Abstract
This chapter presents the NMR technique APSY (automated projection spectroscopy) and its applications for sequence-specific resonance assignments of proteins. The result of an APSY experiment is a list of chemical shift correlations for an N-dimensional NMR spectrum (N≥3). This list is obtained in a fully automated way by the dedicated algorithm GAPRO (geometric analysis of projections) from a geometric analysis of experimentally recorded, low-dimensional projections. Because the positions of corresponding peaks in multiple projections are correlated, thermal noise and other uncorrelated artifacts are efficiently suppressed. We describe the theoretical background of the APSY method and discuss technical aspects that guide its optimal use. Further, applications of APSY-NMR spectroscopy for fully automated sequence-specific backbone and side chain assignments of proteins are described. We discuss the choice of suitable experiments for this purpose and show several examples. APSY is of particular interest for the assignment of soluble unfolded proteins, which is a time-consuming task by conventional means. With this class of proteins, APSY-NMR experiments with up to seven dimensions have been recorded. Sequence-specific assignments of protein side chains in turn are obtained from a 5D TOCSY-APSY-NMR experiment.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- APSY
- Automated peak picking
- Automated resonance assignment
- GAPRO
- NMR
- Projection spectroscopy
- Protein backbone
- Protein side chains
1 Introduction
In NMR studies of biological macromolecules in solution [1–4], multidimensional NMR data are commonly acquired by sampling the time domain in all dimensions equidistantly [5]. With recent advances in sensitivity, such as high field strengths and cryogenic detection devices, the time required to explore the time domain in the conventional way often exceeds the minimal experiment time required by sensitivity considerations, so that the desired resolution determines the duration of the experiment. This situation of the “sampling limit,” is common in three- and higher-dimensional experiments with small and medium-sized proteins [6].
When working in the sampling limit, it is worthwhile to obtain the spectral information by “unconventional” experimental schemes, such as non-uniform sampling of the time domain [7–9] or by combination of two or more indirect dimensions [10–12]. The latter approach is also the basis for projection-reconstruction (PR-) NMR [13–16], where the projection–cross-section theorem [17, 18] is combined with image reconstruction techniques [19, 20] to reconstruct the multidimensional frequency domain spectrum from experimentally recorded projections. Further practical acquisition and processing techniques for unconventional multidimensional NMR experiments have been demonstrated [21–33]. Several of these methods are discussed in the other chapters of this book.
The analysis of NMR spectra involves intensive human intervention, and automation of NMR spectroscopy with macromolecules is thus of general interest. Major challenges are the distinction of real resonance peaks from thermal noise and spectral artifacts, as well as peak overlap [34–36]. On grounds of principle, automated analysis benefits from higher-dimensionality of the spectra [21, 37], since the peaks are then more widely separated, and hence peak overlap is substantially reduced.
APSY (automated projection spectroscopy) combines the technique to record projections of high-dimensional NMR experiments [15] with automated peak-picking of the projections and a subsequent geometric analysis of the peak lists with the algorithm GAPRO (geometric analysis of projections). Based on geometrical considerations, GAPRO identifies peaks in the projections that arise from the same resonance in the N-dimensional frequency space, and subsequently calculates the positions of these peaks in the N-dimensional spectral space. The output of an APSY-experiment is thus an N-dimensional chemical shift correlation list of high quality which allows efficient and reliable subsequent use by computer algorithms. Due to extensive redundancy in the input data for GAPRO, high precision of the chemical shift measurements is achieved. Importantly, APSY is fully automated and operates without the need to reconstruct the high-dimensional spectrum at any point.
In the following sections, the theoretical and practical foundations of APSY are introduced. Several practical aspects are discussed including the sensitivity of APSY experiments. Then, applications of APSY for the assignment of protein resonances are described. For the backbone assignment, the high-quality APSY peak lists are used as the input for a suitable automatic assignment algorithm. For example, the 6D APSY-seq-HNCOCANH experiment connects two sequentially neighboring amide moieties in polypeptide chains via the 13C’ and 13Cα atoms. Further applications are the backbone assignment of unfolded proteins and the side chain assignment of folded proteins.
2 Theoretical Background
2.1 The Projection–Cross-Section Theorem
The projection–cross-section theorem states that an m-dimensional cross section, c m (t), through N-dimensional time domain data (m < N) is related by an m-dimensional Fourier transformation to an m-dimensional orthogonal projection of the N-dimensional NMR spectrum, P m (ω), in the frequency domain [17, 18]. Thereby, P m (ω) and c m (t) are oriented by the same angles with regard to their corresponding coordinate systems (Fig. 1).
Illustration of the projection–cross-section theorem [17–19] for a 2D frequency space with two indirect dimensions k and j. 1D data \( c_1^{jk}(t) \) on a straight line in the 2D time domain (t j , t k ) (left) is related to a 1D orthogonal projection \( P_1^{xy}\left( \omega \right) \) of the spectrum in the 2D frequency domain (ω j , ω k ) (right) by a 1D Fourier transformation, F t , and the inverse transformation, Fω. The projection angle α describing the slope of \( c_1^{jk}(t) \) defines also the slope of \( P_1^{xy}\left( \omega \right) \). The cross peak \( {Q^i} \) (black dot) appears at the position \( Q_f^i \) in the projection. Further indicated are the spectral widths in the two dimensions of the frequency domain, SW j and SW k . and the evolution time increments Δ, Δ k and Δ j (1)–(4). Adapted with permission from [38]
Kupče and Freeman showed that this theorem can be utilized to record projections of multidimensional NMR experiments. The time domain is sampled along a straight line (Fig. 1) and quadrature detection for this cross-section c m (t) is obtained by combing data from corresponding positive and negative projection angles using the trigonometric addition theorem [14, 15]. The subsequent hypercomplex Fourier transformation results in the projections P m (ω) [11, 15, 22]. Projections with a dimensionality of m = 2, with one directly recorded and one indirect dimension, are the most practical case. For such 2D projections, the indirect dimension is a 1D projection of the N − 1 indirect dimensions of the N-dimensional experiment. The orientations of both c 2 (t) and P 2 (ω) are described by N − 2 projections angles.
For example, in a 5D APSY experiment (N = 5), three projection angles α, β and γ, define the orientations of c 2 (t) and P 2 (ω). The two unit vectors \( {\vec{p}_1} \) and \( {\vec{p}_2} \), which span the indirect and the direct dimension, respectively, are given by
An N-dimensional NMR spectrum is spanned by user-defined sweep widths SW i for each of the N dimensions (i = 1,…, N). For a projection spectrum defined by \( {\vec{p}_1} \), an appropriate sweep width SW needs to be calculated (Fig. 1). Considering that the distribution of chemical shifts in a given dimension is well described by a normal distribution [39], the sweep width can be calculated as [40]
where \( p_1^i \) are the coordinates of the vector \( {\vec{p}_1} \) (1). The dwell time for the recording of discrete data points, Δ, is then calculated as
and the resulting increments for the N − 1 evolution times \( {t_i},{\Delta_i} \), in the N − 1 indirect dimensions (Fig. 1), are given by
For an optimal phasing of the projection spectra in the N-dimensional space it is advisable to sample the time domain starting at the origin (Fig. 1). Thus, all APSY pulse sequences should allow sampling access to this time domain point with zero evolution time.
2.2 Projections of Cross Peaks
In a set of j projections with different projection vectors \( {\vec{p}_{1,f}} \), an N-dimensional cross peak \( {Q^i} \) is projected orthogonally to the locations \( Q_f^i \). Here, f is an arbitrary numeration of the set of j projections f = 1, … , j. In the 2D coordinate system of projection f, the projected cross peak has the position vector \( \vec{Q}_f^i \) = [\( \nu_{f,1}^i \), \( \nu_{f,2}^i \)], with \( \nu_{f,1}^i \) and \( \nu_{f,2}^i \) being the chemical shifts along the projected indirect dimension and the direct dimension, respectively. It is convenient to define the origins of both the N-dimensional coordinate system and the 2D coordinate system in all dimensions in the center of the spectral ranges. Then the position vector \( \vec{Q}_f^i \) in the N-dimensional frequency space is given by
The N-dimensional cross peak \( {Q^i} \) is located in an (N − 2)-dimensional subspace, which is orthogonal to the projection plane at the point \( Q_f^i \) (Fig. 1). The “peak subgroup” of an N-dimensional chemical shift correlation \( {Q^i} \) is the set of projected peaks, \( \{ {Q_1^i,\,...,Q_j^i} \} \), that arise from it. It is the key function of the GAPRO algorithm to identify the peak subgroups in the j peak lists of the projections and to calculate the coordinates of \( {Q^i} \) from them.
2.3 The APSY Procedure
The APSY procedure follows the flow-chart shown in Fig. 2. It is illustrated in Fig. 3 and the APSY input parameters are defined in Table 1. At the start, the operator selects the desired N-dimensional NMR experiment, the dimensionality of the projection spectra, and j sets of projection angles. The projection spectra are recorded and automatically peak picked using the GAPRO peak picker, resulting in j peak lists. The GAPRO peak picker identifies all local maxima of the spectrum with a sensitivity (signal-to-noise) larger than a user-defined value R min. The position of the maximum is interpolated for each peak by a symmetrization procedure that involves the intensities of the two neighboring data points in each dimension [40]. The GAPRO peak picker does not attempt to distinguish real peaks from spectral artifacts or random noise; every local maximum is identified as a peak. The j peak lists contain peaks Q gf , where g is an arbitrary numeration of the peaks and f of the projections (f = 1,…, j). GAPRO then arbitrarily selects N − 1 of these peak lists, and generates for each peak Q gf a subspace L gf , which contains the point Q gf and which is orthogonal to the projection f (Fig. 3b). The intersections of the subspaces L gf in the N-dimensional space are candidates for the positions of N-dimensional cross peaks (open circles in Fig. 3b). To account for the imprecision in the picked peak positions due to thermal noise, the calculation of intersections of subspaces allows a user-defined tolerance value in the direct dimension, Δν min. For each of the candidate points, the support, S, is then calculated. S is the number of subspaces from all j projections that contain the candidate point. Thereby at most one subspace from each projection is considered (Fig. 3c) so that N − 1 ≤ S ≤ j. For the calculation of the support, in addition to the user-defined tolerance values for the direct dimension, Δν min, a tolerance for the indirect dimension, r min, is also required to account for the imprecision of picked peak positions. The geometric analysis algorithm can also include aliased peaks in the experimental 2D projections at this point of the calculation. The peaks that contribute to the support of a given candidate point form a “peak subgroup.” The subgroups are ranked for high S-values, and the top-ranked subgroup is selected. In case of degeneracy, one of the top-ranked subgroups is arbitrarily selected. The subspaces contributing to this subgroup are removed from further analysis, and new S-values for the residual candidate points are calculated from the remaining subspaces (Fig. 3c). This procedure is repeated until the value of S for all remaining subgroups falls below a user-defined threshold, S min. At this point in the algorithm a list of the identified peak subgroups is generated. The subgroup identification is repeated with k different, randomly chosen starting combinations of N − 1 projections, and k peak subgroup lists are thus obtained (gray box in Fig. 2). These k lists are merged into a single list, which is again subjected to ranking and elimination of all subgroups with S < S min. From the resulting final list of subgroups, the peak positions in the N-dimensional space are calculated (Fig. 3d). Since the peak positions are redundantly determined by the experimental data, particularly high precision can be obtained for the final result. The computational techniques used for individual steps in Fig. 2 are described in [38, 40].
Flowchart of the APSY procedure. Square boxes indicate processes, and boxes with rounded corners denote intermediate or final results. The steps surrounded with gray are repeated k times, and thus generate k lists of peak subgroups. Adapted with permission from [38]
Illustration of the algorithm GAPRO for N = 3, j = 5, k = 1 and S min = 3. (a) Three dimensional view of the situation with the unknown 3D spectrum shown as a cube in the center and two 2D projections with α = 0° and α = 45°. (b–e) These panels are oriented like the gray ω 1/ω 2-plane in (a). (b) A 2D cross section through the unknown 3D spectrum is surrounded by 1D cross sections through the five experimental 2D projections with projection angles α = 0°, 90°, −30°, 45°, and −65°. The cyan dots mark the result of the automatic peak picking of the 2D projections. The algorithm then arbitrarily selected N − 1 = 2 of the j projections for the first round of spectral analysis, with α = 0° and α = −60°. The intersections of the subspaces corresponding to the peaks in these two projections (green lines) identify eight candidate points in the 3D spectrum (open circles). (c) Using the subspaces from all five projections, the support S (number of intersecting subspaces, see text) is calculated for each candidate point. Yellow and red dots indicate S = 2 and S = 5, respectively. (d) One of the three candidate points with the highest support (S = 5) is arbitrarily selected. All peaks in the projections that contribute to the selected candidate point are identified as a peak subgroup (gray dots in the projections labeled with number 1). The subspaces from this subgroup are removed from the further analysis (gray dashed lines). The support S of remaining candidate points is recalculated (there remains one point with S = 5, and another one with S = 4 is shown in orange). (e) After two more rounds of the procedure indicated in (d), two additional subgroups are identified and labeled with numbers 2 and 3, respectively. From the three subgroups, the positions of three peaks in the 3D spectrum are calculated (black dots). Adapted with permission from [38]
2.4 The Secondary Peak Filter
In APSY, the discrimination between artifacts and noise corresponds to distinguishing between peaks in the projections that stem from an N-dimensional resonance and are thus correlated and those that are uncorrelated. With a sufficiently large number of independently recorded experimental projection spectra only true N-dimensional chemical shift correlations are contained in the final peak list. This separation can be further enhanced by applying an additional, secondary peak filter to the final result of the GAPRO calculation. Thereby the N-dimensional APSY peak list is “back-projected” onto the experimental projections, and the spectral sensitivities at the resulting positions are read out. Based on user-defined criteria, the N-dimensional GAPRO peak list can then be filtered to remove weakly supported peaks or remaining artifacts. All peaks with more than n violations of the threshold R single (Table 1) are deleted. The secondary filter thus provides an efficient additional validation of the GAPRO result and permits the use of less stringent parameters in the GAPRO run.
3 Practical Aspects
3.1 Sensitivity for Signal Detection in APSY-NMR Experiments
The intensity of a given multi-dimensional NMR signal varies in the individual projections of an APSY experiment. In the following the expressions for the signal-to-noise ratio of a given resonance are presented. This formalism can then be used to optimize the performance of APSY-NMR experiments.
By adapting general equations for 2D spectra [5] to APSY-NMR [40], the sensitivity of a signal in a projection of an experiment m with projection angles \( \vec{\varphi } \) = (α, β,…) is given by
where the three terms K A , \( {s_m}(0) \) and \( {f_m}( {\vec{\varphi }} ) \) represent, respectively, the impact of the detected spin type A, the signal intensity at time zero, and the dependence on the projection angles \( \vec{\varphi } \).
K A accounts for the properties of the detected nuclear species A (often protons), including the probe sensitivity, the main polarizing magnetic field strength, and the window function applied before Fourier transformation. Thus, the value of K A can be maximized for a detected given nucleus type A and a given NMR instrument for all experiments that are detected on this nucleus.
\( {s_m}(0) \) is the signal intensity of the experiment m at the time domain origin. This factor enables a comparison of the relative intrinsic sensitivities of different APSY-NMR experiments that are detected on the same nucleus, and can thus help to identify high-sensitivity experiments. Values for \( {s_m}(0) \) can be estimated either experimentally, e.g., from 1D NMR spectra of the time domain origins, or from model calculations [41]. Table 2 lists calculated values for different amide proton-detected experiments.
Finally, \( {f_m}( {\vec{\varphi }} ) \) describes the dependence of the sensitivity of an experiment m on the projection angles \( \vec{\varphi } \) and on the acquisition and processing parameters [40]:
Here, \( s_m^e( {\vec{\varphi },t} ) \) is the signal envelope in the indirect dimension, q is the number of angles that differ from 0° or 90° (the number of subspectra to be combined for quadrature detection is 2q [11, 15]), and \( n( {\vec{\varphi }} ) \) is the operator-chosen number of scans recorded for each of the subspectra. \( {h_m}( {\vec{\varphi },t} ) \) is the applied window function, \( {t_{\max }}( {\vec{\varphi }} ) \) the maximal evolution time, and \( M( {\vec{\varphi }} ) \) the number of indirect points sampled.
\( {f_m}( {\vec{\varphi }} ) \) is largely governed by the envelope function \( s_m^e( {\vec{\varphi },t} ) \). It can be shown that monoexponential relaxation in all indirect dimensions results in \( s_m^e( {\vec{\varphi },t} ) \) being a monoexponential decay with a decay rate constant \( R_{2,m}^*( {\vec{\varphi }} ) \) given by
Here, \( {\vec{R}_{2,m}} \) is an N-dimensional vector containing the transverse relaxation rates along all indirect dimensions, with \( R_{2,m}^i = 0 \) for constant-time evolution elements in the dimension i. Since the standard GAPRO analysis attaches equal weight to each projection spectrum, it is desirable to have similar sensitivities for all individual projection experiments. If the projection angle-dependence of \( s_m^e( {\vec{\varphi },t} ) \) is known, (7) provides a basis for producing similar sensitivities for all the projections used in a given APSY experiment, since the user-defined parameters \( n( {\vec{\varphi }} ) \), \( M( {\vec{\varphi }} ) \), \( {h_m}( {\vec{\varphi },t} ), \) and \( {t_{\max }}( {\vec{\varphi }} ) \) can be individually adjusted for each projection experiment [5, 42].
3.2 Sensitivity and Speed of APSY-NMR Experiments
With a practical example we want to illustrate the performance of APSY in terms of sensitivity and speed. The example is a 4D APSY-HNCOCA experiment with the 12-kDa protein TM1290, of which the sequence-specific resonance assignments are known [43]. A total of 13 2D projections were measured in 13 min (1 min per projection). The 4D APSY-HNCOCA experiment was recorded with [U-13C,15N]-labeled TM1290 at 25 °C on a 600 MHz Bruker Avance III spectrometer with a room temperature probe. The concentration was adjusted to 1.0 ± 0.05 mM, as determined by PULCON [44]. The 13 pairs of projection angles (α, β) comprised: (90°, 0°), (0°, 0°), (0°, 90°), (±60°, 0°), (0°, ±60°), (90°, ±60°), and (±20°, ±70°).
In the 4D peak list generated by the algorithm GAPRO from these 13 projections, all 110 expected 4D (ω1(15N), ω2(13C’), ω3(13Cα), ω4(1HN)) chemical shift correlations were contained. With the selected short measuring time, the intensity of the weakest NMR signals in the projections is comparable to the intensity of the thermal noise (Fig. 4). Nonetheless, even the weakest of the 110 correlation peaks (indicated by arrows in Fig. 4) was recognized by GAPRO as a true correlation, whereas no false 4D correlations appeared. This shows that APSY makes use of the combined sensitivity of all the projections in the input, and that it does not require unambiguous identification of the individual peaks in each projection.
One-dimensional cross-sections through 13 2D projections of a 4D APSY-HNCOCA experiment of the protein TM1290. The data set was recorded in a total measuring time of 13 min on a Bruker Avance III 600 MHz spectrometer equipped with a room temperature probe head, with a 1 mM protein concentration at 25 °C. The cross sections were taken through the ω 4-position the weakest of the 110 peaks of TM1290. In each of the 13 cross-sections, the position of this weakest peak is indicated with an arrow. Asterisks denote two other resonances that are present at this ω 4-position. All other local maxima seen in the cross-sections arise from random spectral noise. Reproduced with permission from [40]
3.3 Selection of Projection Angles
The APSY method does not impose restrictions on the choices of the projection angles or the number of projections, except that, on fundamental grounds, the total number of projections must be at least N − 1 and that each indirect dimension needs to be evolved at least once in the set of projections. On this basis, the selection of projection angles for a given APSY-NMR experiment should be guided by two main considerations. First, the projection angles should be distributed about evenly in the time domain. Second, projections with large q values (number of projection angles that differ from 0° or 90°) are to be disfavored, since the sensitivity for the recording of the 2D projection spectra is proportional to 2−q/2 (7). It is further recommended that the decrease in sensitivity due to higher q-values is compensated by adjusting the number of scans \( n\left( {\vec{\varphi }} \right) \) accordingly (7).
An additional improvement is achieved with the use of dispersion-optimized projection angles, in particular if the sweep widths of the indirect dimensions are significantly different. Dispersion-optimized projection angles adjust the contributions of the indirect dimensions to the same size, and thus contribute to eliminating chemical shift overlap. The dispersion-optimized, or “matching” projection angle α* for two dimensions, i and j, with sweep widths SW i and SW j is given by
For example, if the sweep widths of two dimensions differ 11-fold (as they do for C’ and Cβ), then α* = 84°. A set of three projection angles with values of 60°, 84°, and 87° would thus be a good choice, whereas a seemingly more symmetric selection with angles of 22.5°, 45°, and 67.5° would lead to two basically identical projections [40]. Expressions similar to (9) can be derived for combinations of three or more dimensions.
3.4 Optimizing the GAPRO Parameters for a Given Experiment
Among the input variables of the geometric algorithm GAPRO, three parameters have a dominant effect on the result of the spectral analysis: S min, Δν min, and r min (Table 1). The selection of the minimal support S min is most important, since only candidate signals with a support S ≥ S min will be included in the final peak list. Figure 5a shows the variation of the peak list resulting from a 4D APSY-HNCOCA experiment when different values of S min are used. The data set consisted of the 13 projections recorded with the protein TM1290 mentioned above, for which 110 amino acids are expected. For S min between 3 and 8, the final result contains all the expected peaks. As a general guideline, it is advisable to set S min to about one third of the number of input projections, and to keep S min > (N + 2) for an N-dimensional experiment with 2D projections.
Impact of APSY parameters on the result. (a) Dependence of the total number P of 4D APSY-HNCOCA peaks of the protein TM1290 on the GAPRO parameter S min. Light gray bars represent the number of correct correlation peaks, dark gray bars the number of artifacts, and the dotted line indicates the expected 110 peaks. (b, c) Dependence of the total number P of 4D APSY-HNCOCA peaks of the protein TM1290 on the GAPRO parameters r min and Δν min, respectively. The dotted line indicates the expected 110 peaks. (d) Impact of the number j of 2D projections on the percentage of the expected correlations, D, for three APSY experiments with the protein TM1290. 4D APSY-HACANH (squares), 5D APSY-HACACONH (diamonds) and 5D APSY-CBCACONH (triangles). The data correspond to Table 3. Reproduced with permission from [40]
The two additional key parameters are the intersection tolerances for the direct and indirect dimensions, Δν min and r min. A variation of these parameters shows that each of these tolerances has to be larger than a certain minimal value, which depends on the digital resolution, the signal line widths and the sensitivity of a given experiment (Fig. 5b,c). If the tolerances are chosen too small, corresponding subspaces do not intersect and correct peaks are not found. However, if the parameters are too large, no negative effects occur in the final result except that the computation time increases substantially due to the increasing number of intersection possibilities. As a general guideline, it is advisable to use one to two times the respective digital resolutions in the direct and indirect dimensions of the 2D projection spectrum as values for Δν min and r min, respectively.
3.5 Selection of the Number of Projections
Choosing the minimal number of projections needed for a given experiment is important to minimize the required instrument time. A good decision on the number of projections considers the type of APSY-NMR experiment, the expected number of correlation peaks per amino acid residue, the size and type of protein under study, the choice of the projection angles, and the required quality of the result. Representative examples for the number of projections needed in particular experiments are shown in Fig. 5d and Table 3. For polypeptides with smaller chemical shift dispersion, such as denatured proteins, a higher number of projections is required for obtaining comparable results as for globular proteins. It should be noted that APSY can be run using a convergence scheme which interleaves the recording of new projections with the analysis of the existing data by GAPRO. The convergence scheme stops the data recording as soon as a preset number of peaks have been resolved or when the protein has been assigned.
Note further that in many APSY experiments, some of the projections can be measured with individually optimized, shortened pulse sequences, which omit magnetization transfers that are not required and hence have improved sensitivity [15]. For example, a direct projection of the ω(15N)-dimension in a multidimensional APSY experiment can be replaced by a standard [15N,1H]-HSQC experiment.
4 APSY-Based Automated Resonance Assignments
4.1 Overview
APSY provides peak lists of chemical shift correlations for multidimensional NMR experiments. Due to the averaging of a large number of observed signals in the set of projections, the determination of the N-dimensional chemical shifts becomes very precise. APSY is thus well suited for applications that require precise peak positions. Here we want to concentrate on applications for resonance assignments in protein spectra. APSY-NMR combined with a suitable assignment algorithm enables fully automated sequence-specific assignments for globular and denatured proteins.
4.2 Combinations of 4D and 5D APSY-NMR Experiments
Strategies for sequence-specific backbone resonance assignment of polypeptides usually contain two key elements. First, sequential NMR connectivities lead to the identification of discrete peptide fragments of different lengths. Second, these fragments are mapped onto the known polypeptide sequence, based on the chemical shift statistics of the amino acid types. The vast majority of conventional backbone assignment experiments are detected on the amide proton due to the high experimental sensitivity and other practical aspects. For the same reasons, we also limit the present discussion of APSY experiments to this nucleus. With this selection, the 1H and 15N chemical shifts of the amide moiety are readily contained in each correlation. APSY can connect two sequential amide moieties in a single experiment (see below); however, usually at least two APSY experiments are needed to connect two sequential amide moieties. As illustrated in Fig. 6, the 13Cα atom is always a nucleus available for the sequential connection, and a second matching nucleus can be either 1Hα, 13Cβ, or 13C’. For the mapping of fragments onto the sequence, the 13Cβ chemical shift increases the reliability of sequence-specific assignments, since it allows the unambiguous distinction between different amino acid types [45–48]. With the requirement that two chemical shifts should define the sequential connectivities, five groups of four- and five-dimensional correlation experiments can be devised (Fig. 6). The relative sensitivities of the APSY experiments required for these assignment strategies can be estimated by model calculations (Table 2) [40].
Combinations of intraresidual and sequential chemical shift correlations to be recorded with HN-detected APSY-NMR experiments for polypeptide backbone assignments of 13C,15N-labeled proteins. Each colored shape contains the nuclei correlated by a 4D or 5D experiment. The orange areas contain the nuclei for which the individual correlations overlap. The notations used for the different groups of experiments are indicated in each panel. Reproduced with permission from [40]
As one practical example, we show the application of the HA–CA(CB) strategy to obtain the backbone resonance assignments of the protein TM1290. TM1290 is the same protein as was studied in the experiment of Fig. 5. The HA–CA(CB) strategy is realized with the three experiments 4D APSY-HACANH, 5D APSY-HACACONH, and 5D APSY-CBCACONH. These three experiments were carried out with the same sample of TM1290 as described above and were performed at 25 °C on a 750 MHz Bruker Avance III spectrometer with a room temperature probe. The input for the assignment algorithm GARANT [49] consisted of the three final APSY peak lists and the amino acid sequence [50]. Table 3 presents key parameters used for the recording of these three experiments. The lower part of Table 3 lists the numbers of expected and observed correlations for the three experiments used in the HA–CA(CB) approach. All detected TM1290 backbone resonances were correctly assigned [40]. This example thus shows that in a total instrument time of about 1 day and with minimal human intervention, the complete and correct sequence-specific resonance assignments of a 12-kDa protein were obtained with the APSY-based approach.
For the assignment of larger proteins, the CB–CA and the CO–CA strategies are preferred over the HA–CA strategy, since they are compatible with deuteration, which in turn increases the experimental sensitivity. The CB–CA strategy is realized with the combination of the 4D APSY-HNCACB and the 5D APSY-HNCOCACB experiment [51]. With this approach, the backbones of two human proteins were assigned, the 22-kDa protein kRas at 0.4 mM concentration and a 15-kDa drug target protein (protein A) at 0.3 mM concentration (Fig. 11) [51]. For each of the two proteins, 76 h of experiment time on a 600 MHz spectrometer with a cryogenic probe were used for the 2 backbone experiments (16 projections of the 4D APSY-HNCACB in 48 h and 32 projections of the 5D APSY-HNCOCACB in 48 h) corresponding to a total of 3 days and assigned by the algorithm MATCH. The overall completeness of the backbone resonance assignment was 95% for the 22-kDa protein kRas and 98% for the 15-kDa protein A (Fig. 11), where the missing assignments comprised segments with unfavorable protein dynamics.
4.3 Backbone Assignments with a Single 6D APSY-NMR
The combination of 4D and 5D APSY-NMR experiments thus provides backbone assignments for perdeuterated, [U-13C,15N]-labeled proteins up to at least 25 kDa. For smaller proteins up to 12 kDa an alternative and more elegant approach can be used [50]. The pathway HN–N–C’–Cα–N–HN directly connects two sequentially adjacent amide moieties in a single experiment and with APSY the full potential of this magnetization transfer pathway is exploited [50]. The 6D APSY-seq-HNCOCANH experiment was recorded with a 0.9 mM solution of the protein 434-repressor(1–63) at 30 °C on a Bruker 750 MHz spectrometer with room temperature probe. A total of 25 2D projections were recorded in 40 h. The same experiment was also recorded with a 3.0 mM solution of TM1290 at 35 °C on a Bruker DRX 500 MHz spectrometer with a cryogenic probe. The total spectrometer time used for the recording of 25 2D projections was 20 h.
For 434-repressor(1–63), the resulting APSY peak list contained 56 out of 57 peaks expected from the amino acid sequence and for TM1290 all but three of the expected peaks were obtained [43]. Both lists did not contain any artifact and provided very precise chemical shifts. The precision of chemical shift measurements can be directly assessed in this data set, since the resonance of each amide moiety is part of two different 6D peaks. The amide proton chemical shift is measured in the direct dimension ω6 and in the indirect dimension ω1; the amide 15N chemical shift in ω5 and ω2, respectively. From the 93 amide moieties of TM1290 that contributed to two peaks, the precision (standard deviation) for the proton and nitrogen chemical shift measurements was 0.0014 ppm (0.72 Hz), and 0.0137 ppm (0.69 Hz), respectively. These precise and artifact-free 6D peak lists were used as inputs for the assignment algorithm GARANT [49], yielding the correct sequence-specific assignment for each protein.
4.4 D APSY-NMR Spectroscopy for the Assignment of Non-Globular Proteins
Studies of soluble non-globular polypeptides are of great relevance for protein folding as well as for insight into the structural basis of functional non-globular polypeptides [52–59]. However, the available data on this class of proteins are scarce because they are not amenable to meaningful single-crystal studies, and solution NMR studies have been limited by small dispersion of the chemical shifts [60–62]. Increased interest in detailed structural and dynamic characterization of soluble non-globular polypeptides has, however, more recently been generated by the discovery of a rapidly increasing number of proteins that are intrinsically unfolded in their functional state in solution [54, 55, 58]. APSY provides a fully automated approach to solving this problem with the use of very high-dimensional NMR.
One approach is a combination of the above-mentioned 6D APSY-seq-HNCOCANH with the 5D APSY-HNCOCACB experiment [63]. Thereby, the 6D APSY scheme [50] connects neighboring amide groups sequentially, and the 5D APSY scheme measures the Cβ chemical shifts. These two experiments can also be combined into a single magnetization transfer pathway, the 7D APSY-seq-HNCO(CA)CBCANH (Figs. 7a, and 8) [63]. Magnetization of the amide proton i is transferred with seven subsequent INEPT [64] steps to the amide proton i − 1 (steps a–g in Fig. 7a). Along this pathway, six evolution periods are introduced for the frequencies of the nuclei 1HN i , 15N i , 13C’ i−1, 13Cβ i−1, 13Cα i−1, and 15N i−1. Thus, each seven-atom fragment of residues i and i − 1 gives rise to a single peak, except if a proline residue or a chain end is located at either of the positions i or i − 1.
Magnetization transfer pathways of (a) the 7D APSY-seq-HNCO(CA)CBCANH NMR experiment and (b) the 5D APSY-HC(CC-TOCSY)CONH experiments. The dashed gray arrows indicate INEPT magnetization transfer steps [64]. The thick gray line in (b) represents isotropic mixing. Adapted with permission from [63] and [65]
Pulse sequence of the 7D APSY-HNCO(CA)CBCANH experiment. Radio-frequency pulses are applied at 118.0 ppm for 15N, 173.0 ppm for 13C’, and at 42.0 ppm for 13Cα and 13Cβ. At the start of each transient, the 1H carrier frequency is set at 8.24 ppm, indicated by “HN,” and at the time point “H2O” the carrier is changed to 4.7 ppm. Black and white symbols represent 90°- and 180°-pulses, respectively. Unlabeled bars stand for rectangular pulses applied at maximum power. Pulses marked with capital letters have individually adjusted lengths and shapes, depending on their purpose. All pulse lengths are given for a 1H frequency of 750 MHz. 13C’-pulses: A, 180°, rectangular shape, 38.3 μs; B, 90°, rectangular shape, 42.8 μs; C, 180°, I-Burp [66], 220 μs. 13Cαβ-pulses: D, 180°, I-Burp (applied at 51.0 ppm), 220 μs; E, 90°, Gaussian cascade Q5 [67], 280 μs; F, 180°, Gaussian cascade Q3 [67], 185 μs; H, 180°, rectangular shape, 38.3 μs. The 15N-pulses labeled with an asterisk are centered with respect to \( t_3^{\text{a}} + t_3^{\text{b}} \) and \( t_3^{\text{c}} \), respectively. The 13C’-pulses labeled with an asterisk are centered with respect to ρ + t 5/2 and ρ − t 5/2, respectively. The last six pulses on the 1H line represent a 3–9–19 Watergate pulse train [68]. Decoupling using DIPSI 2 [69] on 1H and WALTZ-16 [70] on 15N is indicated by white rectangles. The triangle with t 7 represents the acquisition period. On the line marked PFG, curved shapes indicate sine bell-shaped, pulsed magnetic field gradients along the z-axis with the following durations and strengths: G1, 600 μs, 13 G/cm; G2, 1,000 μs, 37 G/cm; G3, 800 μs, 16 G/cm; G4, 800 μs, 34 G/cm; G5, 600 μs, 19 G/cm; G6, 600 μs, 27 G/cm; G7, 800 μs, 13 G/cm; G8, 1,000 μs, 37 G/cm; G9, 800 μs, 16 G/cm. Pulse phases different from x are indicated above the pulses. Phase cycling: φ 1 = ψ 2 = φ r = {x, −x}, ψ 4 = {x, x, −x, −x}, ψ 6 = y. The initial delays were \( t_1^{\text{a}} = t_1^{\text{c}} = {2}.{7} {\text{ms,}}\;t_2^{\text{a}} = t_2^{\text{c}} = {14}.0 {\text{ms}},\;t_3^{\text{a}} = t_3^{\text{c}} = {4}.{7}\;{\text{ms}},\;t_6^{\text{a}} = t_6^{\text{c}} = {14}.0\;{\text{ms}} \), and \( t_1^{\text{b}} = t_2^{\text{b}} = t_3^{\text{b}} = {t_4} = {t_5} = t_6^{\text{b}} = 0\;{\text{ms}} \). Further delays were τ = 2.7 ms, ζ = 14.0 ms, η = 6.8 ms, λ = 4.7 ms, and ρ = 20.75 ms. Quadrature detection for the indirect dimensions was achieved using the trigonometric addition theorem [11, 15] with the phases ψ 1, ψ 2, ψ 3, ψ 4 – ψ 6, ψ 6, and ψ 7 for t 1, t 2, t 3, t 4, t 5, and t 6, respectively. Evolution periods were implemented as direct evolution for t 4, and in constant-time fashion for t 1, t 2, t 3, t 5, and t 6. For t 1, t 2, t 3, and t 6, semi-constant time evolution was used for those maximal evolution periods that are too long to be accommodated in constant-time periods. Reproduced with permission from [63]
The 7D APSY-seq-HNCO(CA)CBCANH is illustrated here with the NMR assignment of the 148-residue outer membrane protein X (OmpX) denatured with 8 M urea in aqueous solution [71]. The experiment was recorded with 100 2D projections in a total measuring time of 2 days (50 h) at 15 °C on a Bruker 750 MHz spectrometer with room temperature probe (Fig. 8) [38]. Out of the 142 expected peaks, 139 were actually observed [63]. The three missing peaks connect the residues 98–101, a backbone segment that features unfavorable backbone dynamics.
As for the 6D experiment, high precision of the chemical shift measurements is crucial for the sequential assignments, since these rely on matching of the amide 15N and 1H chemical shifts of sequentially neighboring amide moieties. A precision of 0.46 Hz and 0.44 Hz, respectively for the 1H and 15N chemical shifts was achieved (Fig. 9). Figure 9 also illustrates the significance of peak separation compared to the precision of the chemical shift measurements for unfolded proteins. This high precision enabled automated NMR assignment with the program GARANT [49] in spite of residual chemical shift degeneracy in some of the seven dimensions [63].
Precision of chemical shift measurements by the 7D APSY-seq-HNCO(CA)CBCANH experiment. The data shown was recorded with a 3 mM sample of urea-denatured OmpX in 8 M urea aqueous solution at pH 6.5. (a) Spectral region from the (0°,0°,0°,0°,0°)-projection, which corresponds to a 2D [15N,1H]-correlation spectrum. The black dots are projections of the 7D peak positions determined by GAPRO, as represented by the [ω 6, ω 7]-correlations onto the experimental 2D projection spectrum. Orange squares indicate two clusters of overlapped signals which are displayed on an expanded scale in (b) and (c). (b, c) The two different [15N,1H]-pairs contained in each 7D signal are indicated in red ([ω 2, ω 1]-correlation) and blue ([ω 6, ω 7]-correlation). Contours are drawn at a distance of 1.0 Hz around the peak positions projected from the 7D data set. In (a)–(c), resonance assignments are given using one-letter amino acid symbols and the sequence positions. (d) Histogram of the variance between the measurements of the same amide proton chemical shift from the two 7D signals correlating two sequentially neighboring groups of 7 atoms. (e) Same as (d) for amide nitrogen-15 shifts. Reproduced with permission from [63]
Since the longitudinal and transverse relaxation time constants in soluble non-globular proteins are in first order independent of the length of the polypeptide, similar experimental sensitivities can be expected for much larger unfolded proteins. Further, since the high precision of the APSY experiment of below 1 Hz falls substantially below the occurring distances between pairs of neighboring resonances in high-dimensional spaces [1, 72], similar assignment results to those achieved with the 150 residue OmpX can be expected for other unfolded proteins of much larger size. This has indeed been shown, where, by using the 7D APSY-seq-HNCO(CA)CBCANH experiment, the Zweckstetter group could assign the backbone resonances of the 441 residue Tau, a disordered polypeptide, within 5 days of measurement time, reducing the overall analysis time by more than order of magnitude as compared to a conventional approach [73]. APSY-NMR thus has tremendous potential for new insights into structure–function correlations of natively unfolded proteins, as well as for key contributions to the protein folding problem.
4.5 Automated NMR Assignment of Protein Side Chain Resonances
The precise APSY peak lists can also be the basis for side chain resonance assignments of proteins. A well suited magnetization transfer pathway for this purpose is the HC(CC-TOCSY)CONH pathway, which correlates side chain with backbone nuclei [74–78]. With APSY, the dimensionality of this experiment can be extended to five [65] (Fig. 7b). The pathway starts simultaneously on all aliphatic side chain protons including the Hα. An INEPT element transfers magnetization to the covalently bound carbon. Subsequently, the magnetization is transferred among the aliphatic carbon nuclei by isotropic mixing. At the end of the mixing time the magnetization on the Cα nucleus is transferred via the carbonyl carbon to the amide nitrogen of the successive amino acid residue and finally to the attached amide proton, from which the signal is acquired. For a given amino acid, the resulting 5D APSY correlation peak list thus contains a group of C–H correlations which have identical chemical shifts in the three backbone dimensions ω3(13C’), ω4(15N), and ω5(1HN).
The peak intensities of the correlation peaks in CC-TOCSY experiments depend strongly on the amplitude of the magnetization transfer during the isotropic mixing period and hence on the length of this period [79, 80]. There is no single mixing time for which all C–H moieties of all 20 amino acids have sufficiently large transfer amplitudes. This problem can be elegantly circumvented with APSY, since the TOCSY mixing time can be varied along with the projection angles. The analysis of the set of projection spectra with GAPRO does not require that a given 5D peak is present in all projections. By using a set of mixing times that enables sufficiently high transfer for all aliphatic side chain carbon moieties in some of the projections, it is possible to cover the resonance frequencies of all C–H moieties from all 20 amino acids in the resulting APSY correlation peak list.
Calculations of the transfer amplitudes in CC-TOCSY experiments show that the set of three mixing times – 12 ms, 18 ms, and 28 ms – covers all protons in the 20 amino acids [65]. The mixing time of 18 ms, which is commonly used in classical experiments, transfers magnetization from a majority of carbons in the side chains to the α-carbon nuclei. The mixing time of 12 ms is favorable for signals which have a small transfer at 18 ms. The long mixing time of 28 ms favors signals of long side chains, but also signals of short side chains, which are very weak or not present at the two other mixing times.
With these three mixing times, the 5D APSY-HC(CC-TOCSY)CONH experiment was recorded with a 1 mM solution of the 12.4-kDa globular protein TM1290 in 24 h of spectrometer time using 36 projections (Fig. 10). Based on the reference assignment of this protein, 424 cross peaks are expected in the resulting 5D APSY correlation peak list [43]; 368 thereof were actually found in the present experiment. These 368 correlations contained the chemical shifts of 97% of the aliphatic carbons and 87% of the aliphatic protons in the protein.
2D projection of the 5D ASPY-HC(CC-TOCSY)CONH experiment with TM1290 recorded on a 750 MHz spectrometer using a TOCSY mixing time of = 17.75 ms. The projection with angles α = −46.6°, β = 0°, γ = −17.2° is shown. The region in magenta in the left panel is shown enlarged on the right hand side. The colored dots are the projections of the final 5D APSY peak list. Red dots indicate peaks present at the mixing time τ m = 17.75 ms, blue dots indicate peaks only present in spectra with other mixing times. Reproduced with permission from [65]
The resulting 5D APSY-HC(CC-TOCSY)CONH chemical shift correlation list together with the known backbone assignment are the sole input for the side chain assignment algorithm ALASCA (Algorithm for Local and linear Assignment of Side Chains from APSY data) [65]. In the ALASCA algorithm, each 5D APSY-HC(CC-TOCSY)CONH correlation is attributed to the residue, which has the nearest backbone chemical shifts in the 3D space of the (ω(13C’), ω(15N), ω(1HN)) frequencies. Subsequently, for each amino acid in the protein, the correlations of the TOCSY peak group are assigned to the side chain atoms by matching the chemical shifts of the 5D correlations to statistical values from the BMRB database [39].
As for the applications providing backbone assignments, the precision of the chemical shifts obtained for ω5(1HN), ω4(15N), and ω3(13C’) from the 5D APSY-HC(CC-TOCSY)CONH experiment is crucial for the assignment. It was found to be 0.5 Hz for ω5(1HN), 2.3 Hz for ω4(15N), and 3.6 Hz for ω3(13C’), which is substantially below the digital resolution of the individual projection spectra. With ALASCA all 368 peaks contained in the 5D peak list of TM1290 were correctly assigned.
The 5D APSY-HC(CC-TOCSY)CONH experiment was also used to assign the side chains of the two larger proteins, the 22-kDa protein kRas at 0.4 mM concentration and the 15-kDa drug target protein A at 0.3 mM concentration [51]. A total of 34 projections were recorded in 51 h on a 600 MHz spectrometer equipped with a cryogenic probe at an experiment temperature of 23 °C. The assignment yielded for each protein nearly 90% of the Ala, Ile, Leu, Thr, and Val side chain methyl groups (Fig. 11).
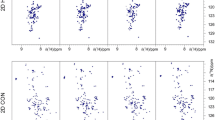
Sequence-specific resonance assignments of two proteins obtained with the APSY CA-CB-CM strategy [51]. Data is shown for two proteins, the 15-kDa “protein A” (panels a–c) and the 22-kDa protein kRas (panels d–f). Selected sample parameters are indicated. The amide resonance assignments are shown in blue on 2D [15N,1H]-HSQC spectra (panels a and d), with their central regions shown enlarged (panels b and e). The methyl group assignments are indicated in orange on 2D [13C,1H]-HMQC spectra (panels c and f). Adapted with permission from [51]
Overall, the high quality of the GAPRO peak list of the 5D APSY-HC(CC-TOCSY)CONH experiment in terms of dimensionality, completeness, precision and very low number of artifacts provides an excellent basis for a reliable automated assignment of aliphatic side-chain atoms. Although the TOCSY mixing does not provide information on the direct covalent connectivities among the carbon nuclei, the 5D peaks can be used for reliable sequence-specific resonance assignment of aliphatic resonances, due to the availability of all five dimensions.
5 Conclusion and Outlook
This chapter presented the foundations of automated projection spectroscopy (APSY) that uses the algorithm GAPRO for automated spectral analysis. We showed applications of APSY for high-dimensional heteronuclear correlation NMR experiments with proteins. Without human intervention after the initial set-up of the experiments, complete peak lists for 4D to 7D NMR spectra, with a chemical shift precision of below 1 Hz, are typically obtained.
The positions of the peaks in the projection spectra that arise from a real N-dimensional peak are correlated among the projection spectra, whereas the positions of random noise are uncorrelated. This different behavior efficiently discriminates projected peaks against artifacts, and artifacts are therefore unlikely to appear in the final peak list. APSY is also well prepared to deal with inaccurate peak positions. Since the final N-dimensional APSY peak list is computed as the average of a large number of independent measurements, inaccurate peak positions in some of the projections have only a small influence on the overall precision. APSY has the advantage of relying exclusively on the analysis of experimental low-dimensional projection spectra, with no need ever to reconstruct the parent high-dimensional spectrum. APSY does not impose restrictions on the selection of the number of projections or the combinations of projection angles. The experience from our work indicates that sensitivity for signal detection rather than overcrowding of the 2D projection spectra is the limiting factor in practical applications of APSY-NMR with proteins.
In addition to providing automated peak picking and computation of the corresponding chemical shift lists, APSY supports automated sequential resonance assignment. Thus, APSY is a valid alternative to related NMR techniques. APSY can be the first step, after sample preparation, in a fully automated process of protein structure determination by NMR with successive automated algorithms for the NOESY spectrum analysis and structure calculation.
APSY software and tools can be downloaded from www.apsy.ch.
Abbreviations
- 1D (2D, 3D, 4D, 5D, 6D, 7D):
-
One- (two-, three-, four-, five-, six-, seven-) dimensional
- ALASCA:
-
Algorithm for local and linear assignment of side chains from APSY data
- APSY:
-
Automated projection spectroscopy
- GAPRO:
-
Geometric analysis of projections
- NMR:
-
Nuclear magnetic resonance
- TOCSY:
-
Total correlation spectroscopy
References
Wüthrich K (1986) NMR of proteins and nucleic acids. Wiley, New York
Bax A, Grzesiek S (1993) Methodological advances in protein NMR. Acc Chem Res 26:131–138
Kay LE, Gardner KH (1997) Solution NMR spectroscopy beyond 25 kDa. Curr Opin Struct Biol 7:722–731
Wüthrich K (2003) NMR studies of structure and function of biological macromolecules (Nobel lecture). Angew Chem Int Ed 42:3340–3363
Ernst RR, Bodenhausen G, Wokaun A (1987) Principles of nuclear magnetic resonance in one and two dimensions. Oxford University Press, Oxford
Szyperski T, Yeh DC, Sukumaran DK, Moseley HNB, Montelione GT (2002) Reduced dimensionality NMR spectroscopy for high-throughput protein resonance assignment. Proc Natl Acad Sci USA 99:8009–8014
Orekhov VY, Ibraghimov I, Billeter M (2003) Optimizing resolution in multidimensional NMR by three-way decomposition. J Biomol NMR 27:165–173
Kozminski W, Zhukov I (2003) Multiple quadrature detection in reduced-dimensionality experiments. J Biomol NMR 26:157–166
Rovnyak D, Frueh DP, Sastry M, Sun ZYJ, Stern AS, Hoch JC, Wagner G (2004) Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. J Magn Reson 170:15–21
Szyperski T, Wider G, Bushweller JH, Wüthrich K (1993) Reduced dimensionality in triple-resonance NMR experiments. J Am Chem Soc 115:9307–9308
Brutscher B, Morelle N, Cordier F, Marion D (1995) Determination of an initial set of NOE-derived distance constraints for the structure determination of 15N/13C-labeled proteins. J Magn Reson B 109:238–242
Kim S, Szyperski T (2003) GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectral information. J Am Chem Soc 125:1385–1393
Kupce E, Freeman R (2003) Projection-reconstruction of three-dimensional NMR spectra. J Am Chem Soc 125:13958–13959
Kupce E, Freeman R (2003) Reconstruction of the three-dimensional NMR spectrum of a protein from a set of plane projections. J Biomol NMR 27:383–387
Kupce E, Freeman R (2004) Projection-reconstruction technique for speeding up multidimensional NMR spectroscopy. J Am Chem Soc 126:6429–6440
Kupce E, Freeman R (2004) Fast reconstruction of four-dimensional NMR spectra from plane projections. J Biomol NMR 28:391–395
Bracewell RN (1956) Strip integration in radio astronomy. Aust J Phys 9:198–217
Nagayama K, Bachmann P, Wüthrich K, Ernst RR (1978) The use of cross-sections and of projections in two-dimensional NMR spectroscopy. J Magn Reson 31:133–148
Mersereau RM, Oppenheim AV (1974) Digital reconstruction of multidimensional signals from their projections. Proc IEEE 62:1319–1338
Lauterbur PC (1973) Image formation by induced local interactions − examples employing nuclear magnetic resonance. Nature 242:190–191
Moseley HNB, Riaz N, Aramini JM, Szyperski T, Montelione GT (2004) A generalized approach to automated NMR peak list editing: application to reduced-dimensionality triple resonance spectra. J Magn Reson 170:263–277
Freeman R, Kupce E (2004) Distant echoes of the accordion: Reduced dimensionality, GFT-NMR, and projection-reconstruction of multidimensional spectra. Concepts Magn Reson A 23:63–75
Eghbalnia HR, Bahrami A, Tonelli M, Hallenga K, Markley JL (2005) High-resolution iterative frequency identification for NMR as a general strategy for multidimensional data collection. J Am Chem Soc 127:12528–12536
Luan T, Jaravine V, Yee A, Arrowsmith CH, Orekhov VY (2005) Optimization of resolution and sensitivity of 4D NOESY using multi-dimensional decomposition. J Biomol NMR 33:1–14
Malmodin D, Billeter M (2005) Multiway decomposition of NMR spectra with coupled evolution periods. J Am Chem Soc 127:13486–13487
Kupce E, Freeman R (2006) Hyperdimensional NMR spectroscopy. J Am Chem Soc 128:6020–6021
Szyperski T, Atreya HS (2006) Principles and applications of GFT projection NMR spectroscopy. Magn Reson Chem 44:S51–S60
Coggins BE, Zhou P (2006) PR-CALC: a program for the reconstruction of NMR spectra from projections. J Biomol NMR 34:179–195
Lescop E, Brutscher B (2007) Hyperdimensional protein NMR spectroscopy in peptide-sequence space. J Am Chem Soc 129:11916–11917
Cornilescu G, Bahrami A, Tonelli M, Markley JL, Eghbalnia HR (2007) HIFI-C: a robust and fast method for determining NMR couplings from adaptive 3D to 2D projections. J Biomol NMR 38:341–351
Mishkovsky M, Kupce E, Frydman L (2007) Ultrafast-based projection-reconstruction three-dimensional nuclear magnetic resonance spectroscopy. J Chem Phys 127:034507
Zhang Q, Atreya HS, Kamen DE, Girvin ME, Szyperski T (2008) GFT projection NMR based resonance assignment of membrane proteins: application to subunit C of E. coli F1F0 ATP synthase in LPPG micelles. J Biomol NMR 40:157–163
Jaravine VA, Zhuravleva AV, Permi P, Ibraghimov I, Orekhov VY (2008) Hyperdimensional NMR spectroscopy with nonlinear sampling. J Am Chem Soc 130:3927–3936
Koradi R, Billeter M, Engeli M, Güntert P, Wüthrich K (1998) Automated peak picking and peak integration in macromolecular NMR spectra using AUTOPSY. J Magn Reson 135:288–297
Herrmann T, Güntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR 24:171–189
Baran MC, Huang YJ, Moseley HNB, Montelione GT (2004) Automated analysis of protein NMR assignments and structures. Chem Rev 104:3541–3555
Xia YL, Zhu G, Veeraraghavan S, Gao XL (2004) (3,2)D GFT-NMR experiments for fast data collection from proteins. J Biomol NMR 29:467–476
Hiller S, Fiorito F, Wüthrich K, Wider G (2005) Automated projection spectroscopy (APSY). Proc Natl Acad Sci USA 102:10876–10881
Seavey BR, Farr EA, Westler WM, Markley JL (1991) A relational database for sequence-specific protein NMR data. J Biomol NMR 1:217–236
Hiller S, Wider G, Wüthrich K (2008) APSY-NMR with proteins: practical aspects and backbone assignment. J Biomol NMR 42:179–195
Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc 34:93–158
Wider G (1998) Technical aspects of NMR spectroscopy with biological macromolecules and studies of hydration in solution. Prog Nucl Magn Reson Spectrosc 32:193–275
Etezady-Esfarjani T, Peti W, Wüthrich K (2003) NMR assignment of the conserved hypothetical protein TM1290 of Thermotoga maritima. J Biomol NMR 25:167–168
Wider G, Dreier L (2006) Measuring protein concentrations by NMR spectroscopy. J Am Chem Soc 128:2571–2576
Richarz R, Wüthrich K (1978) 13C NMR chemical shifts of common amino acid residues measured in aqueous solutions of linear tetrapeptides H−Gly−Gly−X−L-Ala−OH. Biopolymers 17:2133–2141
Oh BH, Westler WM, Darba P, Markley JL (1988) Protein 13C spin systems by a single two-dimensional NMR experiment. Science 240:908–911
Grzesiek S, Bax A (1993) Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J Biomol NMR 3:185–204
Güntert P, Salzmann M, Braun D, Wüthrich K (2000) Sequence-specific NMR assignment of proteins by global fragment mapping with the program MAPPER. J Biomol NMR 18:129–137
Bartels C, Güntert P, Billeter M, Wüthrich K (1997) GARANT − a general algorithm for resonance assignment of multidimensional nuclear magnetic resonance spectra. J Comput Chem 18:139–149
Fiorito F, Hiller S, Wider G, Wüthrich K (2006) Automated resonance assignment of proteins: 6D APSY-NMR. J Biomol NMR 35:27–37
Gossert AD, Hiller S, Fernández C (2011) Automated NMR resonance assignment of large proteins for protein–ligand interaction studies. J Am Chem Soc 133:210–213
Anfinsen CB (1973) Principles that govern folding of protein chains. Science 181:223–230
Shortle D (1993) Denatured states of proteins and their roles in folding and stability. Curr Opin Struct Biol 3:66–74
Plaxco KW, Gross M (1997) Cell biology – the importance of being unfolded. Nature 386:657–659
Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CR, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang CH, Kissinger CR, Bailey RW, Griswold MD, Chiu M, Garner EC, Obradovic Z (2001) Intrinsically disordered protein. J Mol Graph Model 19:26–59
Daggett V, Fersht AR (2003) Is there a unifying mechanism for protein folding? Trends Biochem Sci 28:18–25
Mayor U, Guydosh NR, Johnson CM, Grossmann JG, Sato S, Jas GS, Freund SMV, Alonso DOV, Daggett V, Fersht AR (2003) The complete folding pathway of a protein from nanoseconds to microseconds. Nature 421:863–867
Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208
Lindorff-Larsen K, Rogen P, Paci E, Vendruscolo M, Dobson CM (2005) Protein folding and the organization of the protein topology universe. Trends Biochem Sci 30:13–19
Wüthrich K (1994) NMR assignments as a basis for structural characterization of denatured states of globular proteins. Curr Opin Struct Biol 4:93–99
Schwalbe H, Fiebig KM, Buck M, Jones JA, Grimshaw SB, Spencer A, Glaser SJ, Smith LJ, Dobson CM (1997) Structural and dynamical properties of a denatured protein. Heteronuclear 3D NMR experiments and theoretical simulations of lysozyme in 8 M urea. Biochemistry 36:8977–8991
Dyson HJ, Wright PE (2001) Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol 339:258–270
Hiller S, Wasmer C, Wider G, Wüthrich K (2007) Sequence-specific resonance assignment of soluble nonglobular proteins by 7D APSY-NMR spectroscopy. J Am Chem Soc 129:10823–10828
Morris GA, Freeman R (1979) Enhancement of NMR signals by polarization transfer. J Am Chem Soc 101:760–762
Hiller S, Joss R, Wider G (2008) Automated NMR assignment of protein side chain resonances using automated projection spectroscopy (APSY). J Am Chem Soc 130:12073–12079
Geen H, Freeman R (1991) Band-selective radiofrequency pulses. J Magn Reson 93:93–141
Emsley L, Bodenhausen G (1990) Gaussian pulse cascades – new analytical functions for rectangular selective inversion and in-phase excitation in NMR. Chem Phys Lett 165:469–476
Piotto M, Saudek V, Sklenar V (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR 2:661–665
Shaka AJ, Lee CJ, Pines A (1988) Iterative schemes for bilinear operators − application to spin decoupling. J Magn Reson 77:274–293
Shaka AJ, Keeler J, Frenkiel T, Freeman R (1983) An improved sequence for broad-band decoupling − WALTZ-16. J Magn Reson 52:335–338
Tafer H, Hiller S, Hilty C, Fernández C, Wüthrich K (2004) Nonrandom structure in the urea-unfolded Escherichia coli outer membrane protein X (OmpX). Biochemistry 43:860–869
Braun D, Wider G, Wüthrich K (1994) Sequence-corrected 15N random coil chemical shifts. J Am Chem Soc 116:8466–8469
Narayanan RL, Dürr UHN, Bibow S, Biernat J, Mandelkow E, Zweckstetter M (2010) Automatic assignment of the intrinsically disordered protein Tau with 441-residues. J Am Chem Soc 132:11906–11907
Montelione GT, Lyons BA, Emerson SD, Tashiro M (1992) An efficient triple resonance experiment using carbon-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically enriched proteins. J Am Chem Soc 114:10974–10975
Logan TM, Olejniczak ET, Xu RX, Fesik SW (1992) Side chain and backbone assignments in isotopically labeled proteins from two heteronuclear triple resonance experiments. FEBS Lett 314:413–418
Grzesiek S, Anglister J, Bax A (1993) Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J Magn Reson B 101:114–119
Jiang L, Coggins BE, Zhou P (2005) Rapid assignment of protein side chain resonances using projection-reconstruction of (4,3)D HC(CCO)NH and intra-HC(C)NH experiments. J Magn Reson 175:170–176
Sun ZY, Hyberts SG, Rovnyak D, Park S, Stern AS, Hoch JC, Wagner G (2005) High-resolution aliphatic side-chain assignments in 3D HCcoNH experiments with joint H-C evolution and non-uniform sampling. J Biomol NMR 32:55–60
Braunschweiler L, Ernst RR (1983) Coherence transfer by isotropic mixing − application to proton correlation spectroscopy. J Magn Reson 53:521–528
Clore GM, Bax A, Driscoll PC, Wingfield PT, Gronenborn AM (1990) Assignment of the side-chain 1H and 13C resonances of interleukin-1 beta using double- and triple-resonance heteronuclear three-dimensional NMR spectroscopy. Biochemistry 29:8172–8184
Acknowledgments
Financial support from the Swiss National Science Foundation, the ETH Zürich, the NCCR Structural Biology, and the Biozentrum Basel is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Hiller, S., Wider, G. (2011). Automated Projection Spectroscopy and Its Applications. In: Billeter, M., Orekhov, V. (eds) Novel Sampling Approaches in Higher Dimensional NMR. Topics in Current Chemistry, vol 316. Springer, Berlin, Heidelberg. https://doi.org/10.1007/128_2011_189
Download citation
DOI: https://doi.org/10.1007/128_2011_189
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-27159-5
Online ISBN: 978-3-642-27160-1
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)