsis (Koornneef et al. 1998), maize (Beavis et al. 1991), and Drosophila (Mackay 2004). Recent examination of allelic variation in the Melting flesh gene has uncovered evidence that this phenomenon may also occur for softening rate in peach and nectarine (C. Peace et al., manuscript in prep.).
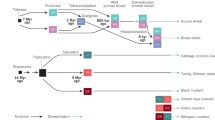
Another revelation from probing the genetic basis of Melting flesh was that for some cultivars, another functional endoPG gene resides same locus (less than 50 kbp upstream of the Melting flesh gene). This second gene encodes an identical amino acid sequence to the Melting flesh gene, but differs slightly in the DNA sequence of its introns and promoter region (Peace et al. 2005b,. 2007; A. Callahan and C. Peace, manuscript in preparation). These differences presumably alter its transcription and/or translation, and subsequent timing and location of enzymatic action, as this second endoPG gene controls the Freestone trait (Peace et al. 2007). Presence of the gene produces freestone fruit, where the flesh fibers are detached from the stone in ripe fruit and the stone comes away freely. Absence of the Freestone endoPG gene is associated with clingstone fruit, where fibers remain attached to the stone. It is possible that minor allelic variants may underlie less extreme phenotypes of flesh fiber adhesion. Together, the two endoPG genes underlie the Freestone-Melting flesh (F-M) locus, and although the two genes are separate, their identical protein product and same genomic location identifies the F-M locus as being pleiotropic. All combinations of presence/absence of the two genes exist in naturally occurring trees, representing the four major functional alleles of F-M (Fig. 1).
Interactions within the F-M locus include additivity, dominance, pleiotropy, and perhaps epistasis. The Freestone and Melting flesh endoPG genes combine additively in the F allele (actually a haplotype as it encompasses two adjacent genes) to give the FMF phenotype (Fig. 2). In diploid combination in peach and nectarine trees, the F allele is dominant over the other three alleles (thus F- = FMF), the f allele is dominant over the f1 and n alleles (ff, ff1, and fn = CMF), and the n allele is recessive to all (f1f1 and f1n = CNMF, nn = CNSF) (Fig. 1). Although primarily affecting the freestone trait, the Freestone gene also appears to pleiotropically influence softening, as the flesh of fruit that are homozygous for the n allele lose very little firmness during ripening whereas the f1 allele (in homozygosity or as f1n) results in gradual softening to a rubbery texture. The f1 allele does not provide the expected freestone non-melting flesh (FNMF) phenotype – instead it is CNMF. The NMF phenotype seems to mask the expression of freestone, as suggested by Bailey and French (1949). Indeed, a stable FNMF phenotype has not been reported. However, the fact that the ff1 allelic combination, which includes both the Freestone and Melting flesh genes but on different chromosomes, produces CMF and not FMF fruit (Peace et al. 2005b) suggests that the Freestone gene of the f1 allele is inherently non-functional rather than being epistatically affected by the lack of the Melting flesh gene.
Genomic approaches for crop improvement use various technologies and techniques to address three general fields of genomic study: functional, structural, and comparative genomics. Genomic research in Rosaceae crops is commonly directed at understanding the genetic control of important agronomic traits with the aim of producing “deliverables” to improve product quality and reduce production costs
In conclusion, while Melting flesh was originally considered a simple Mendelian trait, two copies of the controlling gene were discovered at the locus, with several major phenotypes resulting from their allelic variants caused by presence/absence of functional gene copies. The locus also appears to underlie quantitative variation in fruit softening. The F-M locus occurs at the distal end of Prunus linkage group 4, which genetically links it with nearby quantitative trait loci (QTLs) for other important traits such as bleeding, soluble solids concentration, titratable acidity, and flowering date (Peace et al. 2005a, 2006).
1.1.3 Mealiness in Peach and Nectarine
Mealiness is another texture-related trait that is of major concern to the peach and nectarine industry, due to its broad dislike by consumers. A large proportion of world cultivars of fresh market peach and nectarine produce fruit that often become mealy (dry and soft with a grainy mouthfeel) after cold storage of a few weeks. Such fruit have the outward appearance of good quality, and are often sold to unaware, and ultimately unsatisfied, consumers (Crisosto et al. 1999). Yet cold storage is required to halt softening and bruising while fruit are shipped to distant markets. Some cultivars appear more susceptible than others, suggesting a genetic component.
The endoPG enzyme has long been implicated in the development of mealiness in susceptible cultivars (Buescher and Furmanski 1978). Study of mealiness susceptibility in a peach population segregating at the F-M locus concluded that endoPG plays a qualitative role in the trait’s expression, where a functional Melting flesh endoPG gene must be present for endoPG activity to occur during but not after cold storage (Peace et al. 2006). Although the melting phase does not occur in fruit that become mealy, partial endoPG activity in storage leads to gradual softening after storage, and appears to enable other genes involved in mealiness development to be expressed (Peace et al. 2005a, 2006). CNMF (and CNSF) fruit are effectively resistant to mealiness, in that they remain too firm to be classified as mealy and don’t exhibit the partial expression of endoPG in cold storage. Thus the F-M locus is epistatic in the genetic control of mealiness because some of its alleles mask the expression of other loci conditioning mealiness susceptibility. Avoiding mealiness in the fresh market peach industry can therefore be achieved by using non-melting or non-softening types, but if the buttery texture that consumers tend to prefer is to remain, the genetic basis of mealiness susceptibility must be identified in melting flesh types.
Within melting flesh cultivars, susceptibility to mealiness appears to be a quantitative trait with a significant genetic component (Peace et al. 2005a). Heritability was calculated as 0.25–0.30 within melting flesh progeny of two peach populations, with duplicated trees and observations conducted over three years (Peace et al. 2006), indicating that unless this genetic component is controlled by many small-effect loci, it should be feasible to discover and exploit loci conditioning mealiness susceptibility. Genetic models based on phenotypic segregation in controlled crosses suggest that in melting flesh types, mealiness is controlled by as few as two loci with dominant gene action (Peace et al. 2006). Genome-wide QTL analysis in one population identified at least three stable QTLs collectively accounting for almost 50% of the genotypic variation in melting flesh progeny. These QTLs did not always combine additively; some were compensatory, suggesting that if used in marker-assisted selection, one can be selected for in the absence of another to achieve the same level of resistance (Ogundiwin et al. 2007). The QTLs are being further targeted, via map saturation and verification in larger populations, while simultaneously taking a candidate gene approach (see below) to develop diagnostic genetic tests (Peace et al. 2005c, 2006; Ogundiwin et al. 2007). EndoPG may also have a further role to play. Given the major effect of the absence of the Melting flesh endoPG gene on mealiness, absence of the Freestone gene or other, less extreme, alleles of the F-M locus may be expected to quantitatively affect mealiness susceptibility. Indeed, in a melting flesh population segregating only for presence of the Freestone endoPG gene, a QTL for mealiness susceptibility co-located with the F-M locus (E. Ogundiwin et al., manuscript in prep.). A comprehensive microarray analysis is currently underway to obtain to identify additional candidate genes associated with mealiness and elucidate the functional relationships (Ogundiwin et al. 2008).
In conclusion, mealiness susceptibility is a heritable quantitative trait for which an understanding of its genetic basis would be valuable for crop improvement. Genomic analysis is dissecting its complexity into specific elements, and it appears likely that with available resources and technologies, the controlling genes will soon be identified.
As shown in the examples above, highly heritable single gene traits are the most amenable to revealing their genetic architecture. Dirlewanger et al. (2004) summarized the locations of 28 qualitative traits in the Prunus genome, for example. Closer examination of such loci may reveal further complexity, and thus additional efforts are useful to understand further effects and interactions. Nevertheless, identification of such major loci allows their use in crop improvement in Rosaceae through genotyping and other approaches (described later). Because quantitative traits require an understanding of potentially many interacting genetic and environmental factors, they are more difficult to elucidate. Ideally, quantitative traits are dissected into their individual components, as attempted for susceptibility to cold storage disorders in peaches (Ogundiwin et al. 2007) which led to the identification of a possible controlling gene for a large proportion of the genetic component of susceptibility to browning (Ogundiwin et al. 2008), in addition to a major heritable role for endoPG in mealiness and bleeding (Peace et al. 2006). For many other quantitative traits, their complexity has not yet been untangled. Highly heritable quantitative traits, while easier to dissect, are also readily improved by phenotypic selection, as the better performing individuals in a breeding program tend to carry the alleles for that superiority. However, there still remains much value in elucidating the genetic architecture of these traits, such as in parent and cross selection, gene pyramiding (combining a series of positive alleles from multiple genes, e.g. for durable disease resistance), and in saving time in progeny selection for crops with extended juvenility. Valuable traits with low heritability are the greatest challenge for genetic architecture dissection, and yet would benefit most from such genomic knowledge as phenotypic selection results in slow genetic gain over generations. Advances in genomics tools and technologies may address even these historically recalcitrant traits.
1.2 Genomic Approaches for Crop Improvement
Genomics approaches fall into various categories and go under various names. Structural, functional, and comparative genomics describe three basic categories of knowledge that researchers gather as they ultimately seek to discover the genetic basis of biological processes and important agronomic traits. Within, and often spanning each of these fields of study, are interconnected technologies and techniques that can be brought to bear in such scientific endeavors, and form an expanding toolkit that the modern Rosaceae genomicist (or geneticist) can employ for their fundamental or applied research (Fig. 2). Some of these approaches are described below, with examples of their application in the Rosaceae family.
1.2.1 Fields of Study
1.2.1.1 Structural Genomics
Structural genomics is concerned with the physical structure and organization of individual genomes. Genome maps, both genetic linkage maps and physical maps, provide an informative description of the chromosomes of Rosaceae crops that can be used to localize important loci and determine interactions between them that ultimately produce phenotypes of interest. Genetic linkage maps, which describe the degree of co-inheritance between genetic loci across the genome of an organism, are abundant for Prunus crops (stone fruit and almond), pome fruit crops (apple and pear, and under development for others such as loquat), cane berries (raspberry and blackberry), rose, and diploid strawberry (Dirlewanger et al. 2004; Shulaev et al. 2008; Jung et al. 2008). Such maps are usually produced for the purposes of determining the genetic control of specific traits. However, reference maps of Prunus (Aranzana et al. 2003), apple (Silfverberg-Dilworth et al. 2006), and strawberry (Sargent et al. 2006) have been developed for use as a general resource within crop groups. The bin-mapping approach (described later) can simplify the general placement of any DNA sequence within an existing genetic linkage map, particularly when combined with a heterozygous reference linkage map (Howad et al. 2005).
More recently, physical maps have been created (peach: Zhebentyayeva et al. 2008; apple: Han et al. 2007) to allow more precise genomic placement of any DNA sequence. Physical maps describe the location of hundreds or thousands of overlapping identifiable sequence landmarks, such as BACs (large segments of the genome cloned into bacteria) or ESTs (expressed sequence tags), which have been anchored to a genetic linkage map. In contrast to linkage maps, physical maps measure distance in base pairs rather than relative genetic linkage measured in centiMorgans (cM) and the number of base pairs contained within a cM will vary across the genome. A fully-saturated physical map would contain BACs (or other DNA clones) tiled across the entire genome and allow physical base-pair distances between features of a chromosome to be readily calculated.
The ultimate genome map would fully integrate genetic linkage markers, physical location of cloned genomic DNA and complete genomic sequence for all the chromosomes of an organism. Complete genome sequencing is currently underway for one reference individual each for apple, peach, and strawberry, placing the Rosaceae plant family at the cutting edge of the genomics revolution. These complete maps are expected to function as publicly accessible resources for each crop group by 2010, greatly enhancing the efficiency of identifying gene networks controlling important traits for rosaceous crop production.
Structural genomics is also applicable to a finer scale, in determining the physical structure of individual loci. The structural organization of the self-incompatibility locus of Prunus, for example, as described by Ushijima et al. (2004), deepens our understanding of the functioning, diversity, and manipulation of the evolutionarily and commercially important traits of cross-compatibility and self-fertility.
1.2.1.2 Functional Genomics
Functional genomics addresses biological questions by studying the function of individual genes and the interactions among groups of genes. It uses both “forward” genomic (or genetic) approaches that start from a phenotype or function and work toward the identification of DNA sequence, and “reverse” approaches that start from DNA sequence and work back to a function. Functional genomics typically relies upon a complement of forward approaches to discover genes associated with a trait and reverse approaches to confirm and study the role of specific genes in biological function. Global or genome-wide analysis is used to identify various genes or gene networks associated with a trait. These include methodologies such as phenotypic screening of mutant “libraries” and many types of transcriptional profiling which are described below. BLAST analysis is often relied upon to compare the nucleic acid sequence homology of the genes or transcripts identified in the analysis against large databases of previously annotated sequences from a wide array of organisms. The supposition of function from sequence homology is a valuable shortcut in functional genomics, and effective in many cases. However, BLAST annotation does not determine gene function, it merely suggests it. In addition, many sequences lack significant homology to functionally characterized proteins or domains, and functionality cannot be implied. For example, Horn et al. (2005) reported that 24.3% of almost 10,000 peach EST sequences had no known homology. Functionality is usually determined or verified using “reverse” genomic approaches, such as RNAi, over-expression of a transgene, and some types of transcript profiling. Given the large cost of one-gene-at-a-time functional testing, such studies are usually limited to genes with known or at least strongly suspected horticulturally significant function.
A major constraint in functional genomics is the availability of high-throughput phenotyping and functional assays to facilitate the analysis of hundreds or thousands of mutants and genes. The long juvenility of many Rosaceae species and the importance of flower and fruit traits in these crops make this a significant challenge for Rosaceae genomics. Altering the expression of the MADS box genes regulating juvenility through genetic engineering provides a possible mechanism to overcome this obstacle and has recently been demonstrated to reduce juvenility in apple and plum (Flachowsky et al. 2007; Kotoda et al. 2006).The development of high-throughput gene function testing platforms are also being developed for Rosaceae (http://strawberrygenomics.com/).
1.2.1.3 Comparative Genomics
Comparative genomics connects studies in structural or functional genomics across crops by seeking commonalities within and between Rosaceae genera and subfamilies. Comparative genomics in Rosaceae is based on the assumption, originating from taxonomic classification by morphology, that rosaceous species are connected through a shared ancestry and thus have genomic similarities. Researchers investigating Rosaceae evolution seek to identify how, when, and where the family split from others, and member species and higher taxonomic clades (e.g. subgenera, genera, and subfamilies) differentiated from each other (e.g. Potter et al. 2007). Evolutionary genomics in Rosaceae is beginning to provide a solid foundation for comparative genomics, facilitating the transfer of genetic knowledge between species and shedding light on the genomic basis of the diversity of form and function in this family. Genetic maps constructed for specific experimental populations are readily aligned with others from the same crop where common markers are used, and such efforts between crops have identified an almost identical genome structure between apple and pear (Pierantoni et al. 2004) and between all crop members of Prunus (Dirlewanger et al. 2004). Common elements have also been identified across more distantly related Rosaceae crops, such as strawberry and Prunus (Vilanova et al. 2007) and apple and Prunus (Dirlewanger et al. 2004). Once whole genome sequences are available for apple, peach, and strawberry, as well as others as sequencing and assembly technologies advance and become cheaper, it should be possible to reconstruct the ancestral “Rosaceae genome” – a reference genome map for the Rosaceae family that will greatly facilitate the transfer of genetic knowledge across crops. Large-scale functional annotation of ESTs in Rosaceae relies on comparative genomics extending beyond this family, with gene function described from GenBank sequences that rarely originate from Rosaceae species. Comparative genomics on a gene-specific scale is illustrated in the characterization of a powdery mildew resistance locus across Malus, Prunus, and Rosa (Xu et al. 2007).
1.2.2 Technologies and Techniques
1.2.2.1 Genetic Mapping and QTL Analysis
Genetic loci are assembled into linkage groups and ordered relative to each other within groups using the technique known as genetic linkage mapping. Linkage groups represent chromosomes, initially partial segments and eventually whole chromosomes, once enough loci are included to span intervening gaps between the segments. Distances between loci are described in recombination units, usually cM. Genetic maps are created to serve either of two purposes: (1) to develop a general resource that locates new loci or “markers” on the genome of an organism, frequently referred to as a reference map or (2) to determine the number, location, and effects of loci controlling specific traits of interest. As described in Structural genomics above, genetic maps exist for all of the major rosaceous crops, including reference maps for each of the three crop subfamilies. The public Genome Database for Rosaceae displays many of these maps to allow users to focus in on regions of interest, or align maps for comparative purposes (Jung et al. 2008). Genetic maps in Rosaceae began with qualitative morphological traits under monogenic control, expanded to the use of isozyme, restriction fragment length polymorphism (RFLP), and random amplified polymorphic DNA (RAPD) markers, and now tend to rely on simple sequence repeat (SSR) markers due to their ability to facilitate alignment with other maps. Readily generated, but population-specific, dominant markers such as amplified fragment length polymorphisms (AFLPs) are used to saturate the regions between SSRs. Markers derived from ESTs and candidate genes are increasingly being included in genetic maps (see Candidate gene approach below)
Traditional QTL discovery approaches utilize software that require specific population types, usually F2 or backcross which have relatively simple statistics for QTL analysis underlying them. However, experimental mapping germplasm of Rosaceae crops are typified by F1 populations, because widespread self-incompatibility and high heterozygosity, coupled with long generation times for the tree crops, make it difficult to construct the simpler genetic populations. The development of the double pseudo-testcross strategy (Grattapaglia and Sederoff 1994) allowed researchers to use F1 populations by mimicking the backcross model for each heterozygous parent of the cross. Once software became available for QTL analysis of F1 populations that considered possible effects from all four alleles segregating at a locus in a diploid cross of QTLs (MapQTL, van Ooijen et al. 2005), it quickly became popular in Rosaceae QTL studies. Available QTL mapping software applies to diploid species, and QTL analysis in polyploids remains problematic. Thus, genetic maps in polyploid crops of Rosaceae (strawberry, plum, rose, and tart cherry) are the least developed, and QTL discovery in these crops the least advanced.
To “map” or estimate the location of a marker on a given chromosome, a marker must be polymorphic within the population so that the frequency of recombination among the progeny can be measured. Reference maps therefore benefit from the use of wide crosses between unrelated parents to maximize the chance that any marker is polymorphic. The Prunus reference map relies on a cross between peach and almond and most markers screened on it are polymorphic (Aranzana et al. 2003).
Bin-mapping, described by Howad et al. (2005), is emerging as an efficient approach to locate any marker or DNA sequence in an organism’s genome. By screening markers on a subset of the mapping population pre-selected to represent widely separated cross-over events on all chromosomes (referred to as a bin-set), a general map location can be rapidly estimated for polymorphic markers. The bin-set for the TxE (almond “Texas” x peach “Earlygold”) reference population for Prunus consists of just eight plants (one of the parents, the F1 hybrid, and six of F2 progeny plants), and any polymorphic codominant marker will fall into one of 67 intervals on the map (“bins”), each equivalent to approximately an eighth of a chromosome (Howad et al. 2005). Bin-sets are also being developed for specific mapping populations, such as apple (Celton et al. 2008), aiding in the marker saturation of targeted regions and the rapid placement of candidate gene markers within a genetic map.
To determine the genetic control of specific traits, parents are chosen that contrast in expression of the trait, in order that the mapping population segregates for the trait, and in general genetic background, so that markers are likely to be polymorphic. For example, in genetic investigations of fruit size in sweet cherry, a large-fruited elite cultivar was crossed with a small-fruited wild variety (Olmstead et al. 2008). With genotypic and phenotypic data collected for the population, statistical procedures are then used to associate allelic variation in genetic markers with performance differences, and position influencing loci on the genetic map. Locations of many qualitative and quantitative traits, for quality, productivity, resistance to diseases, pests, environmental stresses, and physiological disorders are known for Rosaceae crops (described in the later crop-specific chapters). Loci controlling single gene traits can be readily located with simple linkage analysis, where the locus is treated as just another marker. Dirlewanger et al. (2004) summarized the locations of numerous qualitative traits in Prunus. The most common method used to identify associations between markers and quantitative traits in Rosaceae is standard QTL analysis, which uses genotypic and phenotypic data collected from large mapping populations. A popular software package used for QTL analysis is MapQTL (van Ooijen 2005). This QTL identification software is compatible with the map construction software JoinMap that enables the joint analysis of markers inherited from each parent. While inheritance from multiple parental sources complicates genetic analyses, this is a common feature for outcrossing Rosaceae species, where F1 experimental populations are used. The use of MapQTL therefore allows the simultaneous detection and joint influence determination of QTLs with multiple alleles from each parent. Association mapping is a technique to establish gene-trait associations that is useful where mapping populations have not been established, but germplasm from a diverse collection of unrelated individuals is available (Oraguzie and Wilcox 2007). Use of association mapping in Rosaceae has only just begun.
1.2.2.2 Transcript Profiling
Transcript profiling has become a cornerstone of functional genomics because it provides a high-throughput forward (function to sequence) genomic approach for gene discovery that does not require high-throughput functional assays. Transcripts are isolated from a group of plant samples and identified by a variety of methods that take advantage of advances in DNA sequencing and/or the accumulated DNA sequence information available for a given species. In general, the genomicist designs treatments to be applied to plants prior to sample harvest and that provide insight into the particular function to be studied. There are currently several good methodologies for transcript profiling and the development of new methodologies is a rapidly evolving field. The most recent advances in transcript profiling may be considered obsolete within a few years. However, all methodologies have their own advantages and disadvantages, and an understanding of these can help the genomicist select the methodology most appropriate for their crop and goals. Often, no single method is “best” for a specific experimental system, and a variety of methods can overcome the disadvantages of any one method, to provide a more complete or robust analysis. The most appropriate methodology is also dependent upon the amount of EST and genomic sequence information available for the crop of interest. Hence, the most appropriate methods will change as more sequence information becomes available and new technologies are developed.
Transcript profiling within a crop often begins with expressed sequence tag (EST) profiling studies. ESTs are transcribed, spliced nucleotide sequences that are derived from a specific tissue under a specific set of conditions that provide a crude inventory of the genes expressed under those conditions. Often, the tissue used for transcript isolation is the sole treatment or condition in the study. ESTs are usually produced by high-throughput single pass sequencing of cDNA resulting in low quality sequence information of relatively short sequence length, with a relatively high sequencing error rate. The major advantage of EST profiling is the low cost generation of coding (gene) sequence information in species with little genomic sequence information. EST profiling studies have been conducted in Rosaceous crops and there are currently 416,000 Rosaceae ESTs sequences available in GenBank with over 255,500 ESTs for Malus x domestica, over 71,100 for Prunus persica over 45,400 for Fragaria vesca and over 5,500 for hybrid Rosa (Lazzari et al. 2005; NCBI EST Database 2008; Newcomb et al. 2006). Park et al. (2006) used publicly available EST data to predict genes expressed in apple during fruit growth and development and to predict biochemical pathways involved in biosynthesis of precursors for volatile esters important to fruit flavor. The primary disadvantages of EST profiling are: (1) the genes identified include both genes associated with experimental treatments and housekeeping genes, (2) rare transcripts are difficult to detect, and (3) because EST sequence information is often derived from several different laboratories using different cultivars grown under different environmental conditions, associating trends in gene expression with specific biological functions can be difficult. To facilitate the identification of rare transcripts, cDNA libraries are often “normalized” to equalize the relative abundance of all transcripts (Soares et al. 1994). Because normalization eliminates information on EST abundance, these libraries should not be included in studies attempting to use EST profiling data to establish trends in gene expression with specific biological functions.
Suppression subtractive hybridization (SSH) and cDNA-amplified fragment length polymorphism (cDNA-AFLP) are transcript profiling techniques that can efficiently identify both abundant and rare transcripts differentially up- or down-regulated under specific experimental conditions. Both techniques are useful in species with little or no sequence information. A limitation of both techniques is that they usually do not yield a complete inventory of gene expression. Genes regulated in response to specific treatments are selected in SSH by sequential nucleic acid hybridizations in which the reference treatment cDNA, designated as the “driver”, is present in a molar excess compared to “tester” cDNA, in which changes in gene expression are being investigated (Diatchenko et al. 1996). As the mechanics of the assay also include amplification of differentially expressed sequences by PCR that favor the normalization of up- and down-regulated sequences, regardless of the relative abundance of the original mRNA in the cell, a primary advantage of SSH is its ability to detect rare transcripts often missed by general EST profiling methods. Disadvantages of SSH are that it is not quantitative and requires careful control of non-treatment variation between samples. SSH analysis has been successfully used to characterize apple’s response to several abiotic and biotic stresses, including short days, cold temperature, UV irradiation, fire blight, apple scab and phyllosphere colonization (Ban et al. 2007; Bassett et al. 2006; Degenhardt et al. 2005; Kuerkcueoglu et al. 2007; Norelli et al. 2008). cDNA-AFLP is another PCR based methodology that uses restriction enzymes to cut cDNA, followed by subsequent ligation of adaptors to facilitate PCR amplification and visualization of fragments on polyacrylamide gels to identify differentially expressed transcript. The primary advantage of cDNA-AFLP is that it facilitates direct, side by side comparison of transcript fragments from different cultivars under different experimental conditions for cross cultivar comparisons. Because cDNA-AFLP requires extraction of individual DNA fragments from gels to obtain sequence information, it is labor intensive and is not amenable to high-throughput data recovery. cDNA-AFLP has been widely used to characterize transcriptional responses in the Rosaceae (Balogh et al. 2005; Campalans et al. 2001; Geuna et al. 2007; Jensen et al. 2003)
The generation of EST and genome sequence information within the Rosaceae makes the use of more comprehensive transcript profiling techniques, such as microarray analysis, possible. Microarray analysis is a hybridization-based technique in which thousands of gene probes designed to match predicted open reading frames are arrayed on a solid surface and hybridized to transcripts labeled with a fluorescent dye. In general, RNA is isolated from biological samples, used as template for the synthesis of cDNA that is either labeled with a fluorescent dye or used as template for the synthesis of RNA that is labeled. After hybridization the array surface is laser scanned to determine the amount of transcript hybridized to each probe. It is a quantitative method that allows the researcher to obtain a “snap shot” of the expression of thousands of gene in specific tissues under a specific set of conditions. There are many different platforms used for microarray analysis due to the numerous options in manufacturer, method of fabrication, probe type and array design. Microarrays can be printed with fine-pointed pins on glass slides, often referred to as “printed arrays”, or produced by various photolithography and electrochemical printing methods, sometimes referred to as “biochips”. Early microarrays often used cDNA probes that do not require extensive genome sequence information for design but tend to have higher cross-hybridization between gene family members, greater difficulty detecting splice variants, require PCR synthesis of hundreds of genes that is time consuming and prone to error, and tend to have lower quality control than oligonucleotide probes. Oligonucleotide probes require extensive genome sequence information and bioinformatics analysis for proper design, but they have largely replaced cDNA probes because they can overcome the problems associated with cDNA probes and they facilitate high density arrays. Short oligonucleotide probes (20–30 mers) are cheaper to produce and facilitate ultra-high density arrays, whereas long oligonucleotide probes (60–70 mers) tend to have greater specificity to individual gene family members. Two-channel arrays allow direct comparison of two different treatment samples on a single array by labeling each cDNA template with a different fluorophore. Although an absolute level of gene expression can be obtained from two-channel designs, results from these arrays are often presented in relative difference or the ratio of gene expression among the various probes. One-channel arrays are designed to estimate the absolute level of gene expression from single-dye hybridization. This makes it easier to compare microarray results from different experiments, but requires twice as many arrays. The design and analysis of microarray experiments is complex due to the multiple variables in microarray technology and the large number of gene probes tested. Multiple levels of replication are necessary to account for the variation among biological samples, arrays, transcript labeling and dye detection, resulting in large, costly experiments utilizing many individual arrays. The vast amount of data generated and multiple comparisons between thousands of probes make statistical analysis and data interpretation challenging. Because the technique is quantitative, statistical analysis is necessary to draw valid conclusions regarding changes in gene expression. Although an array may contain tens of thousands to a couple of hundred thousand probes, only a small percent of them may show statistically significant results. The large number of platforms, the number of independent users, the varying data formats and the varying methods of analysis used in microarray experiments make standardization and comparison of results difficult. Despite these limitations, microarrays are a powerful tool for transcript profiling because they are capable of simultaneously detecting changes in the expression of many genes which facilitates the association of specific signaling and enzymatic pathways to complex biological functions.
One of the first Malus microarrays was a 15,720 oligonucleotide probe, printed array developed at HortResearch in 2005 that was based on a subset of non-redundant EST contigs (unigenes) derived from HortResearch’s apple EST database (Newcomb et al. 2006). This array has been used to study the environmental effects on tree-to-tree variability in the orchard and the genesis of fruit aroma (Pichler et al. 2007; Schaffer et al. 2007). More recently, a 40,000 feature Malus array was developed in the laboratory of Dr. Schuyler Korban at University of Illinois that contains 548 control probes and 39,412 long-oligonucleotide (70 mer) probes designed to Malus unigenes derived from publicly available EST data and approximately184,000 Malus ESTs (154,000 5' reads and 30,000 3' reads) identified in an NSF-funded project from different tissues, genotypes, developmental stages and stress conditions (Gasic et al., submitted). These Malus microarrays have also been successfully used for transcript profiling during stone development in peach fruit (Callahan et al. 2008). Microarrays have also been developed as diagnostic tools to detect pathogen development within Rosaceae host species (Schneider and Sherman 2007; Sholberg et al. 2005).
1.2.2.3 Proteomics and Metabolomics
System-wide technologies in molecular biology extend to detection and analyses of the entire protein and metabolite array in organisms, although often focused on particular tissues at particular developmental stages as for transcriptomics. Proteomics (Pandey and Mann 2000) and metabolomics (Fiehn 2002) are the disciplines concerned with the application of such technologies. They can be employed to better understand the molecular physiological processes underlying traits (complementing forward genetics approaches), or the downstream effects of gene expression (complementing reverse genetics approaches). While a rosaceous plant may have in total tens of thousands of different genes and slightly more transcripts (although only a fraction in any given tissue), it may contain hundreds of thousands of different proteins (including enzymes and structural units) and metabolites (particularly secondary metabolites). The interactive networks of these gene-environment products are therefore likely to be extremely complex. Proteomics and metabolomics have great potential to elucidate biological processes, but are recent arrivals on the molecular biology scene and their associated toolboxes are still mostly under development. Challenges remain in large-scale identification of proteins and metabolites (Fridman and Pichersky 2005), in addition to associating networks and specific proteins and metabolites with horticultural traits. Furthermore, while individual proteins can be readily connected to their encoding gene, connecting specific metabolites with their underlying genetic sources is difficult (Schauer and Fernie 2006). As yet, there are only a few applications of these disciplines in Rosaceae. Grimplet et al. (2004) used proteomics to connect expressed genes with their translated products in apricot. Alm et al. (2007) examined hundreds of proteins in strawberry to study allergen content. In apple flesh, Guarino et al. (2007) detected 303 distinct proteins, of which 44 were identified and associated with 28 different genes. Metabolic profiling of apple peel detected more than 200 components, of which 78 were identified (Rudell et al. 2008).
1.2.2.4 Candidate Gene Approach
Narrowing the vast array of information resulting from genomic research to specific genes is fundamental to the application of genomics for crop improvement. The candidate gene approach attempts to utilize knowledge generated by structural, functional, and/or comparative genomics, as well as classical molecular biology, physiology and genetics, to identify “candidate” genes with a high likelihood of playing an important role in the phenotype of a specific trait. Once candidates are identified, DNA markers, such as simple sequence repeat (SSR) and single nucleotide polymorphism (SNP), are developed for the genes. These gene-specific markers are then mapped, and their locations compared to known loci for the trait of interest. Co-localization of candidate gene markers with either known qualitative or quantitative trait loci identifies candidates that warrant functional verification, and provides a rational approach to maximize limited resources for greatest impact. If further functional analysis determines a causative role for the gene in the trait of interest, functional allele-specific markers for the trait are established that test for the causative DNA sequence differences underlying the functional differences. Allele-specific gene markers are also known as “perfect markers”, as they avoid the possibility of recombination that can occur when a marker is only genetically linked to the gene. Gene-specific markers are usually very robust and can be useful across different genera of the Rosaceae. Etienne et al. (2002) described a candidate gene study of peach to identify genes underlying major loci and QTLs for acidity and sugar content. Eighteen candidate genes were chosen, twelve were mapped, and a gene involved in solute accumulation co-located with a QTL for soluble solids concentration. Other examples of significant progress with the candidate gene approach in Rosaceae include associating the gene for flavanone 3-hydroxylase with yellow fruit color in strawberry (Deng and Davis 2001), genes for ethylene biosynthesis and cell wall modification genes of Md-ACS1, Md-ACO1, and Md-Exp7 (an expansin) with firmness and/or storability in apple (Oraguzie et al. 2004; Costa et al. 2005, 2008), PpLDOX (leucoanthocyanidin dioxygenase) with cold storage-induced browning in peach (Ogundiwin et al. 2008), the transcription factor MdMYB10 with red flesh color in apple (Chagne et al. 2007), and endoPG with Freestone, Melting flesh, and mealiness in peach (Peace et al. 2005b).
In some cases, the identification of specific candidate genes can be based on prior biological research that established the association of specific enzymes or proteins with a biological process. The association of endoPG with fruit softening and the establishment of it as a marker for melting flesh is an example of such a case. In other cases, a specific class of protein may be associated with a trait that can serve as a means of identifying “candidates”. For example, major resistance (R) genes often encode nuclear binding site (NBS) – leucine rich repeat (LRR) protein kinases, and NBS-LRR resistance gene analogs (RGAs) can be used for the identification of candidate disease resistance genes (Baldi et al. 2004; Samuelian et al. 2008). A “resistance gene map” was presented by Lalli et al. (2005) that described the genomic location of such gene sequences putatively involved in pathogen resistance in Prunus.
For many complex traits of importance, genomic analysis can lead to the identification of several hundred genes associated with a specific trait. In such cases, bioinformatics combined with inference drawn from the scientific literature can be used to narrow the focus to a smaller number of candidate genes. For example, transcript profiling of fire blight-challenged apple leaf tissue resulted in the identification of 650 Malus expressed sequence tags (ESTs) associated with fire blight disease (Norelli et al. 2008a; Malnoy et al. 2008). Bioinformatics was used to identify fire blight-associated ESTs that (1) appeared unique when compared with ESTs isolated from apple tissues that were not challenged with the fire blight pathogen (Baldo et al. 2007), (2) had significant BLAST similarities to 2,800 Arabidopsis genes known to be regulated in response to bacterial challenge (Thilmony et al. 2006) or systemic acquired resistance, and (3) had been identified by both suppression subtractive cDNA hybridization (SSH) and cDNA-AFLP transcript profiling. The ESTs identified by bioinformatics were then ranked for their potential importance in resistance based upon inferences from the scientific literature. SSR and SNP markers derived from highly ranked fire blight-associated ESTs were mapped in a “M.9” x “Robusta 5” population in which a major QTL for fire blight resistance has been located on linkage group 3 (Peil et al. 2007). Markers for heat shock protein 90 (Hsp81-2), a secretory class III peroxidase, and a serine/threonine-protein kinase mapped to the LG3 fire blight resistance QTL and reduced the QTL’s size from 12 to 4 cM. Markers for a “putative disease resistance protein” (NCBI AY347778) and Skp1 (SCF-type E3 ubiquitin ligase) mapped to positions corresponding to the location of two QTLs reported in other populations (Calenge et al. 2005; Khan et al. 2006). To date, of 28 candidate fire blight resistance gene markers that have been mapped, six have co-located to or near known fire blight resistance QTLs (Norelli et al. 2008b). As whole genome sequence becomes available for Rosaceae species, similar approaches could be used to scan coding regions within established QTLs for potential candidate genes, further improving the efficiency of the candidate gene approach.
Co-localization of a candidate gene marker and a specific genetic locus does not prove a causative role for the gene in a specific trait phenotype; co-localization could be the result of coincidental linkage. Further functional analysis is necessary to establish a causative role. Furthermore, if the candidate gene does not co-localize in a specific segregating population, it cannot be concluded that the gene is not associated with the trait of interest. Complex traits can be affected by multiple biological mechanisms controlled by genes at several locations of the genome that may be of importance in other populations. To be able to detect all functional alleles of all causative genes for a trait of interest in a crop, or at least in the germplasm to be improved in a breeding program, it is therefore important to survey individuals that fully represent the germplasm and apply appropriate bioinformatics tools such as pedigree based analysis (see A uniting statistical approach below) or association mapping.
1.2.2.5 Transgenic Analysis
Efficient Agrobacterium-mediated plant transformation technology has been developed within the three Rosaceae subfamilies containing the majority of rosaceous crops: Amygdaloideae (Srinivasan et al. 2005), Maloideae (Chevreau and Bell 2005; Dandekar 2002), and Rosoideae (Folta 2006; Martin 2002; Oosumi et al. 2006). However, some important crop species within the Rosaceae, such as peach, remain difficult to transform. In crops where the preservation of cultivar identification is desirable, such as apple and pear, genetic engineering provides a means to correct specific trait defects, such as disease susceptibility, in desirable cultivars of economic importance (Malnoy et al. 2007). Genetic engineering can also produce novel phenotypes that may not occur in nature, such as blue colored roses (Katsumoto et al. 2007). In cases where desired phenotypic variation occurs in wild species with deleterious agronomic traits, such as poor fruit quality, genetic engineering can bypass the several generations of breeding crosses that may be required to incorporate the trait into a favorable genetic background (Malnoy et al. 2008). Gene transfer technology also provides a powerful tool for the analysis of gene function, which has been difficult by classical genetic methods in much of the Rosaceae, due to the extended juvenility, large plant size, and self incompatibility that occurs within the family. Although improved transgenic cultivars can result directly from functional analysis, intellectual property rights associated with the technology often necessitate separate tracks for functional analysis and cultivar improvement.
Observing the effects of altered candidate gene expression on biological processes is a proven approach for using sequence information to study biological function (Dandekar et al. 2004; Malnoy et al. 2007). Candidate gene expression can be increased by transgenic expression (over-expression) or reduced by gene silencing. Over-expression requires cDNA or genomic sequence for the entire coding region, frequently referred to a “full-length” sequence. Because the cauliflower mosaic virus (CaMV) 35S promoter will usually result in high levels of gene transcription in most plant tissues, it is frequently the promoter of choice for over-expression studies. However, somaclonal variation and secondary effects caused by the CaMV 35S promoter can complicate analysis of gene function by this method. The 35S promoter can activate expression from other cis-located promoters (Zheng et al. 2007), complicating analysis by the increased expression of more than one gene and thus making this promoter an especially poor choice for the functional analysis of transcription factors. The high levels of transcription resulting from use of the 35S promoter can also trigger gene silencing (Mishiba et al. 2005). Somaclonal variation, which can arise during transformation and tissue culture procedures, results from several causes including gene inactivation (or activation) mediated by transfer DNA (T-DNA) insertion, polyploidy, chromosomal translocations and physiological changes resulting from tissue culture (Brown et al. 1992; Filipecki and Malepszy 2006;). Observed biological differences between a transgenic line and the parent cultivar are the combined result of transgene expression and line-specific somaclonal events, thus requiring the comparison of many transgenic lines to either (1) establish a statistically significant correlation between the level of gene expression and the level of biological function, or (2) separately estimate the effect of somaclonal variation in several transgenic lines transformed with an empty vector and the effect of transgene expression in several lines containing the transgene. The use of a chemically inducible promoter for over-expression studies (Malnoy et al. 2006; Norelli et al. 2007; Zuo et al. 2000) can overcome most of these problems by allowing comparison of the same transgenic line under conditions of non-induced and induced transgene expression, thus overcoming the problem of line-specific somaclonal effects by direct biological comparisons within a single transgenic background. Additionally, constitutive expression of candidate genes can frequently result in deleterious effects on plant growth and/or pleiotropic phenotypes. For example, expression of an endochitinase gene in apple under the control of the 35S promoter both increased resistance to apple scab and stunted growth, making it difficult to conclude what part of the change in resistance was due to endochitinase versus reduced growth (Bolar et al. 2000). An inducible promoter system can overcome these problems by limiting gene expression to a short period of time during biological analysis. A drawback to chemically inducible promoters is that they are usually not applicable for cultivar improvement, thus necessitating separate development tracks for functional analysis and cultivar improvement.
Gene expression can be down-regulated by both transcriptional gene silencing mechanisms, such as transposon insertion, and post-transcriptional gene silencing (PTGS) that is RNA-mediated and also known as RNA interference (RNAi). RNAi is mediated by double-stranded RNA (dsRNA) and is homology-dependent gene silencing (Eamens et al. 2008). RNAi does not require full length coding sequence for gene silencing, thus facilitating functional analysis from EST data or in situations where full-length cDNA clones are not available. RNAi also has some advantages over knock-out, or insertional, mutants when conducting reverse analysis (sequence to function). First, RNAi constructs directly target a specific gene, which overcomes the problem of having to generate a large population of lines with knockout mutations in order to have a high degree of certainty of disrupting the function of any given gene (Helliwill et al. 2002). RNAi constructs will also give rise to plants with different degrees of gene silencing; this can result in viable, partially-silenced lines for genes that are lethal when completely knocked out. RNAi can be induced by transgenes containing an inverted repeat of DNA sequence that will result in the direct synthesis of dsRNA. It can also be induced by aberrant RNA molecules, either native or transgenic, that are converted to dsRNA by an RNA-dependent RNA polymerase (RdRP). Although the requirement for RdRP in initiating RNAi is undisputed, the substrate for this enzyme and what defines an “aberrant’ mRNA is not fully understood (Eamens et al. 2008). RNAi can therefore be induced by various transgene designs in a sense (5’ to 3’), antisense (complementary DNA strand) or inverted-repeat orientation. Because sense and antisense constructs are dependent upon RdRP activity for the production of dsRNA, they tend to be less efficient in inducing RNAi than transgenes containing inverted repeats (Wesley et al. 2001). Inverted repeats occurring in the 3’ untranslated region of the transcript can also efficiently induce RNAi (Brummell et al. 2003), which may allow the use of RNAi in forward genomic approaches (function to sequence) (RNAi News, 2005). Currently, the most efficient transgene design for the induction of RNAi are “hairpin” constructs in which the 2 inverted repeat DNA sequences are separated by a transposable element. Hairpin designs are difficult to construct de novo, however several vectors have been developed to facilitate these designs (Wesley et al. 2001). Additionally, hairpin RNAi vectors that utilize lambda phage recombination, or GATEWAY™ technology (Helliwill et al. 2002; Mathews 2004), are amenable to high-throughput approaches.
The high cost of developing transgenic over-expressing and RNAi-silenced lines limits their use to the genomics analysis of a limited number of candidate genes of horticultural importance. Transient RNAi expression by agroinfiltration has been demonstrated in Fragaria and provides a rapid, low-cost alternative to the selection of stable transgenic lines for the analysis of gene function (Hoffmann et al. 2006). High-throughput reverse genetic analysis would also be greatly facilitated by the development of a virus-induced gene silencing (VIGS) system for the Rosaceae (Constantin et al. 2008; Godge et al. 2008). In comparison to agroinfiltration, in which gene silencing is restricted to the area of infiltration, VIGS can provide systemic gene silencing. A VIGS system is being developed for apple (Li et al. 2004; Yaegashi et al. 2007).
Random insertional mutagenesis has been a powerful tool for the analysis of complex biological traits in model systems because it allows a forward approach (function to sequences) that makes no a priori assumptions regarding the genetic control of a trait. Transformation technology facilitates random mutagenesis by transposon or T-DNA insertion and a large collection of T-DNA insertion mutants and AcDs activation tag lines are under development for Fragaria vesca (Oosumi et al. 2006; Shulaev et al. 2008). Mutagenesis by transposon or T-DNA insertion in the genome provide DNA “tags” to facilitate rapid identification of disrupted sequence, thus eliminating the need for extensive genetic analysis in gene identification. Because of the large number of mutants that must be screened in this approach, forward genomic analysis by mutagenesis relies upon efficient high-throughput phenotypic assays. Random insertional mutagenesis is less effective in organisms with large genomes, such as apple, due to a larger amount of non-coding DNA and therefore a lower frequency of gene disruption per insertion. Although the insertion of some transposons in non-coding regions can alter the regulation of down- and up-stream coding regions, these regulatory mutants can be difficult to analyze and complexity greatly increases with larger amounts on non-coding DNA. Similarly, the methodology has not been very effective in polyploid species.
1.2.2.6 Genotyping and Marker-Assisted Breeding
The most commonly touted channel for using genomics in crop improvement is through genotyping (i.e. the application of genetic tests) of cultivars and breeding germplasm. Genotyping can be applied to existing cultivars in production and advanced selections in breeding programs to better understand and monitor their field performance (diagnostics), to potential breeding parents to better understand their breeding value (parent selection), and to seedling populations in breeding programs to improve efficiency of selection (seedling selection). Genotyping requires the development of marker “tool kits”, which are sets of robust markers that can be readily screened on the germplasm of interest. Robustness refers to verification that the marker-trait associations are maintained over a wide range of germplasm and production conditions, or at least verification in the germplasm and conditions for which the markers are to be specifically applied. Ready screening refers to the availability of genotyping protocols and technologies suited to the number and condition of plants to be tested. Markers flanking a QTL region following QTL analysis, or functional markers representing the genes themselves following candidate gene analysis, are used as the predictive genetic tests of performance by screening DNA obtained from the plants of interest. Marker-assisted breeding (MAB) refers to the use of markers to assist in one or more operations of breeding programs, such as parent selection, family size planning, parentage verification, seedling selection, performance evaluation of advanced selections, and cultivar commercialization. Marker-assisted selection (MAS) refers just to the use of markers for selection in breeding – both of parents and seedlings, but usually referring to seedlings. The development of marker-trait associations, i.e. the experimental stage, is often erroneously included in MAB and MAS, often through the ambiguous term “marker development” which can mean the generation of new markers such as for map construction, the search for marker-trait associations, or the conversion of an experimental association into a robust marker for practical application.
Marker-trait associations must be verified to ensure they are applicable in the material to be tested. There are several reasons that associations may be lost during this verification step. First, the association may be a false positive arising from experimental conditions. Second, linkage disequilibrium (the association between a particular marker allele and trait allele) may be lost due to too much historical recombination. The more closely linked a marker is to the functional sequence difference itself (e.g. a specific mutation in a gene), the greater the likelihood that functional association is maintained. Researchers therefore seek these functional sequence differences even if linked markers are available, although the latter are often able to adequately serve breeding purposes. Use of flanking markers for a QTL increases the likelihood of successful performance prediction, as the specific QTL allele targeted will only lose its association with marker alleles if the very rare case of recombination between both markers and the QTL has occurred. Third, functional alleles identified in an experimental population may not be frequent in wider germplasm and therefore not detected in unrelated plants, limiting the extent of germplasm to which the genetic test is applicable. For example, markers for a newly-introgressed resistance allele are not applicable in the bulk of crosses where neither parent carries the resistance allele. However, verification of marker-trait associations may detect additional functional alleles that do not exist in the experimental population. The purposeful search for available functional alleles is known as allele mining. Allele mining includes describing the alleles present in the plants of interest such as the parents of a breeding program, and may extend to wider genepools such as germplasm collections.
For diagnostics and parent selection, methods of DNA extraction and genotyping can be low-throughput, i.e. at the scale of tens to hundreds of samples at time, as the numbers of plants under investigation are correspondingly limited. The actual marker types used for these low-throughput purposes can be isozymes and RFLPs through to the latest automated technologies. Genotyping for diagnostics and parent testing have therefore advanced the furthest in Rosaceae, and soon after marker-trait associations are discovered in experimental material, the genotypic profiles of cultivars are often reported to indicate the robustness of the associations and to describe the genetic character of each cultivar. Self-incompatibility (SI) groups to which cultivars belong greatly influences orchard design for most Rosaceae tree crops. In almond, cherry, plum, and apricot, uncovering the genes controlling this trait at the S locus enabled the development of simple PCR tests to place cultivars into SI groups, which are used to determine cross-compatible combinations and identify self-fertile cultivars (Tamura et al. 2000; Sonneveld et al. 2003; Sutherland et al. 2004; Halász et al. 2005). Discovery of an allele of an ACS (1-aminocyclopropane-1-carboxylic acid [ACC] synthase) gene in apple conferring low ethylene production and longer storage life led to genotyping of cultivars to characterize their ethylene genotype (Oraguzie et al. 2007). This ACS gene was also genotyped in combination with another gene in the ethylene biosynthetic pathway, ACO (ACC oxidase), to characterize cultivars and advanced selections of an apple breeding program (Zhu and Barritt 2008).
Application of genetic tests in breeding programs to reduce the squandering of resources on low-value seedlings requires the implementation of high-throughput DNA extraction and genotyping. Every year, Rosaceae breeders produce hundreds to many thousands of seeds, which are germinated, grown, field-planted, and eventually mostly eliminated, all the while undergoing phenotypic evaluation for traits of importance, to arrive at a tiny proportion of selected individuals (“selections”) that are worthy of proceeding to more intensive performance evaluations. Marker-assisted seedling selection (MASS) involves integrating genotyping into these routine operations, augmenting the selection process by substituting genetic marker tests for sensory or instrumental phenotypic tests wherever it is determined to be more efficient in cost and/or time. Implementation of MASS for thousands of seedlings in a season requires the development of a streamlined process for sampling, extracting DNA, genotyping, and timely supply and application of results that is relevant to the idiosyncrasies of a breeding program. This infrastructure is an obstacle that most public breeding programs of Rosaceae crops have yet to overcome, as robust markers for numerous traits exist but very few are in operation. MASS for resistance to the diseases of scab and powdery mildew in apple, reported by Kellerhals et al. (2004), represents a rare case of real world implementation.
1.3 Outlook
Unlike field crops such as wheat, corn, or soybean, most individual Rosaceae crops are supported by relatively small industries. The dozen major rosaceous crops represent a very diverse group of plants with assorted attributes and challenges for genetic improvement. Yet this diversity is also the strength of the family. Having a shared ancestral “Rosaceae genome” predicts that the controlling genes of common traits will often be the same, and underlying biological mechanisms may not be as different as appearances suggest. Comparisons between Rosaceae crops provide contrasts that can reveal the controlling gene networks and speed genetic improvement. For example, comparisons between cherry and plum or strawberry and raspberry may reveal the genetic basis of fruit size, apple and pear for fruit shape, and across Rosaceae for disease resistance mechanisms. Similarly, basic biological processes can be uncovered within Rosaceae, for example by comparing plant form between strawberry and apple, fruit ethylene response between climacteric peach and non-climacteric cherry, and fruit development between strawberry, raspberry, rose, apple, almond, and peach. Research funds offered by individual industries are both inappropriate and insufficient to address such fundamental yet far-reaching issues. Studies spanning two or more Rosaceae crops, particularly those across subfamily borders, will require an unprecedented level of coordination and collaboration. Fortunately, the international Rosaceae genomics, genetics, and breeding community has taken enormous strides in this direction, exemplified by several exciting initiatives.
1.3.1 A Centralized Web Portal and Database
The Genome Database for Rosaceae (GDR) was created in 2003 in response to rapidly expanding volumes of genomic data in the public domain. EST libraries and genetic maps were the first genomic resources to be hosted on the GDR, followed by transcript and physical maps. Frequent access to such resources has made the GDR an information hub for the Rosaceae network of scientists, breeders, and allied professionals, displaying community announcements, highlighting projects, providing bioinformatics tools for data analyses, and storing ever-increasing genomic data in a readily-accessible and public database. By collecting and processing these structural, functional, and comparative genomics data in one open location, the GDR has enabled the development of an active Rosaceae genomics community in which members operate beyond the limits of single crops. The GDR continues to take on a greater role as a community information hub. A series of twelve USDA-funded Rosaceae genomics projects that started in 2005 uploaded their data and other project outcomes to the GDR, ensuring wide community dissemination. The diversity of genomic and genetic information represented by these twelve projects is large, and beyond that traditionally housed at this site. Indeed, the GDR is expanding to incorporate genotypic, phenotypic, QTL, pedigree, and gene expression data. Plans are underway to develop education and extension modules, to better inform stakeholders – from researchers and breeders to policy makers, industry, and the general public – about the activities, concerns, breakthroughs, and promise of Rosaceae genetics and genomics.
1.3.2 Shared Mapping Resources
The Rosaceae Consortium of Mapping Populations (RosPOP, www.bioinfo.wsu.edu/gdr/community/international/rospop.php) is an initiative designed to facilitate access to plant materials and information from segregating progeny populations of Rosaceae for researchers other than the population owners. Participation in RosPOP requires a formal, although essentially a goodwill, agreement between consortium members that specifies the resources to be shared. Material supplied includes access to the plants themselves and derived materials (e.g. budwood, pollen, fruit, leaves, DNA, and RNA) and data collected from those individuals (phenotypic and genotypic). Traditionally, individual researchers create and study their own experimental mapping populations, focusing on the traits they are most interested in and have the resources to collect data for. Constructing mapping populations is a time-consuming and expensive endeavor in itself, requiring for the tree crops four or more years from making the crosses between the desired parents until fruit production from the resulting seedlings. RosPOP intends to make maximum use of these valuable genetic resources by bringing to bear additional funds, labor, and scientific expertise for a broader scope and increased efficiency of genetic analyses. This approach fosters new and strengthened collaborations between researchers and institutions, and reduces redundancy in worldwide efforts toward Rosaceae genetic improvement. The advent of RosPOP represents a new era in international collaboration for Rosaceae genetic mapping and gene-trait association research.
1.3.3 Standardized Phenotyping
Another recent advance in community coordination is the concept of standardized phenotyping across Rosaceae genetic resources. Various studied sets of Rosaceae germplasm, such as individuals from genetic experiments, cultivars, breeding populations, or ex situ germplasm collections, tend to be phenotypically characterized according to immediate needs of individual investigations. Lack of consistency between studies limits the utility of collected phenotypic data. In contrast, DNA genotypes can be readily compared between studies. Standardized phenotyping offers an opportunity to align the characterization of germplasm collection accessions, such as those of the USDA’s National Plant Germplasm System, more closely with the needs of breeding programs and the interests of genomics researchers. In a wider context, the ability to directly compare both genotypic and phenotypic data across germplasm sets will greatly enhance Rosaceae-wide efforts to establish gene-trait associations by increasing the size of datasets available for analyses. For example, standardized phenotyping could facilitate direct comparisons between populations from two or more breeding programs to obtain more accurate estimates of heritability and genotype x environment interaction for priority traits. Another example application could be determining similarities in genetic factors underlying phytonutrient composition within and among apple, cherry, and raspberry cultivars.
Standardized phenotyping is a challenging approach, requiring considerable coordination and agreement between researchers to establish both a comprehensive set of descriptors and trust in the validity of the resulting data. Descriptors used must be fairly heritable, efficient to use, and relevant to both industry priorities and biological questions. Although challenging, standardized phenotyping will be necessary to address the extent to which the same gene networks control similar traits across Rosaceae crops or their functional divergence from the ancestral genome. As such, this approach will require and foster unity in international Rosaceae genomics, genetics, and breeding.
1.3.4 A Uniting Statistical Approach
Pedigree Based Analysis (PBA) is a powerful statistical approach able to simultaneously identify marker-trait associations, validate their robustness and applicability to individual breeding programs, and mine alleles for functional diversity. While traditional QTL discovery approaches rely on experimental populations which are usually created for the specific purpose of identifying or fine-mapping QTLs, the PBA approach avoids the need for such dedicated populations. Furthermore, although genomics-assisted breeding requires validation and allele mining in breeding germplasm, traditional QTL approaches stop at the discovery stage. The versatility of PBA is achieved by analyzing genotypic and phenotypic data of breeding germplasm itself. This approach is well suited to the multiple pedigree-linked populations of variable size that typify Rosaceae breeding germplasm (van de Weg 2004). PBA identifies networks of major genes and QTLs that determine genetic variation in horticulturally important traits, elucidating their interactions and mining their functional allelic diversity (van de Weg 2004). The strategy integrates marker and phenotypic data over past, current, and future generations within and across breeding programs, thus creating a flexible and continuously expanding platform for marker identification, validation, and use (van de Weg 2004). The PBA approach is based on two complementary statistical approaches. The first identifies QTL regions based on Markov chain Monte Carlo simulations and Bayesian statistics. The second is based on “Identity By Descent” values of each allele of a genotype, taking the different alleles of founding cultivars as factors in statistical analysis (Bink et al. 2008). PBA was the underlying and unifying scheme for the European HiDRAS project, concluding in late 2007, that aimed to identify genetic factors controlling apple fruit quality (including texture components) for increasing the acceptability of disease resistant apples (Gianfranceschi and Soglio 2004; Kellerhals and Eigenmann 2006).
1.3.5 Team Building
Because the journey from investment in genomic science to profitable fruit production spans a tremendous range of expertise, teams of specialists functioning as collaborative units are necessary to ensure that genomic research will impact crop improvement. Effective team building starts with direct and two-way communication between the scientific community and the fruit industry (Fig. 3A). Communication goals include: (1) making the project more responsive to industry needs, (2) improving the dissemination of genomic research information to the industry community, and (3) fostering the efficient integration of industry needs, research objectives, and the development of new cultivars. Industry-research communication should take place during both project planning and execution, receiving input on industry needs during project planning and identifying possible extension “deliverables” resulting from the research during project execution.
Team building is necessary for investment in genomic research to lead to increased profitability for Rosaceae industries and improved products for consumers. A) Effective team building starts with direct and two-way communication between the scientific community and Rosaceae industries. B) The development of community resources in each specific field of genomics fosters the development of knowledge in all fields. C) Collaboration with computational biologists strengthens projects and leads to development of useful bioinformatic tools. D) Collaboration and two-way communication between scientists working in cultivar development, genomics, and bioinformatics fosters the timely development of new cultivars that meet the needs of industry. E) Similarly, collaborations between biotechnologists and genomicists lead to the development of genetically engineered cultivars, therapeutics, and diagnostic tests that meet industry needs
All three fields of genomics will not necessarily be involved in all projects, particularly in smaller projects with limited resources. However, communication and collaboration among researchers in the various fields of genomics will facilitate the project’s ability to capitalize on new community resources developed in other fields of genomics as they become available (Fig. 3B). Because of the tremendous size of many genomic databases and the need to connect them into effective matrixes, the inclusion of computational biologists or bioinformaticists will strengthened the project team. Their involvement as a collaborating scientist, rather than a support consultant, increases the likelihood that the project will result in innovative computational approaches and useful bioinformatic tools (Fig. 3C).
Similarly, when scientists that will apply advances in genomics to specific horticultural practices are involved in project planning and execution, the likelihood that a project will have a significant impact on crop improvement is increased (Fig. 3D and E). Potential collaborators include geneticists (molecular mapping), plant breeders, horticulturalists, plant physiologists, plant pathologists, entomologists, genetic engineers, and chemists. Respectful two-way communication between scientists, rather than an arrogant assumption that genomics research is superior, will facilitate a synergistic collaboration between disciplines. In summary, the vertical integration of genomic research with industry needs and other scientific disciplines increases the likelihood that funds invested in genomic research will result in significant impact on crop improvement and increased profitability for the fruit industry.
2 Conclusions
The potential impact of genomics on Rosaceae crop improvement is enormous. Just as past breeding and research has delivered varied and valuable genetic products, the science of genomics will contribute to the ongoing advances in cultivar improvement necessary to keep up with new challenges to production and the demands of the marketplace. In-depth understanding of Rosaceae genomes and their functional components will not only impact cultivar improvement, but also foster the development of new diagnostics, therapeutics, and cultural practices. Genomic advances will need to address agricultural sustainability by reducing environmental impact, reducing land and water use requirements, and reducing chemical and energy input. It will need to address consumer desires for high quality products that are beautiful, tasty, healthy, consistent, and convenient, and enhance our quality of life. It will need to address the needs of the Rosaceae crop industries to reduce production costs in order to remain viable in the world market. The genomic tools, technologies, and basic knowledge developed in the short term will provide the foundation for addressing many of these challenges in the long term as they are directed to practical benefit. Advances in Rosaceae systems will also aid under-researched fruit, nut, and perennial flower crops for which Rosaceae crops often provide the unofficial model. Working collaboratively with industry and other scientific disciplines, the opportunity exists to anticipate future needs and, with current genomics capabilities, to pro-actively develop solutions for sustained supply of the many Rosaceae products that improve human health and well-being.
References
Alm R, Ekefjärd A, Krogh M, Häkkinen J, Emanuelsson C (2007). Proteomic variation is as large within as between strawberry varieties. J. Proteome Res. 6 : 3011–3020
Aranzana MJ, Carbo J, Arus P (2003). Microsatellite variability in peach [Prunus persica (L.) Batsch]: cultivar identification, marker mutation, pedigree inferences and population structure. Theor. Appl. Genet. 106:1341–1352
Bailey JS, French AP (1949). The inheritance of certain fruit and foliage characteristics in the peach. Mass. Agr. Expt. Sta. Bul. 452
Baldi P, Patocchi A, Zini E, Toller C, Velaso R, Komjanc M (2004). Cloning and linkage mapping of resistance gene homologues in apple. Theor. Appl. Genet. 109:231–239
Balogh A, Koncz T, Tisza V, Kiss E, Heszky L (2005). The effect of 1-MCP on the expression of several ripening-related genes in strawberries. HortScience 40:2088–2090
Ban Y, Honda C, Bessho H, Pang X-M, Moriguchi T (2007). Suppression subtractive hybridization identifies genes induced in response to UV-13 irradiation in apple skin: isolation of a putative UDP-glucose 4-epimerase. J. Exp. Bot. 58:1825–1834
Bassett CA, Wisniewski ME, Artlip TS, Norelli JL, Renaut J, Farrell RE Jr (2006). Global analysis of genes regulated by low temperature and photoperiod in peach bark. J. Am. Soc. Hort. Sci. 131:551–563
Beavis, W.D., Grant, D. Albertsen, M. and Fincher, R. 1991. Quantitative trait loci for plant height in four maize populations and their associations with qualitative genetic loci. Theor. Appl. Genet. 83: 141–145
Bink MCAM, Boer MP, ter Braak CJF, Jansen J, Voorrips RE, van de Weg WE (2008). Bayesian analysis of complex traits in pedigreed plant populations. Euphytica 161: 85–96
Bolar JP, Norelli JL, Wong K-W, Hayes CK, Harman GE, Aldwinckle HS (2000). Expression of endochitinase from Trichoderma harzianum in transgenic apple increases resistance to apple scab and stunts growth. Phytopathology 90: 72–77
Brown PTH, Lang FD, Lorz H (1992) Molecular changes in tissue culture-derived plants. Adv. Mol. Genet. 5:171–195
Brummell DA, Balint-Kurti, Harpster MH, Payly JM, Oeller PW, Gutterson N (2003) Inverted repeat of a heterologous 3’-untranslated region for high-efficiency, high-throughput gene silencing. Plant J. 33:793–880
Calenge F, Drouet D, Denance C, Van de Weg WE, Brisset M-N, Paulin JP, Durel CE (2005). Identification of a major QTL together with several minor additive or epistatic QTLs for resistance to fire blight in apple in two related progenies. Theor. Appl. Genet. 111:128–135
Callahan AM, Scorza R, Bassett C, Nickerson M, Abeles FB (2004). Deletions in an endopolygalacturonase gene cluster correlate with non-melting flesh texture in peach. Func. Plant Biol. 31:159–168
Callahan AM, Dardick CD, Chiozzotto R, Schaffer RJ, Piagnani MC, Scorza R (2008) Gene Expression Profiling Of Peach Fruit During Stone Development. Plant & Animal Genomes XVI Conference, 2008 http://www.intl-pag.org/16/abstracts/PAG16_P07a_716.html
Campalans A, Pages M, Nesseguer R (2001) Identification of differentially expressed genes by the cDNA-AFLP technique during dehydration of almond (Prunus amygdalus). Tree Physiol. 21:633–643
Chagne D, Carlisle CM, Blond C, Volz RK, Whitworth CJ, Oraguzie NC, Crowhurst RN, Allan AC, Espley RV, Hellens RP, Gardiner SE (2007). Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics 8:212
Chevreau E, Bell R (2005) Rosaceae: Pyrus spp. Pear and Cydonia spp. Quince. In: Litz RE (ed) Biotechnology of fruit and nut crops, CABI Publishing, Wallingford and Cambridge, MA, pp 543–565
Constantin GD, Gronlund M, Johansen IE, Stougaard J, Lund OS (2008) Virus-induced gene silencing (VIGS) as a reverse genetic tool to study development of symbiotic root nodules. Mol. Plant Microbe Interact. 21:720–727
Costa F, Stella S, Van de Weg WE, Guerra W, Cecchinel M, Dallavia J, Koller B, Sansavini S (2005). Role of the genes Md-ACO1 and Md-ACS1 in ethylene production and shelf life of apple (Malus domestica Borkh). Euphytica 141:181–190
Costa F, van de Weg WE., Stella S, Dondini L, Pratesi D, Musacchi S, Sansavini S (2008). Map position and functional allelic diversity of Md-Exp7, a new putative expansin gene associated with fruit softening in apple (Malus x domestica Borkh.) and pear (Pyrus communis). Tree Genet. Genomes 4:575–586
Crisosto CH, Mitchell FG, Ju Z (1999) Susceptibility to chilling injury of peach, nectarine, and plum cultivars grown in California. HortScience 34: 1116–1118
Dandekar AM (2002) Introduction and expression of transgenes in apples. In: Khachatourians GC, McHughen A, Scorza R, Nip W-K, Hui YH (eds) Transgenic plants and crops, Marcel Dekker, Inc., New York and Basel, pp 327–344
Dandekar AM, Teo G, Defilippi BG, Uratsu SL, Passey AJ, Kader AA, Stow JR, Colgan RJ, James DJ (2004). Effect of down-regulation of ethylene biosynthesis on fruit flavor complex in apple fruit. Transgenic Res. 13:373–384
Degenhardt J, Al-Masri AN, Kurkcuoglu S, Szankowski I, Gau AE (2005). Characterization by suppression subtractive hybridization of transcripts that are differentially expressed in leaves of apple scab-resistant and susceptible cultivars of Malus domestica. Mol. Gen. Genet. 273: 326–335
Deng C, Davis TM (2001) Molecular identification of the yellow fruit color (c) locus in diploid strawberry: a candidate gene approach. Theor. Appl. Genet. 103:316–322
Diatchenko L, Lau YFC, Campbel AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E, Siebert PD (1996). Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. 93: 6025–6030
Dirlewanger E, Graziano E, Joobeur T, Garriga-Caldere F, Cosson P, Howard W, Arus P (2004). Comparative mapping and marker assisted selection in Rosaceae fruit crops. Proc. Natl. Acad. Sci. USA 101:9891–9896
Eamens A, Wang M-B, Smith NA, Waterhouse PM (2008) RNA silencing in plants: yesterday, today and tomorrow. Plant Physiol. 147:456–468
Etienne C, Rothan C, Moing A, Plomion C, Bodenes C, Svanella-Dumas L, Cosson P, Pronier V, Monet R, Dirlewanger E (2002). Candidate genes and QTLs for sugar and organic acid content in peach [Prunus persica (L.) Batsch]. Theor. Appl. Genet. 105: 145–159
Fiehn O (2002). Metabolomics – the link between genotype and phenotype. Plant Mol. Biol. 48: 155–171
Filipecki M and Malepszy S (2006) Unintended consequences of plant transformation: A molecular insight. J. Appl. Genet. 47:277–286
Flachowsky H, Peil A, Sopanen T, Elo A, Hanke V (2007). Overexpression of BpMADS4 from silver birch (Betula pendula Roth.) induces early-flowering in apple (Malus x domestica Borkh.). Plant Breeding 126:137–145
Folta KM (2006). Transformation of strawberry: the basis from translation genomics in Rosaceae. In Vitro Cell. Devel. Biol. Plant 42:482–490
Fridman E, Pichersky E (2005). Metabolomics, genomics, proteomics, and the identification of enzymes and their substrates and products. Curr. Opin. Plant Biol. 8: 242–248
Gasic K, Gonzales DO, Thimmapuram J, Malnoy M, Gong G, Han Y, Vodkin LO, Liu L, Aldwinckle HS, Carroll N, Orvis K, Goldsbrough P, Clifton S, Pape D, Fulton L, Martin J, Theising B, Wisniewski ME, Fazio G, Korban SS (submitted) Comparative analysis and functional annotation of a large expressed sequence taf collection of apple (Malus x domestica)
Geuna F, Banfi R, Bassi D (2007). Identification and characterization of transcripts differentially expressed during development of apricot (Prunus armeniaca L.) fruit. Tree Genet Genomes 1:69–78
Gianfranceschi L, Soglio V (2004). The European project HiDRAS: innovative multidisciplinary approaches to breeding high quality disease resistant apples. Acta Horticult. 663:327–330
Godge MR, Purkayastha A, Dasgupta I, Kumar PP (2008) Virus-induced gene silencing for functional analysis of selected genes. Plant Cell. Rep. 27:209–219
Grattapaglia D, Sederoff R (1994). Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross mapping strategy and RAPD markers. Genetics 137: 1121–1137
Grimplet J, Romieu C, Sauvage FX, Lambert P, Audergon JM, Terrier N (2004). Transcriptomics and proteomics tools towards ripening markers for assisted selection in apricot. Acta Horticult. 663: 291–296
Guarino C, Arena S, De Simone L, D’ambrosio C, Santoro S, Rocco M, Scaloni A, Marra M (2007). Proteomic analysis of the major soluble components in Annurca apple flesh. Mol. Nut. Food Res. 51 : 255–262
Hadfield KA, Bennett AB (1998). Polygalacturonases: many genes in search of a function. Plant Physiol. 117:337–343
Halász J, Hegedus A, Hermán R, Stefanovits-Bányai É, Pedryc A (2005). New self-incompatibility alleles in apricot (Prunus armeniaca L.) revealed by stylar ribonuclease assay and S-PCR analysis. Euphytica 145:57–66
Han Y, Gasic K, Marron B, Beever JE, Korban S (2007). A BAC-based physical map of the apple genome. Genomics 89:630–637
Helliwill CA, Wesoey SV, Wielopolska AJ, Waterhouse PM (2002) High-throughput vectors for efficient gene silencing in plants. Funct. Plant Biol. 29:1217–1225
Hoffmann T, Kalinowski G, Schwab W (2006) RNAi-induced silencing of gene expression in strawberry fruit (Fragaria x ananassa) by agroinfiltration: a rapid assay for gene function analysis. Plant J. 48:818–826
Horn R, Lecouls AC, Callahan A, Dandekar A, Garay L, McCord P, Howad W, Chan H, Verde I, Main D, Jung S, Georgi L, Forrest S, Mook J, Zhebentyayeva T, Yu Y, Kim HR, Jesudurai C, Sosinski B, Arus P, Baird V, Parfitt D, Reighard G, Scorza R, Tomkins J, Wing R, Abbott AG (2005). Candidate gene database and transcript map for peach, a model species for fruit trees. Theor. Appl. Genet. 110:1419–1428
Howad W, Yamamoto T, Dirlewanger E, Testolin R, Cosson P, Cipriani G, Monforte AJ, Georgi L, Abbott AG (2005). Mapping with a few plants: using selective mapping for microsatellite saturation of the Prunus reference map. Genetics 171:1305–1309
Jensen P, Rytter J, Detwiler EA, Travis JW, NcNellis TW (2003). Rootstock effects on gene expression patterns in apple tree scions. Plant Mol. Biol. 53:493–511
Jung S, Staton M, Lee T, Blenda A, Svancara R, Abbott A, Main D (2008). GDR (Genome Database for Rosaceae): integrated web-database for Rosaceae genomics and genetics data. Nucleic Acids Res., 36(Database issue): D1034–D1040
Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, Brugliera F, Holton TA, Karan M, Nakamura N, Yonekura-Sakakibara K, Togami J, Pigeaire A, Tao G-Q, Nehra NS Lu CY, Dyson BK, Tsuda S, Ashikar R, Kusumi T, Mason JG, Tanaka Y (2007) Engineering of the rose flavanoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 48:1589–1600
Kellerhals M, Eigenmann C (2006). Evaluation of apple fruit quality within the EU project HiDRAS. In: M Boos (ed) Ecofruit – 12th International Conference on Cultivation Technique and Phytopathological Problems in Organic Fruit-Growing: Proceedings to the Conference, pp 165–171
Kellerhals M, Sauer C, Guggenbuehl B, Gantner S, Frey B, Frey JE, Patocchi A, Gessler C (2004). Apple breeding for high fruit quality and durable disease resistance. Acta Horticult. 663: 751–756
Khan MA, Duffy B, Gessler C, Patocchi A (2006). QTL mapping of fire blight resistance in apple. Mol. Breed 17:299–306
Koornneef M, Alonso-Blanco C, Peeters AJM, Soppe W (1998). Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:345–370
Kotoda N, Iwanami H, Takahashi S, Abe K (2006). Antisense expression of MdTFL1-like gene, reduces the juvenile phase in apple. J. Am. Soc. Horticult. Sci. 131:74–81
Kuerkcueoglu S, Degenhardt J, Lensing J, Al-Masri A-N, Gua AE (2007). Identification of differentially expressed genes in Malus domestica after application of the non-pathogenic bacterium Pseudomonas fluorescens Bk3 to the phyllosphere. J. Exp. Bot. 58:733–741
Lalli DA, Decroocq V, Blenda AV, Schurdi-Levraud V, Garay L, Le Gall O, Damsteegt V, Reighard GL, Abbott AG (2005). Identification and mapping of resistance gene analogs (RGAs) in Prunus: a resistance map for Prunus. Theor. Appl. Genet. 111:1504–1513
Lazzari B, Caprera A, Vecchietti A, Stella A, Milanesi L, Pozzi C (2005). ESTree db: a tool for peach functional genomics. BMC Bioinformatics 6(Supple 4):S16 doi:10.1186/1471-2105-6-S4–S16
Lester DR, Speirs J, Orr G, Brady CJ (1994). Peach (Prunus persica) endopolygalacturonase cDNA isolation and mRNA analysis in melting and nonmelting peach cultivars. Plant Physiol. 105: 225–231
Lester DR, Sherman WB, Atwell BJ (1996). Endopolygalacturonase and the melting flesh (M) locus in peach. J. Am. Soc. Hort. Sci. 121: 231–235
Li C, Sasaki N, Isogai M, Yoshikawa N (2004). Stable expression of foreign proteins in herbaceous and apple plants using Apple latent spherical virus RNA2 vectors. Arch Virol. 149:1541–1558
Mackay TFC (2004). The genetic architecture of quantitative traits: lessons from Drosophila. Curr. Opin. Genet. Dev. 14:253–257
Malnoy M, Reynoird JP, Borejsza-Wysocka E, Aldwinckle HS (2006) Activation of the pathogen-inducible Gst1 Promoter of potato after elicitation by Venturia inaequalis and Erwinia amylovora in transgenic apple (Malus x domestica). Transgenic Res. 15:83–93
Malnoy M, Jin Q, Borejsza-Wysocka E, He SY, Aldwinckle HS (2007) Overexpresssion of the apple MpNRP1 gene confers increased disease resistance in Malus x domestica
Malnoy M, Xu M, Borejsza-Wysocka E, Korban S, Aldwinckle HS (2008) Two receptor-like genes, Vfa1 and Vfa2, confer resistance to the fungal pathogen Venturia inaequlais inciting apple scab disease. Mol. Plant Microbe Interact 21: 448–458
Martin RR (2002). Genetic engineering of strawberries and raspberries. In: Khachatourians GC, McHughen A, Scorza R, Nip W-K, Hui YH (eds) Transgenic plants and crops, Marcel Dekker, Inc., New York and Basel, pp 449–464
Mathews L (2004). RNAi for plant functional genomics. Comp. Funct. Genom. 5: 240–244
Mishiba KI, Nishihara M, Nakatsuka T, Abe Y, Hirano H, Yokoi T, Kikuchi A, Yamamura S (2005) Consistent transcriptional silencing of 35S-driven transgenes in gentian. Plant J. 44:541–556
NCBI EST Database (2008). National Center for Biotechnology Information, Bethesda. http://www.ncbi.nlm.nih.gov/sites/entrez?db=nucest Cited 6 Jul 2008
Newcomb RD, CrowHurst RN, Gleave AP, Rikkerink HA, Allan AC, Beuning LL, Bowen JH, Gera E, Jamieson KR, Janssen BJ, Laing WA, McArtney S, Nain B, Ross, GS, Snowden KC, Souleyre EJF, Walton EF Yauk Y-K (2006). Analyses of expressed sequence tags from apple. Plant Physiol. 141: 147–166
Norelli JL, Bassett, C, Artlip T, Aldwinckle HS, Malnoy M, Borejsza-Wysocka EE, Gidoni D, Glaishman M (2007) Inducible DNA promoters for use in apple. Acta Horticult. 738: 329–334
Norelli JL, Farrell RE Jr, Bassett CL, Baldo AM, Lalli DA, Aldwinckle HS, Wisniewski ME (2008a) Rapid transcriptional response of apple to fire blight disease revealed by cDNA suppression subtractive hybridization analysis. Tree Genetics and Genomes: doi: 10.1007/sl1295-008-0164-y
Norelli JL, Gardiner SE, Malnoy M, Aldwinckle HS, Baldo AM, Borejsza-Wysocka E, Farrell RE Jr, Lalli DA, Celton J-M, Bassett CL, Korban SS, Wisniewki ME (2008b). Using functional genomics to develop tools to breed fire blight resistant apple. Plant & Animal Genomes XVI Conference 2008 http://intl-pag.org/16/abstracts/PAG16_P05h_455.html
Ogundiwin EA, Peace CP, Gradziel TM, Dandekar AM, Bliss FA, Crisosto CH (2007). Molecular genetic dissection of chilling injury in peach fruit. Acta Horticult. 738:633–638
Ogundiwin EA, Peace CP, Nicolet CM, Rashbrook VK, Gradziel TM, Bliss FA, Parfitt D, Crisosto CH (2008). Leucoanthocyanidin dioxygenase gene (PpLDOX): a potential functional marker for cold storage flesh browning in peach. Tree Genet. Genom. 4:543–554
Olmstead JW, Sebolt AM, Cabrera A, Sooriyapathirana SS, Hammar S, Iriarte G, Wang D, Chen CY, van der Knaap E, Iezzoni AF (2008). Construction of an intra-specific sweet cherry (Prunus avium L.) genetic linkage map and synteny analysis with the Prunus reference map Tree Genet. Genom. DOI 10.1007/s11295-008-0161-1
Oosumi T, Gruszewski HA, Blischak LA, Baxter AJ, Wadl PA, Shuman JL, Veilleux RE, Shulaev V (2006) High-efficiency transformation of the diploid strawberry (Fragaria vesca) for functional genomics. Planta 223:1219–1230
Oraguzie NC, Wilcox PL (2007). An overview of association mapping. In: N Oraguzie, EHA Rikkerink, SE Gardiner, and HN De Silva (eds) Association Mapping in Plants. Springer Verlag, New York, pp. 1–9
Oraguzie NC, Volz RK, Whitworth CJ, Bassett HCM, Hall AJ, Gardiner SE (2007). Influence of Md-ACS1 allelotype and harvest season within an apple germplasm collection on fruit softening during cold air storage. Postharvest Biol. Technol. 44:212–219
Pandey A, Mann M (2000). Proteomics to study genes and genomes. Nature 405: 837–846
Park S, Sugimoto N, Larson MD, Beaudry R, van Nocker S (2006). Identification of genes with potential roles in apple fruit development and biochemistry through large-scale statistical analysis of expressed sequence tags. Plant Physiol. 141: 811–824
Peace CP, Ahmad R, Gradziel TM, Dandekar AM, Crisosto CH (2005a). The use of molecular genetics to improve peach and nectarine post-storage quality. Acta Horticult. 682:403–410
Peace CP, Crisosto CH, Gradziel TM (2005b). Endopolygalacturonase: a candidate gene for Freestone and Melting flesh in peach. Mol. Breeding 16:21–31
Peace CP, Crisosto CH, Garner DT, Dandekar AM, Gradziel TM, Bliss FA (2006). Genetic control of internal breakdown in peach. Acta Horticult. 713:489–496
Peace CP, Callahan AM, Ogundiwin EA, Potter D, Gradziel TM, Bliss FA, Crisosto CH (2007). Endopolygalacturonase genotypic variation in Prunus. Acta Horticult. 738:639–646
Pichler FB, Walton EF, Day M, Triggs C, Janssen B, Wunsche JN, Putterill J, Schaffer RJ (2007). Relative developmental, environmental, and tree-to-tree variability in buds from field-grown apple trees. Tree Genet Genomes 3:329–339
Peil A, Garcia-Libreros T, Rickter K, Trognitz FC, Trognitz B, Hanke M-V, Flachowsky H (2007). Strong evidence for a fire blight resistance gene of Malus robusta located on linkage group 3. Plant Breeding 126:470–475
Pierantoni L, Cho KH, Shin IS, Chiodini R, Tartarini S, Dondini L, Kang SJ, Sansavini S (2004). Characterisation and transferability of apple SSRs to two European pear F1 populations. Theor. Appl. Genet. 109:1519–1524
Pflieger S, Lefebvre V, Causse M (2001). The candidate gene approach in plant genetics: a review. Mol. Breeding 7:275–291
Potter D, Eriksson T, Evans RC, Oh S, Smedmark JEE, Morgan DR, Kerr M, Robertson KR, Arsenault M, Dickinson TA, Campbell CS (2007). Phylogeny and classification of Rosaceae. Plant Syst. Evol. 266:5–43
RNAi News, Technology for Functional Genomics (2005). Rich Jorgensen discusses sense RNAi and forward genetics. http://www.rnainews.com/issues/3_23/rnainsider/112622-1.html. Cited June 17, 2008
Robertson DS (1989). Understanding the relationship between qualitative and quantitative genetics. p. 81–87. In: Helentjaris, T. and Burr, B. (eds), Development and Application of Molecular Markers to Problems in Plant Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Rudell DR, Mattheis JP, Curry EA (2008). Prestorage ultraviolet-white light irradiation alters apple peel metabolome. J. Agr. Food Chem. 56:1138–1147
Samuelian SK, Baldo AM, Pattison JA, Weber CA (2008). Isolation and linkage mapping of NBS-LRR resistance gene analogs in red raspberry (Rubus idaeus L.) and classification among 270 Rosaceae NBS-LRR genes. Tree Genet Genomes doi: 10.1007/s11295-008-0160-2
Sargent DJ, Clarke J, Simpson DW, Tobutt KR, Arús P, Monfort A, Vilanova S, Denoyes-Rothan B, Rousseau M, Folta KM, Bassil NV, Battey NH (2006). An enhanced microsatellite map of diploid Fragaria. Theor. Appl. Genet. 112:1349–1359
Schaffer RJ, Friel EN, Souleyre EJF, Bolitho K, Thodey K, Ledger S, Bowen JH, Ma J-H, Nain B, Cohen D, Gleave AP, Crowhurst RN, Janssen BJ, Yao J-L, Newcomb RD (2007). A Genomics Approach Reveals That Aroma Production in Apple Is Controlled by Ethylene Predominantly at the Final Step in Each Biosynthetic Pathway. Plant Physiol 144: 1899–1912
Schauer N, Fernie AR (2006). Plant metabolomics: towards biological function and mechanism. Trends Plant Sci. 11:508–516
Schneider WL, Sherman DJ (2007). Acomprehensive Prunus pathogen detection array. Phytopath 97:S105
Sholberg P, O’Gorman D, Bedford K, Leveswue CA (2005). Development of a DNA macroarray for detection and monitoring of economically important apple diseases. Plant Disease 89: 1143–1150
Shulaev V, Korban SS, Sosinski B, Abbott AG, Aldwinckle HS, Folta KM, Iezzoni A, Main D, Arus P, Dandekar AM, Lewers K, Brown SK, Davis TM, Gardiner SE, Potter D, Veilleux RE (2008). Multiple models for Rosaceae genomics. Plant Physiol. 147:985–1003
Silfverberg-Dilworth E, Matasci CL, van de Weg WE, van Kaauwen MPW, Walser M, Kodde LP, Soglio V, Gianfranceschi L, Durel CE, Costa F, Yamamoto T, Koller B, Gessler C, Patocchi A (2006). Microsatellite markers spanning the apple (Malus x domestica Borkh.) genome. Tree Genet. Genomes 2:202–224
Soares M, Bonaldo M, Jelene P, Su L Lawton L (1994). Construction and characterization of a normalization cDNA library. Proc. Natl. Acad. Sci. USA 91:9228–9232
Sonneveld T, Tobutt KR, Robbins TP (2003). Allele-specific PCR detection of sweet cherry self-incompatibility (S) alleles S1 to S16 using consensus and allele-specific primers. Theor. Appl. Genet. 107:1059–1070
Srinivasan C, Padilla IMG, Scorza R (2005) Rosaceae: Prunus spp. almond, apricot, cherry, nectarine, peach and plum In: Litz RE (ed) Biotechnology of fruit and nut crops, CABI Publishing, Wallingford and Cambridge, MA, pp 512–542
Sutherland BG, Tobutt KR, Robbins TP (2004). Molecular genetics of self-incompatibility in plums. Acta Horticult. 663:557–562
Tamura M, Ushijima K, Sassa H, Hirano H, Tao R, Gradziel TM, Dandekar AM (2000). Identification of self-incompatibility genotypes of almond by allele-specific PCR analysis. Theor. Appl. Genet. 101:344–349
The United States Rosaceae Genomics, Genetics, and Breeding Initiative White Paper (2006). http://www.rosaceaewhitepaper.com Cited June 18, 2008
Thilmony R, Underwood W, He SY (2006) Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 46:34–53
Ushijima K, Yamane H, Watari A, Kakehi E, Ikeda K, Hauck NR, Iezzoni AI, Tao R (2004). The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J. 39:573–586
van de Weg WE, Voorrips RE, Finkers HJ, Kodde LP, Jansen J, Bink MCAM (2004). Pedigree genotyping: a new pedigree-based approach of QTL identification and allele mining. Acta Horticult. 663:45–50
van Ooijen J (2005). MapQTLÒ5, Software for the mapping of quantitative trait loci in experimental populations. Kyazma B.V., Wageningen, The Netherlands
Vilanova S, Arús P, Sargent DJ, Monfort A (2007). Map comparison between two distant genomes of the Rosaceae: Prunus and Fragaria. Poster 454 in Plant & Animal Genome XV, San Diego, CA
Wesley SA, Helliwell CA, Neil AS, Wang M-B, Liu Q, Gooding PS, Singh, SP, Abbott D, Stourtjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590
Xu Q, Wen X, Deng X (2007). Phylogenetic and evolutionary analysis of NBS-encoding genes in Rosaceae fruit crops. Mol. Phylogeneti Evol. 44:315–324
Yaegashi H, Yamatsuta T, Takahashi T, Li C, Isogai M, Kobori T, Ohki S, Yoshikawa N (2007). Characterization of virus-induced gene silencing in tobacco plants infected with apple latent spherical virus. Arch Virol. 152:1839–1849
Zhebentyayeva TN, Swire-Clark G, Georgi LL, Garay L, Jung S, Forrest S, Blenda AV, Blackmon B, Mook J, Horn R, Howad W, Arús P, Main D, Tomkins JP, Sosinski B, Baird WV, Reighard GL, Abbott AG (2008). A framework physical map for peach, a model Rosaceae species. Tree Genet. Genomes DOI 10.1007/s11295-008-0147-z
Zheng, X, Deng W, Luo K, Duan H, Chen Y, McAvoy R, Song, S, Pei Y, Li Y (2007). The cauliflower mosaic virus (CaMV) 35 S promoter sequence alters the level and patterns of activity of adjacent tissue- and organ-specific gene promoters. Plant Cell. Rep. 26:1195–1203
Zhu Y, Barritt B (2008). Md-ACS1 and Md-ACO1 genotyping of apple (Malus x domestica Borkh.) breeding parents and suitability for marker-assisted selection. Tree Genet. Genomes DOI 10.1007/s11295-007-0131-z
Zuo J, Qi-Wen N, Chua N-H (2000). An estrogen receptor-based transctivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24:265–273
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2009 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Peace, C., Norelli, J. (2009). Genomics Approaches to Crop Improvement in the Rosaceae. In: Folta, K.M., Gardiner, S.E. (eds) Genetics and Genomics of Rosaceae. Plant Genetics and Genomics: Crops and Models, vol 6. Springer, New York, NY. https://doi.org/10.1007/978-0-387-77491-6_2
Download citation
DOI: https://doi.org/10.1007/978-0-387-77491-6_2
Publisher Name: Springer, New York, NY
Print ISBN: 978-0-387-77490-9
Online ISBN: 978-0-387-77491-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)