Abstract
Four olfactory receptor gene families, all of them G protein-coupled receptors, have been identified and characterized in mammals – the odorant (OR), vomeronasal (V1R and V2R) and trace amine-associated (TAARs) receptors. Much less attention has been directed towards non-mammalian members of these families. Since a hallmark of mammalian olfactory receptors is their remarkable species specificity, an evaluation of the non-mammalian olfactory receptors is instructive both for comparative purposes and in its own right. In this review I have compiled the results currently available for all four olfactory gene families and discuss their phylogenomic properties in relation to their mammalian counterparts. Representatives of all four families are found in cartilaginous fish and/or jawless fish, allowing a minimal estimate for the evolutionary origin as preceding the segregation between cartilaginous and bony fish or cartilaginous and jawless fish, respectively. Gene repertoires of teleost olfactory receptors are smaller in size (OR, ORA), comparable (olfC), or even larger (TAAR) than the corresponding mammalian gene repertoires. Despite their smaller repertoire size, the teleost OR and ORA families show much larger divergence than their mammalian counterparts. Evolutionary rates vary greatly between families, with evidence for positive selection in teleost OR genes, whereas the ora genes are subject to strong negative selection, and in fact are being conserved among all teleost species investigated. With one exception, ligands are unknown for any of the four teleost olfactory receptor gene families, and so the considerable knowledge about the odor responses of the olfactory epithelium and the olfactory bulb can only be linked indirectly to the receptor repertoires.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 1 Background
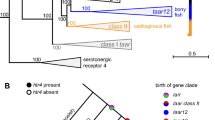
Information about the environment is to a large extent carried by the chemical senses, and in particular the olfactory sense. Thousands of structurally diverse odor molecules perceived and discriminated by vertebrates supply them with a wide range of essential information, ranging from prey localization, predator avoidance, social communication to mating behavior. The receptoire of olfactory receptor genes currently comprises four different gene families, the odorant receptors proper (ORs), vomeronasal receptor genes (V1Rs and V2Rs), and trace amine-associated receptor genes (TAARs), all of them G protein-coupled receptors (GPCRs) (Fig. 1). ORs and TAARs belong to the rhodopsin-like subclass of GPCRs, class A, with short N- and C-termini outside the seven-transmembrane domain, whereas V2Rs belong to class C, and are similar in structure to the metabotropic glutamate receptor, with an additional large N-terminal, extracellular domain. V1Rs have not been formally classified, but are closest to class A receptors.
The four olfactory receptor gene families. ORs, ORAs, and TAARs belong to the class A of GPCRs, with a short N-terminus and a ligand binding site within the TM domains, whereas OlfCs are class C GPCRs, similar to the metabotropic glutamate receptor, with the ligand binding pocket in the large N-terminal extracellular domain
Olfactory receptor gene families in mammals can be rather large, around 1000 OR genes in rodents (Buck and Axel 1991; Mombaerts 2004), and over 100 genes for rodent V1R and V2R genes (Dulac and Axel 1995; Matsunami and Buck 1997). A hallmark of these families is their high species specificity and rapid evolution. Species-specific expansion and loss of genes and even whole subfamilies is a recurrent theme in the mammalian receptor families (Grus et al. 2005; Lane et al. 2004; Zhang et al. 2004). It has been hypothesized that these features provide for efficient adaptation of the olfactory sense to changing environmental conditions. Several recent publications have established the respective properties of the corresponding fish receptor gene families. Here I delineate the four fish receptor gene repertoires and compare their evolutionary properties. Currently the genomes of five teleost species are available (zebrafish, Danio rerio, http://www.sanger.ac.uk; three-spined stickleback, Gasterosteus aculeatus, http://www.broad.mit.edu; medaka, Oryzias latipes, http://medaka.utgenome.org/, http://dolphin.lab.nig.ac.jp/medaka/; tetraodon, Tetraodon nigroviridis, http://www.broad.mit.edu/, http://www.genoscope.cns.fr/spip/ ; fugu, Takifugu rubripes, http://www.fugu-sg.org/project/info.html), as well as unfinished versions of shark and lamprey genomes (http://esharkgenome.imcb.a-star.edu.sg/, http://genome.wustl.edu/, respectively) (Fig. 2).
Zebrafish is the most popular aquatic model organism for molecular studies (Sprague et al. 2008) because of its easy (and cost-efficient) handling, short reproductive cycle, small diploid genome (less than two billion base pairs) and established genetic and genomic methods (which include forward genetic screens, transgenesis, mutagenesis-based knock-out and transient knock-down techniques, but unfortunately no homologous recombination-based modification of endogenous genes). A fast ontogenesis (only five days from fertilization to onset of feeding behavior) has turned out to be a major advantage and not only for developmental studies. Medaka has most of these advantages as well, but is not as widely studied so far. Also, zebrafish does not belong to the neoteleosts like the other four species and, as a more primitive fish, it may be considered a better model organism for tetrapods. The advantage of stickleback lies in a wealth of behavioral observations made in this species. The initial advantage of the two pufferfish species, a several-fold smaller genome, has lost its significance due to recent advances in sequencing techniques.
It will be seen that generally the divergence of the teleost receptor repertoires is larger than that of the corresponding tetrapod receptor repertoires. Repertoire size, however, can be both larger and smaller in teleost vs the corresponding tetrapod receptor families. With one exception, rapid evolution is seen in teleost receptor families, but again, the rate of evolution may be both larger and smaller in the teleost gene families compared to the corresponding tetrapod families.
The olfactory receptor genes can be considered the first layer of olfactory information processing and in fact they define the nature of odorants, since any molecule becomes an odorant solely by virtue of its interaction with an olfactory receptor. However, not many olfactory receptor genes are currently deorphanized, due to the sheer complexity of the task, and because heterologous expression is inefficient for many olfactory receptors. Consequently, detailed ligand response spectra so far only exist for a handful of mammalian and a single fish olfactory receptor (Luu et al. 2004).
Like the opsins, another sensory GPCR family, mammalian OR, TAAR, V1R, and V2R genes are expressed in a monogenic fashion, i.e., a particular receptor neuron expresses only a single gene from a single receptor family (Liberles and Buck 2006; Mombaerts 2004). Initial data from the teleost olfactory system seem to indicate essentially the same, if somewhat relaxed, monogenic expression (Sato et al. 2007). In mammals the neurons expressing the same receptor converge into a single glomerulus per hemisphere (main olfactory bulb, ORs, (Mombaerts 2004) expected for TAARs) or a handful of microglomeruli (accessory olfactory bulb, V1Rs, V2Rs (Mombaerts 2004)). Thus, each receptor gene specifies a separate input channel of the olfactory system and the olfactory bulb constitutes a receptotopic map of odor sensitivities, an odor map (Fried et al. 2002). Both genetic and imaging studies are consistent with such a receptotopic organization in the teleost olfactory bulb (Friedrich and Korsching 1998; Fuss and Korsching 2001; Sato et al. 2005 , 2007); for a more detailed discussion see Yoshihara, this volume. Individual odorants generally bind to several receptors with different affinities and individual receptors generally bind more than one odorant (Buck 2000; Kajiya et al. 2001) – the result is a combinatorial representation by unique, albeit partly overlapping subsets of receptors, ensuring near limitless coding capability of the system. However, highly specific and possibly unique receptors may be expected for pheromones (cf. Friedrich and Korsching 1998; Kajiya et al. 2001).
2 2 Phylogenomic Properties of Four Olfactory Receptor Gene Families
2.1 2.1 Teleost OR Repertoires: Higher Divergence and Smaller Extent than in Mammals
In initial studies a much higher divergence of teleost OR genes as compared to their mammalian counterparts had already been noted, with homologies as low as 20% in pairwise comparisons (Ngai et al. 1993; Weth et al. 1996). Since acceptable quality genomic databases have became available, two groups have made the effort to establish the complete OR repertoire in several fish species, among them zebrafish. Niimura and Nei (2005) have identified 102 intact zebrafish OR genes (plus 35 pseudogenes), which they subdivided by phylogeny into nine groups (Fig. 3). With respect to the relatively large number of pseudogenes it should be noted that the reported prevalence of pseudogenes depends very much on the quality of the databases, i.e. many ostensible pseudogenes are due to errors in the databases. Indeed, in the second large study by Alioto and Ngai only 4 pseudogenes were found, but 143 intact genes plus at least 7 additional OR genes, for which only partial sequence was available (Alioto and Ngai 2005). In this publication, six of the groups from the first study were confirmed; one, not being monophyletic, was split into two groups, and two were considered as outside the scope of the OR family. Traditionally, OR genes have been subdivided into two classes, a so-called fish-specific class I, for which some mammalian family members were also later identified, and a class II, which contains most of the mammalian OR genes. Mammals have lost all but two of the eight to nine groups present in teleost fish, but large gene expansions in these two groups generated OR repertoires an order of magnitude larger than those found in teleost fish.
Phylogenetic tree of four olfactory receptor gene families in zebrafish and fugu. Sequences were retrieved from Niimura and Nei (2005), zebrafish and fugu ORs; Saraiva and Korsching (2007), zebrafish and fugu ORAs; Hashiguchi and Nishida (2007), zebrafish and fugu TAARs; Alioto and Ngai (2006), zebrafish OlfCs; Hashiguchi and Nishida (2006), fugu OlfCs, and aligned using MAFFT version 6 (Katoh and Toh 2008). Outgroups used are metabotropic glutamate receptors, zebrafish opsins, calcium sensor, and T1R receptors. A handful of genes segregating with the outgroups were removed. The tree was constructed using the NJ algorithm (Thompson et al. 1997) and displayed using Hypertree (Bingham and Sudarsanam 2000). Black lines, zebrafish genes; light grey lines, fugu; dark grey lines, outgroups
The zebrafish OR repertoire turns out to be several-fold larger than that of two pufferfish species, which have less than 50 OR genes (Alioto and Ngai 2005; Niimura and Nei 2005). Cloning efforts have been made for catfish, medaka, and loach OR genes (Irie-Kushiyama et al. 2004; Ngai et al. 1993; Yasuoka et al. 1999), but no genome-wide search has been performed for OR genes from other fish species and so it remains to be seen whether these differences in repertoire size reflect the difference between early and late diverging teleost fish, the genome compaction in the pufferfish genus, or, conceivably, the relative importance of the olfactory sense in these species.
The evolutionary origin of the OR gene family was elucidated by a comparison of teleost fish, amphibian, and mammalian OR repertoires. It appears that most, if not all, of the eight or nine teleost OR groups were already present in the common ancestor of teleosts and tetrapods (Alioto and Ngai 2005; Niimura and Nei 2005), and some OR genes even go back to the common ancestor of jawed and jawless fish (Freitag et al. 1999). The latter may be the ancestral genes of class I and II. However, a more thorough analysis of lamprey receptor genes will be necessary to obtain sustainable information about the evolutionary origin of ORs, considering that in a first attempt (Berghard and Dryer 1998) aminergic receptor lamprey genes had been missassigned as ORs.
Despite a slower rate of evolution compared to tetrapod OR families, teleost OR genes do show signs of positive selection (Alioto and Ngai 2005; Ngai et al. 1993). The selective pressure acting on a gene is quantifiable by the ratio of nonsynomous to synonomous base exchanges (dN/dS), which can be calculated for a group of related genes, either as a global value or for each sequence position individually. Positive selection refers to the feature that amino acid-changing mutations are retained to a larger extent than neutral mutations, which results in a dN/dS value larger than one. The frequency of positive selection in the genome is controversial (Studer et al. 2008), but it is generally assumed to occur in transcription factors and some receptor families, including olfactory receptors (Bustamante et al. 2005). In a comparison of the whole zebrafish OR family, four sites showing positive selection were found, two of them within the transmembrane regions expected to contain the ligand binding site (Alioto and Ngai 2005).
The genomic location of teleost OR genes, like their mammalian counterparts, is characterized by the presence of several gene clusters, as well as some isolated genes (Alioto and Ngai 2005). Within the gene clusters, subfamilies are largely contiguous and subfamily members usually exhibit the same transcriptional orientation, suggesting tandem duplication as a mechanism of gene expansion. Teleost OR genes are generally expected to be monoexonic, at least for the coding region (Alioto and Ngai 2005; Niimura and Nei 2005) like their mammalian counterparts, although no dedicated analyses have been performed to substantiate this point.
2.2 2.2 The Teleost ORA Family: A Small, Invariant Gene Repertoire
The ora genes are the teleost homologs of the mammalian V1R genes. The ora designation stands for olfactory receptor gene related to class A of G protein-coupled receptors (the mammalian designation VR stands for vomeronasal receptor, referring to the expression of VRs in a specialized olfactory organ of tetrapods not present in fish). Ora genes have been the latest of the four teleost olfactory receptor families to be found. The first member of this family was uncovered as late as 2005 (Pfister and Rodriguez 2005), followed by the identification of a second gene (Shi and Zhang 2007) and only in 2007 did the full extent of the family become known (Saraiva and Korsching 2007).
With respect to other teleost chemosensory receptor gene families, all fish ora genes form a monophyletic clade, supporting their identification as a single family separate from the other chemosensory receptor families. The ORA clade includes all mammalian V1R receptors; thus the ORA family can be considered paraphyletic, with the mammalian V1Rs originating as a single subclade within the ORA family.
The ora receptor gene family is very small (Fig. 3), with only 6 members compared to over 100 genes in the corresponding rodent V1R gene family. Compared to the other four families, ora genes exhibit several peculiar characteristics, all of them unique for olfactory receptor gene families both in teleosts and tetrapods. Most strikingly, ora genes are highly conserved between all teleost species analyzed so far, such that individual orthologs for all six genes can be detected in all five teleost species analyzed so far (bar a single gene loss in the pufferfish genus) (Saraiva and Korsching 2007). Ortholog ora genes (closest homologs between species) are without exception more closely related to each other than any paralog ora genes (closest homologs within species), indicating that all six family members are evolutionarily much older than the speciation events in the teleost lineage. In fact, some ora genes can even be found in lamprey, a jawless fish (Saraiva and Korsching 2007).
Consistent with this very slow evolution, ora genes show no evidence for positive selection, in contrast to the other olfactory receptor families including the mammalian V1R family. The global dN/dS values for all six ora genes are very small, around 0.2, indicative of strong selective pressure on these genes. When dN/dS values are determined for each codon individually, no single instance of positive selection is found (Saraiva and Korsching 2007). For one of the ora genes an analysis of sequence variation was performed in a group of closely related fish species within the Danio genus; of several algorithms used all but one did not find evidence of positive selection (Pfister et al. 2007).
An unexpected feature of ora genes concerns their genomic location. Olfactory receptor genes often occur in clusters presumably arising from repeated local gene duplication and are often arranged in head-to-tail fashion. In contrast, four of the six ora genes are arranged in closely linked gene pairs across all fish species studied. These gene pairs are asymmetrical, head-to-head for ora1/ora2 and tail-to-tail for ora3/ora4. A pairwise configuration in the phylogenetic tree suggests the existence of three ancestral ORA subclades, all of which are present in lamprey (Saraiva and Korsching 2007), one of which has been lost in amphibia, and a further one in mammals. Two subclades correspond to the above-mentioned gene pairs, the third one consisting of two isolated genes, ora5 and ora6.
Another unexpected feature of the ora genes is the presence of introns in two of six genes. The ancestral genomic structure appears to be monoexonic for ORA1, ORA2, ORA5, and ORA6. This structure is maintained in the mammalian relatives of the ORA1–ORA2 clade (Dulac and Axel 1995; Grus et al. 2005). In marked contrast, ORA4 possesses two exons, and ORA3 four approximately equal-sized exons. ORA3 and ORA4 intron/exon borders are exactly conserved between teleost species, and the sole intron/exon border in ORA4 does not correspond to any intron/exon border in ORA3. Thus, the corresponding intron gains must have occurred after the genesis of the three subclades and indeed after the genomic rearrangements giving rise to the gene pairs, i.e., after the complete ora gene family was established.
All six ora genes are expressed specifically in the olfactory organ of zebrafish, in sparse cells within the sensory surface (Saraiva and Korsching 2007), consistent with the expectation for olfactory receptors and similar to the expression of the tetrapod subclade V1R. Taken together, the high conservation of the ora gene repertoire across teleosts, in striking contrast to the frequent species-specific expansions observed in tetrapods, especially mammalian V1Rs, possibly reflects a major shift in gene regulation as well as gene function upon the transition to tetrapods.
2.3 2.3 The Teleost TAAR Family Evolves Rapidly
Trace amine-associated receptors (TAARs) are related to G protein-coupled aminergic neurotransmitter receptors such as dopamine and serotonine receptors and recognize derivatives of the classical monoamines such as ß-phenylethylamine, octopamine, tryptamine, and tyramine. Initially, TAARs had been considered neurotransmitter receptors (Borowsky et al. 2001), but recently an expression in olfactory sensory neurons was shown for several mammalian taar genes, with expression characteristics very similar to odorant receptors (Liberles and Buck 2006). Thus the taar genes were recognized as a fourth GPCR family of olfactory receptors (Buck 2000).
Following the cloning of the first TAAR receptors in mammals (Borowsky et al. 2001), TAAR genes have been found in several genomes from lower vertebrates (Gloriam et al. 2005) including lamprey (Hashiguchi and Nishida 2007), but not in invertebrates. However, the delineation from classical aminergic neurotransmitter receptors has not been investigated thoroughly so far, and indeed the lamprey genes appear to represent aminergic receptors, not TAARs (Korsching, unpublished observation). The first study evaluating teleost taar genes by Gloriam et al. (2005) made use of very incomplete databases, and thus many of its conclusions, including the size of the family, the phylogenetic reconstruction, the genomic location, the frequency of pseudogenes, the absence of introns, and the suggested nomenclature are now outdated. Still valid are its observations that the TAAR gene family exhibits rapid evolution and correspondingly remarkably species-specific repertoires. A follow-up study confirmed these observations using a more complete data set (Hashiguchi and Nishida 2007). Particularly remarkable is the large taar gene repertoire of zebrafish (Fig. 3); over 100 genes are found, about 5 times the number of genes in the largest mammalian family, and double the number of taar genes found in stickleback (Hashiguchi and Nishida 2007). It will be interesting to study whether the selective pressure acting on teleost taar genes takes the form of positive selection, of which incidences have been observed in the OR, V1R, and V2R families. Currently, taar gene repertoires have been established for fugu, stickleback, medaka, and zebrafish. Fugu has the smallest repertoire, less than 20 genes, followed by medaka with 25 genes, stickleback with 49 genes, and zebrafish with 109 genes (Hashiguchi and Nishida 2007). Ligands for teleost TAARs have not been identified so far, but may include polyamines, which are specifically and sensitively detected by teleost fish (Michel et al. 2003; Rolen et al. 2003).
taar genes occur in a single cluster in tetrapods, evidence of a genesis from local gene duplications, possibly via illegitimate crossover during meiotic recombination. In teleosts, taar genes form two large clusters (Hashiguchi and Nishida 2007), presumably resulting from the whole genome duplication occurring early in the teleost lineage (Nakatani et al. 2007). Additionally, several isolated genes and small groups are found; however, due to the still unfinished genome build in zebrafish, this may not be the final distribution. The most recent common ancestor of tetrapods and teleosts (of lobe-finned and ray-finned fishes) presumably already had a small cluster of taar genes.
Whereas all mammalian and all zebrafish taar genes are monoexonic, an intron was found in many medaka, fugu, and stickleback genes (Hashiguchi and Nishida 2007), consistent with an intron gain early in the evolution of neoteleosts, i.e., relatively late in vertebrate evolution. This is rather remarkable since several whole genome scanning studies found very little evidence for any intron gains during all of vertebrate evolution (Coulombe-Huntington and Majewski 2007; Loh et al. 2008) and may be related to the apparently low selective pressure in the taar gene family.
2.4 2.4 The Teleost OlfC Family is Paraphyletic
OLfC receptors belong to the class C of GPCRs like the mammalian V2Rs, which are their closest chemosensory relatives. Receptors of this class are characterized by their large N-terminal extracellular region, and their similarity to the metobotropic glutamate receptor. Unlike the other three olfactory receptor gene families they are not monophyletic, as three distinct clades have been lumped together under the olfC heading (cf. Fig. 3), following the lead of the mammalian nomenclature (Alioto and Ngai 2006). However, by far the largest clade, group I (Alioto and Ngai 2006), is monophyletic, and the closest phylogenetic neighbor of the largest mammalian V2R clade, albeit both clades are strictly segregated. Its repertoire size varies several-fold between teleost species, but is well within the range of mammalian V2R repertoires – extreme species-specific specialization has led to the complete loss of this family in several mammalian species (Young and Trask 2007). Again, zebrafish exhibits the largest of all teleost OlfC repertoires (Alioto and Ngai 2006; Hashiguchi and Nishida 2006). Two small clades of one to two genes per species appear to be more closely related to the T1R taste receptors and the calcium sensor in some phylogenetic reconstructions (Alioto and Ngai 2006). In contrast to genes from the large clade, genes from the small clades are expressed broadly in the olfactory epithelium (Sato et al. 2005; Speca et al. 1999), conceivably suggesting a modulatory or chaperone function for these genes (cf. Larsson et al. 2004; Silvotti et al. 2005).
OlfC genes exhibit five conserved intron/exon borders resulting in six exons in a characteristic short-short-long-short-short-long arrangement (Alioto and Ngai 2006). None of these intron/exon borders occur in the metabotropic glutamate receptors, but all are shared with the mammalian V2Rs and the closest outgroups, the calcium sensor and the T1R receptors (Korsching, unpublished observation), confirming a common evolutionary origin. To pinpoint the evolutionary origin, data mining in cartilaginous and jawless fish genomes will be required. However, the high species specificity suggests that OlfC/V2R genes constitute an evolutionary recent family, and accordingly may not have been present in the common ancestor of jawed and jawless fish.
Whole subfamilies of OlfC genes are present in zebrafish, but not in neoteleosts, and many instances of species-specific gene expansions are observed. The evolution of the OlfC family appears to be driven to a large extent by local gene duplication, as suggested by the arrangement of most OlfC genes in clusters of phylogenetically related genes (Alioto and Ngai 2006; Hashiguchi and Nishida 2006). Nevertheless, albeit relaxed negative selection is observed at distal ligand binding sites, there is no evidence of positive selection, and in particular the core residues of the amino acid binding motif characteristic for this family are under negative selection (Alioto and Ngai 2006). Although currently no ligands are known for any member of the largest group of OlfC genes (group 1), modelling suggests that many of them have amino acids as ligands like the one well-investigated OlfC member from one of the small groups, OlfC a1 (Luu et al. 2004). Thus, OlfC receptors may constitute the molecular basis to explain odor response studies, which predict many independent receptors for amino acids (Fuss and Korsching 2001).
3 3 A Comparison of Teleost and Tetrapod Olfactory Receptor Repertoires
3.1 3.1 Evolutionary Origin of the Four Olfactory Receptor Gene Families
The estimates for the evolutionary origin of the four olfactory receptor gene families OR, ORA, OlfC, and TAAR have been discussed in the preceding paragraphs. Notably there is no evidence for any of them occurring outside the chordate phylum and, in fact, so far members have only been found in vertebrates. Thus the chemosensory receptors of other phyla such as arthropods or nematodes should have an independent origin within or outside (cf. Sato et al. 2008) the G protein-coupled heptahelical receptor family – which itself dates back to the earliest eucaryotes.
The ora gene family appears to be the oldest family, close to the final repertoire size in lamprey, while the OR family occurs in lamprey but apparently with a much smaller gene repertoire than either later diverging teleost or tetrapod species. Neither OlfC nor TAAR receptor families have been detected in lamprey. It should be noted that some publications concerning lamprey receptor gene assignment have been performed without sufficient information about phylogenetically neighboring gene families, which has led to the erroneous assignment of whole receptor gene families (Berghard and Dryer 1998; Hashiguchi and Nishida 2007). Thus two of the four vertebrate-specific olfactory receptor gene families have their origin in jawless vertebrates, while two families appear to have emerged later in cartilaginous vertebrates.
3.2 3.2 Repertoire Size Varies Considerably Within and Between Teleost Species
Between different teleost species, pufferfish generally have the smallest olfactory receptor gene family sizes, followed by medaka (Oryzias latipes), whereas stickleback (Gasterosteus aculeatus) and zebrafish (Danio rerio) have several-fold larger repertoires than the two pufferfish species analyzed (Takifugu rubripes and Tetraodon nigroviridis). Apart from zebrafish, all these fish species belong to the neoteleostei, a modern group of teleosts, while zebrafish is a more primitive teleost, which belongs to the ostariophysii. Interestingly, it is the zebrafish which always has the largest olfactory receptor gene repertoires. The minimal repertoire sizes found in the pufferfish (Fig. 3) may conceivably be caused by gene loss related to the extreme genome compaction characteristic for this genus (Elgar 1996). Generally, lineage-specific gene gains as well as losses have shaped the teleost OR, TAAR, and V2R-related OlfC repertoires. On the other hand, the V1R-related ora genes have remained a small, rigidly conserved family throughout teleost evolution. Apart from a single gene loss in the pufferfish genus, not a single gene loss or gain event appears to have occurred throughout teleost evolution (Fig. 3).
3.3 3.3 Opposing Shifts in Family Characteristics upon Teleost/Tetrapod Transition in Two Olfactory Receptor Gene Families
Until a short time ago, conventional wisdom held that mammalian species possessed much larger olfactory receptor gene repertoires than those found in earlier diverged vertebrates such as fish. The recent assignment of TAARs as an olfactory receptor gene family (Liberles and Buck 2006) changed this narrative. By far the largest gene family in mammals is constituted by the OR gene repertoire, followed by the V1R and V2R families, and finally the rather limited TAAR gene repertoire. The evolutionary path followed in bony fish development has been distinctly different, with massive diversification in the TAAR family towards sizes close to those observed for teleost OR repertoires. As a result the teleost taar gene repertoires far surpass the mammalian repertoires. On the other hand, the lack of any expansion in the teleost ORA family results in the most extreme contrast in teleost vs tetrapod family size of all four olfactory receptor families.
3.4 3.4 Selective Pressure Among the Four Teleost Olfactory Gene Repertoires
A first indication of selective pressure or absence thereof is given by the species specificity of gene families, which is high for OR, OlfC, and TAARs. Quantitative determination of the selective pressure using dN/dS analysis has shown some incidences of positive selection in OR genes, and relaxed negative selection in OlfC genes, while TAARs still need to be analyzed in that respect. In contrast, the ORA genes show the lowest dN/dS values and a complete absence of positively selected sites (but see Pfister et al. 2007). A rapid evolution in chemosensory receptor gene families is consistent with either pheromonal functions which require species specificity or rapid adaptation to changing environmental specializations.
4 4 Beyond the Phylogenomic Analysis: Current Status of Expression Patterns, Connectivity, and Ligands
From an evolutionary point of view it is interesting to what extent the molecular and cellular logic of the mammalian system is similar to the corresponding teleost receptor gene families. For example, do the same cell types express the teleost receptors? Are the signal transduction pathways conserved? Is the logic of the axonal projection similar to the mammalian situation? With respect to the ligands, major differences between mammalian and teleost olfactory receptors might be expected due to the shift from aquatic to terrestrial life style and a corresponding shift in the biological relevance of many or most odors.
4.1 4.1 Expression Frequency and Spatial Expression Patterns of Teleost Olfactory Receptor Genes
Limited data are available concerning the tissue specificity of olfactory receptor gene expression in teleosts. For mammalian OR an extra-olfactory expression of quite a few receptor genes is known, and for two receptors a function in sperm cell chemotaxis has been postulated (Fukuda et al. 2004; Spehr et al. 2003).
Odorant receptor genes are expressed according to the one receptor neuron – one receptor gene rule, often referred to as monogenic expression. This has generally been true for the other three olfactory receptor families as well, with the exception of a few very broadly expressed receptor genes which may function as chaperones for other receptor genes. The statistics of teleost OR, ORA, and TAAR gene expression appears to conform to the monogenic expression, as most receptor genes, for which reasonably specific probes can be obtained, are expressed in very sparse cell populations, e.g., Weth et al. (1996). Double-labeling studies mostly show exclusive expression of different olfactory receptor gene combinations (Barth et al. 1997), with the exception of a few very closely related receptor gene pairs (Sato et al. 2007).
Within the main olfactory epithelium of mammals, receptor neurons expressing the same OR gene or the same TAAR gene are found at apparently random positions within medial-to-lateral subdivisions of the sensory surface, so-called expression zones (cf. Mombaerts 2004). A small subgroup of genes is expressed in perpendicular expression domains (Hoppe et al. 2006). Within the accessory or vomeronasal olfactory epithelium V1R and V2R-expressing receptor neurons segregate into an apical and a basal layer, respectively. Expression of individual VR genes appears to be restricted to apical-to-basal subdivisions within these layers (Ryba and Tirindelli 1997), although no systematic study has been performed to analyze segregation in this dimension.
Fish OR genes are expressed in spatial expression domains that are broader and overlap more than the mammalian expression zones, but otherwise have the same characteristics of several receptors intermingling in one domain (Weth et al. 1996).
4.2 4.2 Ligands for Teleost Olfactory Receptor Molecules
The olfactory receptor genes can be considered the first layer of olfactory information processing and in fact they define the nature of odorants, since any molecule becomes an odorant solely by virtue of its interaction with an olfactory receptor. Indeed, the olfactory receptors can be thought of as the dimensions of the multidimensional odor space, with each odorant or mixture of odorants defined by a unique set of coordinates in this odor space. Thus a thorough understanding of ligand/receptor interactions is an essential component to fully decipher the logic of olfactory coding.
Not many olfactory receptor genes are currently deorphanized, first due to the sheer complexity of the task, and second because heterologous expression is inefficient for many olfactory receptors. Consequently, detailed ligand response spectra so far only exist for a handful of mammalian olfactory receptors. The general conclusion is that of a relaxed specificity, i.e., several mostly related compounds can excite a particular receptor. With respect to the physicochemical nature of the ligands, one may expect drastic differences between teleost and tetrapod OR and TAAR receptors, since the latter ought to recognize hydrophobic, volatile substances. Teleost and tetrapod V1R and V2R could in principle have similar sets of ligands, because their ligands are transported in mucus towards the receptors, and thus are expected to be hydrophilic. However, the available data do not support this hypothesis.
The only fish olfactory receptor with identified ligands is a member of the OlfC family, OlfC a1 (Alioto and Ngai 2006). It recognizes several amino acids with graded affinity in an heterologous expression system. Interestingly, the optimal ligands for the goldfish receptor are basic amino acids, whereas the zebrafish receptor reacts most strongly to acidic amino acids. Mutation studies have identified the likely residues responsible for this shift in ligand binding characteristics (Luu et al. 2004). It is possible that most OlfC receptors will turn out to bind amino acids, since they share a predicted amino acid-binding motif (Alioto and Ngai 2006).
4.3 4.3 Odor Responses from the Cellular to the Behavioral Level
Known physiologically relevant odorants for teleost fish comprise amino acids, polyamines and nucleotides (food signals), bile acids, steroids and prostaglandins (pheromones) and alarm substances (of graded species-specificity, and so far unidentified molecular nature). These odor signals are detected and processed with high resolution; for example, fish can be trained to distinguish nearly all amino acids from one another (Valentincic et al. 2000 , 2005). Fish possess three types of olfactory receptor neurons – ciliated and microvillous receptor neurons, and so-called crypt cells possessing both cilia and microvilli (Hansen and Zielinski 2005), which so far have not been observed in mammalian olfactory epithelia. In contrast to mammals with at least four segregated olfactory organs (Ma 2007), all fish olfactory neurons are intermingled in a single sensory surface (Hamdani and Doving 2007; Hansen et al. 2004), the olfactory epithelium. The projection areas of ciliated and microvillous receptor neurons have been established by dye tracing and genetic studies to be segregated, if somewhat intertwined (Hamdani and Doving 2002; Hansen et al. 2003; Morita and Finger 1998; Sato et al. 2005); for a detailed discussion see Yoshihara, this volume). This segregation seems to be carried over to the functionally different medial and lateral tracts of projection neurons innervating ciliated and microvillous receptor neurons, respectively (Hamdani and Doving 2007). Interestingly, in contrast to the mammalian system, ciliated neurons and microvillous receptor neurons terminate both in morphologically distinct glomeruli about 50–100 μm in diameter (cf. Baier and Korsching 1994; Sato et al. 2005), and smaller microglomeruli, sometimes referred to as aglomerular plexus (e.g., Baier and Korsching 1994). Anatomical studies have linked crypt cells to small groups of centrally lying ventral glomeruli (Hamdani and Doving 2006; Hansen et al. 2003), which may be innervated by a lateral subdivision of the medial olfactory tract (Hamdani and Doving 2007).
Electrophysiological studies of isolated receptor neurons (Nikonov and Caprio 2007; Restrepo et al. 1990) and projection neurons in the olfactory bulb (Lastein et al. 2006), as well as imaging of odor responses in the olfactory bulb (Friedrich and Korsching 1998) in conjunction with anatomical studies (Baier and Korsching 1994; Sato et al. 2005) have allowed some correlations of receptor neuron type with odor responses on the one hand and olfactory receptor classes on the other. The receptor families seem to be restricted to particular receptor neuron populations, ORs expressed in ciliated neurons, and OlfC in microvillous receptor neurons; see, e.g., Hansen et al. (2004) and Speca et al. (1999). It will be interesting to see whether ORAs are expressed in crypt cells, a sparse cell population, for which so far no receptors have been described (cf. Pfister and Rodriguez 2005; Saraiva and Korsching 2007). The orthologous mammalian V1R genes are expressed in microvillous cells. TAARs may be expressed in ciliated neurons like their mammalian counterparts. Fish microvillous receptor neurons appear to react to amino acids and nucleotides, whereas ciliated neurons may carry the response to bile acids, steroids and polyamines via ORs and TAARs, respectively. Crypt cells of a mackerel species have been shown to respond to amino acids (Schmachtenberg 2006; Vielma et al. 2008). However, this result appears to conflict with a combination of electrophysiological studies (Lastein et al. 2006) and backtracing experiments (Hamdani and Doving 2006) that show a response to steroids in the target region of crypt cells in the olfactory bulb of crucian carp.
Taken together, the fish olfactory bulb provides for the opportunity to study functionally segregated responses of all olfactory receptor neurons in a receptotopic map. Due to the small size and semi-transparent nature of the zebrafish olfactory bulb, it is to be expected that odor responses of all three receptor neuron populations could be measured simultaneously and possibly identified by spatial position. Indeed, in the zebrafish olfactory bulb it has been possible to measure odor responses in lateral, medial, and ventral glomeruli (Friedrich and Korsching 1997, 1998).
5 5 Outlook
The recent discovery of yet another olfactory receptor gene family (the TAARs) invites the speculation that the current repertoire of four different teleost olfactory receptor gene families may still not be complete. Indeed, an olfactory function has been shown for a mammalian member of the membrane-bound guanylate cyclase family (Hu et al. 2007; Leinders-Zufall et al. 2007). The corresponding gene family in teleost fish is known to be larger than in mammals (Yamagami and Suzuki 2005), but a systematic genome-wide study still needs to be done. For the four known families, the repertoires published for several teleost fish species appear reasonably complete. However, for none of these families is evolution in early vertebrates clearly understood. Future access to more and higher quality genome sequences of jawless and cartilaginous fish will enable such studies to be performed more thoroughly than currently possible.
The evolutionary path followed in teleost fish development has been distinctly different from that pursued in tetrapods, with massive diversification in the TAAR family but with a total lack of any expansion in the ORA family. It is expected that the respective ligand repertoires and corresponding biological function will turn out to be distinctly different as well. However, extensive progress in deorphanizing teleost olfactory receptors will be necessary to understand the evolution of ligand repertoires for the olfactory receptor gene families.
References
Alioto TS, Ngai J (2005) The odorant receptor repertoire of teleost fish. BMC Genomics 6:173
Alioto TS, Ngai J (2006) The repertoire of olfactory C family G protein-coupled receptors in zebrafish: candidate chemosensory receptors for amino acids. BMC Genomics 7:309
Baier H, Korsching S (1994) Olfactory glomeruli in the zebrafish form an invariant pattern and are identifiable across animals. J Neurosci 14:219–230
Barth AL, Dugas JC, Ngai J (1997) Noncoordinate expression of odorant receptor genes tightly linked in the zebrafish genome. Neuron 19:359–369
Berghard A, Dryer L (1998) A novel family of ancient vertebrate odorant receptors. J Neurobiol 37:383–392
Bingham J, Sudarsanam S (2000) Visualizing large hierarchical clusters in hyperbolic space. Bioinformatics 16:660–661
Borowsky B, Adham N, Jones KA, Raddatz R, Artymyshyn R, Ogozalek KL, Durkin MM, Lakhlani PP, Bonini JA, Pathirana S et al. (2001) Trace amines: identification of a family of mammalian G protein-coupled receptors. Proc Natl Acad Sci USA 98:8966–8971
Buck LB (2000) The molecular architecture of odor and pheromone sensing in mammals. Cell 100:611–618
Buck L, Axel R (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65:175–187
Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD et al. (2005) Natural selection on protein-coding genes in the human genome. Nature 437:1153–1157
Coulombe-Huntington J, Majewski J (2007) Characterization of intron loss events in mammals. Genome Res 17:23–32
Dulac C, Axel R (1995) A novel family of genes encoding putative pheromone receptors in mammals. Cell 83:195–206
Elgar G (1996) Quality not quantity: the pufferfish genome. Hum Mol Genet 5:(Spec No)1437–1442
Freitag J, Beck A, Ludwig G, von Buchholtz L, Breer H (1999) On the origin of the olfactory receptor family: receptor genes of the jawless fish (Lampetra fluviatilis). Gene 226:165–174
Fried HU, Fuss SH, Korsching SI (2002) Selective imaging of presynaptic activity in the mouse olfactory bulb shows concentration and structure dependence of odor responses in identified glomeruli. Proc Natl Acad Sci USA 99:3222–3227
Friedrich RW, Korsching SI (1997) Combinatorial and chemotopic odorant coding in the zebrafish olfactory bulb visualized by optical imaging. Neuron 18:737–752
Friedrich RW, Korsching SI (1998) Chemotopic, combinatorial, and noncombinatorial odorant representations in the olfactory bulb revealed using a voltage-sensitive axon tracer. J Neurosci 18:9977–9988
Fukuda N, Yomogida K, Okabe M, Touhara K (2004) Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J Cell Sci 117:5835–5845
Fuss SH, Korsching SI (2001) Odorant feature detection: activity mapping of structure response relationships in the zebrafish olfactory bulb. J Neurosci 21:8396–8407
Gloriam DE, Bjarnadottir TK, Yan YL, Postlethwait JH, Schioth HB, Fredriksson R (2005) The repertoire of trace amine G-protein-coupled receptors: large expansion in zebrafish. Mol Phylogenet Evol 35:470–482
Grus WE, Shi P, Zhang YP, Zhang J (2005) Dramatic variation of the vomeronasal pheromone receptor gene repertoire among five orders of placental and marsupial mammals. Proc Natl Acad Sci USA 102:5767–5772
Hamdani El H, Doving KB (2002) The alarm reaction in crucian carp is mediated by olfactory neurons with long dendrites. Chem Senses 27:395–398
Hamdani El H, Doving KB (2006) Specific projection of the sensory crypt cells in the olfactory system in crucian carp, Carassius carassius. Chem Senses 31:63–67
Hamdani El H, Doving KB (2007) The functional organization of the fish olfactory system. Prog Neurobiol 82:80–86
Hansen A, Zielinski BS (2005) Diversity in the olfactory epithelium of bony fishes: development, lamellar arrangement, sensory neuron cell types and transduction components. J Neurocytol 34:183–208
Hansen A, Rolen SH, Anderson K, Morita Y, Caprio J, Finger TE (2003) Correlation between olfactory receptor cell type and function in the channel catfish. J Neurosci 23:9328–9339
Hansen A, Anderson KT, Finger TE (2004) Differential distribution of olfactory receptor neurons in goldfish: structural and molecular correlates. J Comp Neurol 477:347–359
Hashiguchi Y, Nishida M (2006) Evolution and origin of vomeronasal-type odorant receptor gene repertoire in fishes. BMC Evol Biol 6:76
Hashiguchi Y, Nishida M (2007) Evolution of trace amine associated receptor (TAAR) gene family in vertebrates: lineage-specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium. Mol Biol Evol 24:2099–2107
Hoppe R, Lambert TD, Samollow PB, Breer H, Strotmann J (2006) Evolution of the “OR37” subfamily of olfactory receptors: a cross-species comparison. J Mol Evol 62:460–472
Hu J, Zhong C, Ding C, Chi Q, Walz A, Mombaerts P, Matsunami H, Luo M (2007) Detection of near-atmospheric concentrations of CO2 by an olfactory subsystem in the mouse. Science 317:953–957
Irie-Kushiyama S, Asano-Miyoshi M, Suda T, Abe K, Emori Y (2004) Identification of 24 genes and two pseudogenes coding for olfactory receptors in Japanese loach, classified into four subfamilies: a putative evolutionary process for fish olfactory receptor genes by comprehensive phylogenetic analysis. Gene 325:123–135
Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K (2001) Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci 21:6018–6025
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298
Lane RP, Young J, Newman T, Trask BJ (2004) Species specificity in rodent pheromone receptor repertoires. Genome Res 14:603–608
Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB (2004) Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43:703–714
Lastein S, Hamdani El H, Doving KB (2006) Gender distinction in neural discrimination of sex pheromones in the olfactory bulb of crucian carp, Carassius carassius. Chem Senses 31:69–77
Leinders-Zufall T, Cockerham RE, Michalakis S, Biel M, Garbers DL, Reed RR, Zufall F, Munger SD (2007) Contribution of the receptor guanylyl cyclase GC-D to chemosensory function in the olfactory epithelium. Proc Natl Acad Sci USA 104:14507–14512
Liberles SD, Buck LB (2006) A second class of chemosensory receptors in the olfactory epithelium. Nature 442:645–650
Loh YH, Brenner S, Venkatesh B (2008) Investigation of loss and gain of introns in the compact genomes of pufferfishes (Fugu and Tetraodon). Mol Biol Evol 25:526–535
Luu P, Acher F, Bertrand HO, Fan J, Ngai J (2004) Molecular determinants of ligand selectivity in a vertebrate odorant receptor. J Neurosci 24:10128–10137
Ma M (2007) Encoding olfactory signals via multiple chemosensory systems. Crit Rev Biochem Mol Biol 42:463–480
Matsunami H, Buck LB (1997) A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell 90:775–784
Michel WC, Sanderson MJ, Olson JK, Lipschitz DL (2003) Evidence of a novel transduction pathway mediating detection of polyamines by the zebrafish olfactory system. J Exp Biol 206:1697–1706
Mombaerts P (2004) Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci 5:263–278
Morita Y, Finger TE (1998) Differential projections of ciliated and microvillous olfactory receptor cells in the catfish, Ictalurus punctatus. J Comp Neurol 398:539–550
Nakatani Y, Takeda H, Kohara Y, Morishita S (2007) Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res 17:1254–1265
Ngai J, Dowling MM, Buck L, Axel R, Chess A (1993) The family of genes encoding odorant receptors in the channel catfish. Cell 72:657–666
Niimura Y, Nei M (2005) Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci USA 102:6039–6044
Nikonov AA, Caprio J (2007) Highly specific olfactory receptor neurons for types of amino acids in the channel catfish. J Neurophysiol 98:1909–1918
Pfister P, Rodriguez I (2005) Olfactory expression of a single and highly variable V1r pheromone receptor-like gene in fish species. Proc Natl Acad Sci USA 102:5489–5494
Pfister P, Randall J, Montoya-Burgos JI, Rodriguez I (2007) Divergent evolution among teleost V1r receptor genes. PLoS ONE 2:e379
Restrepo D, Miyamoto T, Bryant BP, Teeter JH (1990) Odor stimuli trigger influx of calcium into olfactory neurons of the channel catfish. Science 249:1166–1168
Rolen SH, Sorensen PW, Mattson D, Caprio J (2003) Polyamines as olfactory stimuli in the goldfish Carassius auratus. J Exp Biol 206:1683–1696
Ryba NJ, Tirindelli R (1997) A new multigene family of putative pheromone receptors. Neuron 19:371–379
Saraiva LR, Korsching SI (2007) A novel olfactory receptor gene family in teleost fish. Genome Res 17:1448–1457
Sato Y, Miyasaka N, Yoshihara Y (2005) Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J Neurosci 25:4889–4897
Sato Y, Miyasaka N, Yoshihara Y (2007) Hierarchical regulation of odorant receptor gene choice and subsequent axonal projection of olfactory sensory neurons in zebrafish. J Neurosci 27:1606–1615
Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K (2008) Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452:1002–1006
Schmachtenberg O (2006) Histological and electrophysiological properties of crypt cells from the olfactory epithelium of the marine teleost Trachurus symmetricus. J Comp Neurol 495:113–121
Shi P, Zhang J (2007) Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res 17:166–174
Silvotti L, Giannini G, Tirindelli R (2005) The vomeronasal receptor V2R2 does not require escort molecules for expression in heterologous systems. Chem Senses 30:1–8
Speca DJ, Lin DM, Sorensen PW, Isacoff EY, Ngai J, Dittman AH (1999) Functional identification of a goldfish odorant receptor. Neuron 23:487–498
Spehr M, Gisselmann G, Poplawski A, Riffell JA, Wetzel CH, Zimmer RK, Hatt H (2003) Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 299:2054–2058
Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe DG, Knight J et al. (2008) The zebrafish information network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res 36:D768–D772
Studer RA, Penel S, Duret L, Robinson-Rechavi M (2008) Pervasive positive selection on duplicated and nonduplicated vertebrate protein coding genes. Genome Res 11:11
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Valentincic T, Kralj J, Stenovec M, Koce A, Caprio J (2000) The behavioral detection of binary mixtures of amino acids and their individual components by catfish. J Exp Biol 203:3307–3317
Valentincic T, Miklavc P, Dolenek J, Pliberek K (2005) Correlations between olfactory discrimination, olfactory receptor neuron responses and chemotopy of amino acids in fishes. Chem Senses 30(Suppl 1):i312–i314
Vielma A, Ardiles A, Delgado L, Schmachtenberg O (2008) The elusive crypt olfactory receptor neuron: evidence for its stimulation by amino acids and cAMP pathway agonists. J Exp Biol 211:2417–2422
Weth F, Nadler W, Korsching S (1996) Nested expression domains for odorant receptors in zebrafish olfactory epithelium. Proc Natl Acad Sci USA 93:13321–13326
Yamagami S, Suzuki N (2005) Diverse forms of guanylyl cyclases in medaka fish – their genomic structure and phylogenetic relationships to those in vertebrates and invertebrates. Zoolog Sci 22:819–835
Yasuoka A, Endo K, Asano-Miyoshi M, Abe K, Emori Y (1999) Two subfamilies of olfactory receptor genes in medaka fish, Oryzias latipes. : genomic organization and differential expression in olfactory epithelium. J Biochem 126:866–873
Young JM, Trask BJ (2007) V2R gene families degenerated in primates, dog and cow, but expanded in opossum. Trends Genet 23:212–215
Zhang X, Rodriguez I, Mombaerts P, Firestein S (2004) Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics 83:802–811
Acknowledgement
I would like to thank Kim Robin Korsching for helping with the figures.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2008 Springer-Verlag London
About this chapter
Cite this chapter
Korsching, S. (2008). The Molecular Evolution of Teleost Olfactory Receptor Gene Families. In: Korsching, S., Meyerhof, W. (eds) Chemosensory Systems in Mammals, Fishes, and Insects. Results and Problems in Cell Differentiation, vol 47. Springer, Berlin, Heidelberg. https://doi.org/10.1007/400_2008_11
Download citation
DOI: https://doi.org/10.1007/400_2008_11
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-69918-7
Online ISBN: 978-3-540-69919-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)