Abstract
Microbial polyhydroxyalkanoates (PHA) are a family of biodegradable and biocompatible polyesters which have been extensively studied using synthetic biology and metabolic engineering methods for improving production and for widening its diversity. Synthetic biology has allowed PHA to become composition controllable random copolymers, homopolymers, and block copolymers. Recent developments showed that it is possible to establish a microbial platform for producing not only random copolymers with controllable monomers and their ratios but also structurally defined homopolymers and block copolymers. This was achieved by engineering the genome of Pseudomonas putida or Pseudomonas entomophiles to weaken the β-oxidation and in situ fatty acid synthesis pathways, so that a fatty acid fed to the bacteria maintains its original chain length and structures when incorporated into the PHA chains. The engineered bacterium allows functional groups in a fatty acid to be introduced into PHA, forming functional PHA, which, upon grafting, generates endless PHA variety. Recombinant Escherichia coli also succeeded in producing efficiently poly(3-hydroxypropionate) or P3HP, the strongest member of PHA. Synthesis pathways of P3HP and its copolymer P3HB3HP of 3-hydroxybutyrate and 3-hydroxypropionate were assembled respectively to allow their synthesis from glucose. CRISPRi was also successfully used to manipulate simultaneously multiple genes and control metabolic flux in E. coli to obtain a series of copolymer P3HB4HB of 3-hydroxybutyrate (3HB) and 4-hydroxybutyrate (4HB). The bacterial shapes were successfully engineered for enhanced PHA accumulation.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Polyhydroxyalkanoates (PHA) are a family of structurally diverse intracellular biopolyesters accumulated by many microorganisms [1,2,3]. Because of their similar properties with traditional petroleum-based plastics, PHA have been developed for applications in the packaging, medicine, pharmacy, agriculture, and food industries [4,5,6]. Compared with other well-known biodegradable or biobased polymers with less CO2 emission, such as polylactide (PLA), PHA have much wider diversity in monomers with over 150 structural variations reported [7, 8].
Based on monomer lengths, PHA monomers are divided into short-chain-length (scl) consisting of 3–5 carbon atoms, and medium-chain-length (mcl) of 6–14 carbon atoms (Fig. 1) [8, 9]. Based on the composition of the monomers and their arrangements, PHA have been classified into homopolymers consisting of one monomer, random copolymers of two or more different monomers, and block copolymers of at least two homopolymers connected by covalent bond(s) (Fig. 2) [9, 10]. The microstructures of PHA and monomer compositions affect the thermal and physical properties of PHA, which affects their applications (Table 1) [11, 12]. For example, the most studied PHA family member, poly(3-hydroxybutyrate) or P3HB, first reported in 1926 [13], is very brittle with high crystallinity which limits its applications [14]. In many cases, it is not easy to achieve precise control of PHA structure. For example, random copolymers consisting of 3-hydroxyhexanoate (3HHx or C6), 3-hydroxyoctanoate (3HO or C8), 3-hydroxydecanoate (3HD or C10), and 3-hydroxydodecanoate (3HDD or C12) are always formed when a fatty acid is added to cultures of Pseudomonads belonging to the rRNA homology group I, as β-oxidation in Pseudomonas spp. always shorten the C12 to C10, C8, and C6 [15]. On the other hand, the in situ fatty acid synthesis pathway, although lower in fatty acid synthesis rate for supplying PHA monomers than β-oxidation, also supplies various monomers for PHA synthesis [16], leading to PHA consisting of various monomers in random copolymers. The traditional PHA, such as PHB, PHBV, and PHBHHx, produced by wild-type microorganisms, are still facing problems of high cost and poor properties, and scientists are developing novel methods to lower the cost of PHA or discover novel PHA with high value-added applications or better properties using synthetic biology and metabolic engineering [17]. In many cases, precursors such as fatty acids, alcohols, or functional monomers are expensive, and new pathways are being established to synthesize PHA monomers in vivo from low cost glucose [16, 18]. This approach is very important if the PHA is to be produced on an industrial scale [19]. Recent advances in systems biology have improved the amount of information that can be collected, and synthetic biology tools are developing modeling and molecular implementation methods, promising to move microbial engineering from the iterative approach to a design-oriented paradigm [20].
PHA molecular structures [9]
2 Metabolic Pathways of PHA Synthesis
Many bacteria have been found to produce various polyhydroxyalkanoate (PHA) biopolyesters [8]. For example, Ralstonia eutropha was mostly studied in producing PHB and PHBV [21] and Pseudomonas putida is well-known for synthesizing mcl-PHA [22, 23]. The specificity of a PHA synthase (PhaC) is the most important element determining PHA monomer compositions in different microorganisms [24,25,26]. PhaC from Ralstonia eutropha has been known to be able to polymerize PHA monomers consisting of three (C3) to five (C5) carbon chain lengths termed short-chain-length PHA or scl PHA [27], including poly(3-hydroxypropionate) (P3HP) [28, 29], poly(3-hydroxybutyrate) (PHB) [30], poly(4-hydroxybutyrate) (P4HB) [31, 32], poly(3-hydroxyvalerate) (PHV) [33], and copolymers of 3-hydroxypropionate and 4-hydroxybutyrate (P3HP4HB) [11], as well as similar copolymers of P3HB4HB [18], P3HP3HB [34], and PHBV [5, 8]. Many Pseudomonas spp. contain PhaCs that can polymerize monomers of six (C6) to fourteen (C14) carbon-chain-length to form medium-chain-length PHA (or mcl PHA) [35]. Very few bacteria were found to have PhaCs that can polymerize C4 to C14 to form scl-mcl copolymers [36, 37]. Wild-type Ralstonia eutropha H16 can only produce scl PHA, when introducing PHA synthase gene phaC2 Ps from Pseudomonas stutzeri strain 1317 into PHA synthase gene phbC Re negative mutant R. eutropha PHB-4, the recombinant R. eutropha having the ability to synthesize mcl PHA. During the cultivation on gluconate, the presence of phaC2 Ps in R. eutropha PHB-4 led to the accumulation of PHB homopolymer at 40.9 wt% in dry cells. When using fatty acids as carbon sources, the recombinant successfully produced PHA copolyesters containing both scl PHA and mcl PHA of 4–12 carbon atoms in length. When cultivated on a mixture of gluconate and a fatty acid, the monomer composition of accumulated PHA was strongly affected and the monomer content was easily regulated by the addition of fatty acids in the cultivation medium [36]. A series of optimization strategies were reported on the PHA synthase PhaC2Ps in E. coli, codon optimization of the gene and mRNA stabilization with a hairpin structure were conducted, and the function of the optimized PHA synthase was tested in E. coli. The transcript was more stable after the hairpin structure was introduced, both codon optimization and hairpin introduction increasing the protein expression level compared with the wild-type PhaC2Ps. The optimized PhaC2Ps increased PHB production by approximately 16-fold to 30% of the cell dry weight. When grown on dodecanoate, the recombinant E. coli harboring the optimized gene phaC2 Ps O with a hairpin structure in the 5′ untranslated region was able to synthesize fourfold more PHA, consisting of 3HB and mcl 3HA, compared to the recombinant harboring the wild-type phaC2 Ps [38].
The authors’ group summarized a metabolic pathways map leading to PHA formation (Fig. 3). The most studied PHA synthesis pathways are discussed in the following. Pathway I, starting from sugar to scl PHA, especially PHB, glucose was used as carbon source to produce acetyl-CoA first, followed by metabolism to acetoacetyl-CoA and 3-hydroxybutyryl-CoA, entering the polymerization process to form PHB. The recombinant E. coli also showed high productivity of PHA after introducing the phaCAB operon from Ralstonia eutropha. Based on this pathway, more synthetic pathways were developed to produce more PHA with other structures [18, 34]. Pathway II begins from fatty acid(s) as substrate to enter the β-oxidation cycle, leading to formation of R-3-hydroxyacyl-CoA monomers for mostly mcl PHA synthesis [39]. Pathway III directs acetyl-CoA to malonyl-CoA to 3-ketoacyl-ACP for forming R-3-hydroxyacyl-CoA monomers [40, 41]. Glucose was also used as carbon source to produce novel PHA with high value-added products, such as P3HP, which is discussed later [34]. The types of PHA formed depend not only on monomer supply pathways, but also on specificity of PHA synthases. Generally, a low specificity of a PhaC allows formation of diverse PHA structures [36]. As the properties of copolymer of scl-PHA and mcl-PHA are drawing more attention, a lot of work is focusing on the production of scl-co-mcl PHA using a low specificity of a PhaC [25, 42, 43].
Metabolic pathways for PHA synthesis [9]. 1: PhaA, β-ketothiolase; 2: PhaB, NADP dependent acetoacetyl-CoA reductase; 3: PhaC, PHA synthase; 4: CimA, citramalate synthase; 5: LeuCD, 3-isopropylmalate isomerase; 6: 3-isopropylmalate isomerase; 7: LeuB, 3-isopropylmalate dehydrogenase; 8: PanE, 2HB dehydrogenase; 9: Pct5404: propionyl-CoA transferase; 10: IdhA, lactate dehydrogenase; 11: Pct Cp: propionate CoA transferase; 12: GPD1, glycerol-3-P dehydrogenase, GPP2, glycerol-3-P phosphatase; 13: DhaB, glycerol dehydratase; 14: PduP, propionaldehyde dehydrogenase; 15: AldD, aldehyde dehydrogenase; 16: PCS’, propanoyl-CoA synthetase; 17: DhaT, 1,3-propanediol dehydrogenase; 18: ThrA, aspartokinase I, ThrB, homoserine kinase; ThrAC, threonine synthase; 19: IlvA, threonine deaminase; 20: PhaA, β-ketothiolase; 21: PhaB, NADP-dependent acetoacetyl-CoA reductase; 22: SucD: succinic semialdehyde dehydrogenase; 23: 4hbD, 4-hydroxybutyrate dehydrogenase; 24: OrfZ, 4-hydroxybutyrate-CoA transferase; 25: PhaC1Ps6-19, PHA synthase from Pseudomonas sp. MBEL 6-19; 26: fadB, S-3-hydroxyacyl-CoA dehydrogenase; 27: fadA, 3-ketothiolase; 28: PhaJ, enoyl-CoA hydratase; 29: epimerase; 30: YqeF/FadA, thiolase; 31: FadB, hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase; 32: YdiO, enoyl-CoA reductase, Ter, trans-2-enoyl-CoA reductase from Treponema denticola; 33: PhaC, type II PHA synthase; 34: β-ketoacyl-ACP synthase; 35: β-ketoacyl-ACP reductase; 36: β-hydroxyacyl-ACP dehydrase; 37: enoyl-ACP reductase; 38: PhaG, 3-hydroxyacyl-acyl carrier protein-coenzyme A transferase; 39: PhaC1(STQK), PHA synthase derived from Pseudomonas sp. 61-3 PHA synthase; 40: engineered PhaC1Ps6-19, PHA synthase
3 Diversity of PHA
Diversity of PHA has been focused not only on monomer variations but also on the composition of PHA, especially on PHA main chain structures (Table 2). PHA was first discovered in the form of poly-3-hydroxybutyrate (PHB) in the last century [13]. New monomers 3-hydroxyvalerate (3HV) and 3-hydroxyhexanoate (3HHx) were detected as components of PHA in bacteria in activated sewage sludge in the 1970s [44]. Then, 10–15 years afterward, Pseudomonas oleovorans was found to be able to produce a series of PHA containing 3-hydroxyhexanoate (3HHx), 3-hydroxyoctanoate (3HO), 3-hydroxydecanoate (3HD), and 3-hydroxydodecanoate (3HDD) when grown on different alkanes or fatty acids as substrates [16, 33]. In the following years, scientists started to modify the PHA pathways or introduce the PHA pathways into a better host. For example, the phaCAB operon for PHB production was transformed into E. coli, and the non-PHA producing bacteria also showed high PHB productivity with the heterologous PHB pathway [30]. An increasing number of novel PHA were synthesized using mostly structure-related substrates and, in 1995, 91 different hydroxyalkanoic acids were reported as monomers in PHA [8]. PHA diversity was further increased by producing functional PHA, grafted with other chemicals and polymers [45,46,47]. From then on, diversity of PHA was further expanded to include PHA polymer chains with various microstructures, such as homopolymers, random copolymers, block copolymers, block-random copolymers, functional polymers, graft polymers, and thiopolyesters, as well as their various combinations [10, 11, 45]. Among the diverse PHA, grafted PHA polymers can be most easily extended to a wider diversity, and this is a topic that requires further elucidation [46, 48]. However, only very few PHA are commercially available for application developments, including PHB, PHBV, P3HB4HB, and PHBHHx. All other PHA have been prepared by individual laboratories across the world in very small amounts out of academic curiosity. How to accelerate the pace of discovery and deployment of advanced PHA materials has been a central question for all PHA researchers and stakeholders. All these depend on the availability of the diverse PHA in sufficient quantities for studies of their thermal and mechanical properties, as well as other application potentials. It should be a global effort to establish platforms to supply diverse PHA in sufficient quantities for various developments.
3.1 Homopolymers
So far, only limited homopolymers have been reported, including scl PHA: PHB [49], P3HP [34], P4HB [32], microbial polylactic acid (PLA) [42], and PHV [33], mcl PHA: P3HHx, P3HHp (poly3-hydroxyheptanoate) [50], PHO, P3HD, P3HDD, and P3HTD or poly(3-hydroxytetradecanoate) [35, 51], as well as functional PHA: poly(3-hydroxy-5-phenylvalerate) or P(3HPhV) [47], poly(3-hydroxy-4-pentenoate) [46], poly(3-hydroxy-10-undecenoate) [52], and poly(3-hydroxy-6-phenylhexanoate) [48]. With the success of engineering the β-oxidation pathway, more and more homopolymers can be synthesized. Pseudomonas putida KT2442 often produces mcl PHA consisting of 3HHx, 3HO, 3HD, 3HDD, and 3HTD, and when it was knocked out with its β-oxidation related genes fadA, fadB, fadB2x, fadAx, and phaG, the mutant P. putida KTQQ20 synthesized homopolymer poly-3-hydroxydecanoate (PHD) when grown on decanoic acid [35]. Mcl PHA producer Pseudomonas entomophila L48 was also studied for homopolymer production, when genes encoding 3-hydroxyacyl-CoA dehydrogenase, 3-ketoacyl-CoA thiolase, and acetyl-CoA acetyltransferase in the β-oxidation pathway were knocked out. The mutant P. entomophila LAC26 accumulated over 90 wt% PHA consisting of 99 mol% 3HDD using dodecanoic acid as a carbon source. The β-oxidation-inhibited mutant of P. entomophila was also studied to produce benzene containing PHA, poly(3-hydroxy-5-phenylvalerate) using 5-phenylvaleric acid as carbon source and homopolymer P(3-hydroxy-9-decenoate) using 9-decenol as carbon source [45, 47]. Synthetic biology also makes it possible to create novel PHA with designed structures and compositions.
3.2 Random Copolymers
Most of the commercially produced PHA are random copolymers, including P(3HB-co-3HV) or PHBV, P(3HB-co-4HB) or P3HB4HB, and P(3HB-co-3HHx) or PHBHHx, which have been produced on an industrial scale [2]. Copolymers of mcl PHA termed P(3HHx-co-3HO-co-3HD-co-3HDD) are commonly synthesized by many Pseudomonads belonging to the rRNA homology group I [39], but it is too soft for any application [53]. Recently, random copolymers of P(3HP-co-4HB) [11], P(3HB-co-3HP) [34], poly(3HB-co-3MP) [54], and P(3HB-co-LA) [42] were found to be accumulated by recombinant E. coli, and these copolymers demonstrated improved properties over the existing ones. However, the yield of PHA production needs to be improved for further industrial scale production.
3.3 Block Copolymers
Pederson et al. [10] reported the first PHA block copolymer of PHB-b-PHBV, and the material was found to have anti-ageing property. Block copolymerization is a method of controlling the thermodynamic nature of a polymer, and it is able to withstand the ageing effect that leads to the brittleness of a polymer material [10]. Starting in 2011, the authors’ lab and other groups have succeeded in making a series of diblock copolymers, including PHB-b-P3HVHHp [55], PHB-b-P4HB [56], PHB-b-PHHx [57], P3HB-b-P3HP [58], P3HP-b-P4HB [59], and P3HHx-b-P(3HD-co-3HDD) [60]. The sequential feeding of two or more structurally related carbon substrates led to biosynthesis of block copolymers. For example, by first feeding 1,3-propanediol and late addition of 1,4-butanediol to cultures, the engineered E. coli synthesized block copolymers of P3HP-b-P4HB [59]. All the diblock copolymers were found to have one or more improved properties over their two relative homopolymers, random copolymers or blend polymers. Compositions of diblock copolymers can be adjusted based on monomer substrate ratios in the feeds, leading to adjustable polymer properties. Although multiple-block PHA are still difficult to synthesize, with the development of synthetic biology it should become possible to realize the accurate control of monomer composition and then production of block PHA with diverse structures on a larger scale.
3.4 Graft Polymers
As it is possible to introduce functional groups into PHA chains, such as double or triple bonds, epoxy, carbonyl, cyano, phenyl, and halogen [46], graft PHA polymers can be formed by inserting small molecules or larger polymers into the PHA side chains, leading to dramatic changes PHA properties. So far, successful PHA graft polymers include poly(styrene peroxide)-g-PHA or PS-g-PHA [61], poly(methyl methacrylate peroxide)-g-PHA or PMMA-g-PHA [62], PHA-g-poly(acrylic acid) or PHA-g-PAA [63], PHA-g-AA-chitooligosaccharide or PHA-g-AA-COS [64], PHA-g-Cellulose [65], poly(ethylene glycol)-g-PHA or PEG-g-PHA [66], monoacrylate-poly(ethylene glycol)-g-PHO or PEGMA-g-PHO [67], poly(lactic acid)-g-PHA or PLA-g-PHA [68], vinylimidazole-g-PHO or VI-g-PHO [69], glycerol-1,3-diglycerol diacrylate-g-PHO or GDD-g-PHO [70], (Jeffamine®)-g-PHOU or PHOU-g-Jeffamine, PHOU-g-α-amino-ω-methoxy poly(oxyethylene-co-oxypropylene) [71], PHOU-g-polyhedral oligomeric silsesquioxane or PHOU-g-POSS [72], PHBV-g-poly(phenyl vinyl ketone) or PHBV-g-PVK [73], and PHBV-g-poly(acrylamide) or PHBV-g-PA [74]. Graft copolymers were mostly synthesized by chemical modification. For example, side carboxylic groups of the PHA were coupled with end hydroxyl groups of methoxy-poly(ethylene glycol) (MePEG) or methoxy-poly(lactic acid) (MePLA) in the presence of N,N′-dicylohexylcarbodiimide (DCC) [68]. There are endless possibilities to create new graft PHA homo- or copolymers.
4 Engineering Pathways for Controlling PHA Biosynthesis
4.1 Pathways for scl PHA
Microbial metabolic engineering has been exploited as a powerful approach for enhanced production of novel polyesters. A designed pathway assembled using a synthetic biology approach could also precisely control the PHA composition. The use of recombinant E. coli enabled an efficient production of poly(4-hydroxybutyrate) or P4HB using glucose as a sole carbon source when a pathway was established containing genes encoding succinic semialdehyde dehydrogenase of Clostridium kluyveri and PHB synthase of Ralstonia eutropha combined with inactivation of native succinate semialdehyde dehydrogenase genes sad and gabD to enhance the carbon flux toward P4HB biosynthesis [32]. When the PHB accumulation pathway of Ralstonia eutropha was co-expressed with the P4HB synthesis pathway, the recombinant E. coli produces P(3HB-co-4HB) from glucose [18].
Aeromonas hydrophila 4AK4 normally produces copolyesters PHBHHx. Recombinant A. hydrophila 4AK4 expressing vgb and fadD genes encoding Vitreoscilla hemoglobin and E. coli acyl-CoA synthase, respectively, was found to produce homopolymer poly(3-hydroxyvalerate) (PHV) (C5) using undecanoic acid as a solo carbon source [75]. At the same time, 3-hydroxyvalerate monomer can also be supplied via the threonine degradation pathway. Recently, it became possible to produce PHA containing 2-hydroxybutyrate [76] or lactate [42]. In addition, P3HP can be produced from 1,3-propandiol [29], glycerol alone [77], and glucose as sole carbon source [34].
PHA synthesis genes phbC and orfZ cloned from Ralstonia eutropha H16 and Clostridium kluyveri, respectively, were transformed into a β-oxidation weakened Pseudomonas putida KTOY08ΔGC, a mutant of P. putida KT2442, and the resulting mutant termed KTHH06 was able to produce P3HB-b-P4HB diblock copolymer [56].
4.2 Synthesis of Poly(3-hydroxypropionate-co-4-hydroxybutyrate) with Fully Controllable Structures by Recombinant Escherichia coli Containing an Engineered Pathway
Recently, microbial copolyesters containing 3HP have become increasingly interesting because of the ultrahigh strength brought about by 3HP, and these include P(3HB-co-3HP), P(3HP-co-3HB-co-3HH-co-3HO), P(4HB-co-3HP-co-Lactate), P(4HB-co-3HP-co-2HP), P(3HB-co-3HP-4HB-co-Lactate), and P(3HB-co-3HP-co-4HB-co-2HP) [78]. Natural bacteria are unable to produce 3-hydroxyproionate (3HP) and 4-hydroxybutyrate (4HB) as building blocks for PHA synthase to make the unnatural biopolyester P(3HP-co-4HB) [11]. However, precursors of 3HP and 4HB can come from 1,3-propanediol (PDO) [29] and 1,4-butanediol (BDO) [79], respectively. Copolyesters of 3-hydroxypropionate (3HP) and 4-hydroxybutyrate (4HB), abbreviated as P(3HP-co-4HB), were synthesized by E. coli harboring a synthetic pathway consisting of five heterologous genes including orfZ encoding 4-hydroxybutyrate-coenzyme A transferase from Clostridium kluyveri [80, 81], pcs’ encoding the ACS domain of tri-functional propionyl-CoA ligase (PCS) from Chloroflexus aurantiacus [82], dhaT and aldD encoding dehydratase and aldehyde dehydrogenase from Pseudomonas putida KT2442 [83], and phaC1 encoding PHA synthase from Ralstonia eutropha (Fig. 4) [11, 29]. When grown on mixtures of 1,3-propanediol (PDO) and 1,4-butanediol (BDO), compositions of 4HB in microbial P(3HP-co-4HB) were controllable ranging from 12 mol% to 82 mol% depending on PDO:BDO ratios. Their mechanical and thermal properties showed obvious changes depending on the monomer ratios (Table 3). Morphologically, P(3HP-co-4HB) films only became fully transparent when monomer 4HB content was around 67 mol% (Fig. 5) [11].
Construction of P(3HP-co-4HB) biosynthetic pathways in recombinant Escherichia coli [11]. Enzymes for each numbered step are as follows: (1) 1,3-propanediol dehydrogenase; (2) aldehyde dehydrogenase; (3) propanoyl-CoA synthetase; (4) 4-hydroxybutyrate coenzyme A transferase; (5) PHA synthase
Transparency of P(3HP-co-4HB) consisting of different monomer compositions [11]. From left to right: P(3HP), P(3HP-co-12 mol% 4HB), P(3HP-co-25 mol% 4HB), P(3HP-co-38 mol% 4HB), P(3HP-co-67 mol% 4HB), P(3HP-co-82 mol% 4HB), P(4HB)
Several key enzymes were considered as important for making P(3HP-co-4HB) copolymers with flexible 4HB content: propionyl-CoA synthetase (PCS’) from the 3-hydroxypropionate cycle of phototrophic green non-sulfur eubacterium Chloroflexus aurantiacus is very likely to convert 3HP to 3HP-CoA, and 4HB-coenzyme, a transferase gene orfz from Clostridium kluyveri, was found to turn 4HB into 4HB-CoA effectively [82]. Genes dhaT and aldD were found to turn 1,4-butanediol (BDO) or/and 1,3-propanediol (PDO) into 4HB or/and 3HP, respectively. The enzyme encoded by dhaT was mostly active with substrates containing two primary alcohol groups separated by one or two carbon atoms such as 1,3-propanediol or 1,4-butanediol, and 3HP or/and 4HB yield were affected by expression levels of these two genes [29, 79, 83]. Promoter of PHA synthesis genes phaCAB operon from Ralstonia eutropha (P Re ) was demonstrated to be more active than lac promoter or T7 promoter transcriptionally in E. coli. Finally, PHA synthase PhaC1 of R. eutropha has sufficient activity for polymerizing SCL PHA monomers [11, 29].
A mixture of PDO and BDO in cultures of the recombinant E. coli S17-1 resulted in formation of copolyesters P(3HP-co-4HB) consisting of 3HP and 4HB. Compositions of the 3HP and 4HB in P(3HP-co-4HB) could be adjusted by changing the ratios of PDO to BDO. For example, 63 wt% P(17 mol% 3HP-co-83 mol% 4HB) was accumulated when the PDO:BDO ratio was 1/10; whereas a ratio of 1:1 led to the formation of P(70 mol% 3HP-co-30 mol% 4HB). When PDO/BDO was equal to 10/15 (or 2/3), only 2.3 wt% P(88 mol% 3HP-co-12 mol% 4HB) was synthesized, indicating the toxicity of high BDO or PDO concentration. Especially when the total concentration of BDO and PDO were over 20 g/L, the toxicity became very obvious, as indicated by significant reduction on CDW and PHA production. Obviously, a copolymer consisting of a defined 3HP:4HB ratio can be produced by adjusting the ratios of PDO:BDO. In this study, P(3HP-co-4HB) consisting of 17 mol% 3HP–88 mol% 3HP were obtained. Interestingly, the transparency of P(3HP-co-4HB) was also found to be dependent on monomer compositions. Only P(3HP-co-67 mol% 4HB) was a totally transparent material, whereas other PHA including P(3HP), P(3HP-co-12 mol% 4HB), P(3HP-co-25 mol% 4HB), P(3HP-co-38 mol% 4HB), P(3HP-co-82 mol% 4HB), and P(4HB) were observed to be less transparent [11].
The addition of 4HB monomer into P3HP led to the formation of P(3HP-co-4HB) which clearly lowered the P3HP melting temperatures (T m ) and the glass transition temperature (T g ) from 78°C and −18°C to 61–65°C and −24°C to –41°C with the 4HB ratio increased from 12 mol% to 67 mol% (Table 3). Interestingly, P(3HP-co-82 mol% 4HB) was revealed to have a much lower T m of 36°C and a higher T g of −29°C compared to other copolymers. T m seemed to stabilize at around 63°C in copolymers consisting of 12–67 mol% 4HB. T g decreased from −24°C to −42°C with 4HB content increasing from 12 mol% to 67 mol%. Homopolymer P4HB had the lowest T g of −47°C with a T m of 61°C (Table 3).
Copolymerization reduced yield strengths and Young’s modulus of both P3HP and P4HB (Table 3). However, the elongation at breaks showed an improvement for P(3HP-co-4HB) consisting of 12–38 mol% 4HB over P3HP and P4HB. On the other hand, only P(3HP-co-12 mol% 4HB) had an increase on maximum tension strength over other homo- and copolymers. In terms of thermal and mechanical properties, P(3HP-co-4HB) seems to be unique in combined properties compared with commercial PHA such as PHB, PHBV, and PHBHHx.
As PDO and BDO can be respectively biosynthesized from glucose [84, 85], it becomes possible to establish an engineering pathway for production of P(3HP-co-4HB). Block copolymers of P3HB-b-P3HP could also be produced [59]. The two pathways supplied 3HP and 4HB monomers independently, leading to the formation of homopolymer P3HP in the absence of 4HB, of P4HB in the absence of 3HP, or to random copolymers of P(3HP-co-4HB) when 3HP and 4HB were both available.
4.3 Poly(3-hydroxybutyrate-co-3-hydroxypropionate) from Glucose by Engineering Escherichia coli
Poly(3-hydroxypropionate) (P3HP), an scl-PHA containing three carbon atoms without side chain, shows the best combined mechanical properties, including an elongation at break of more than 600%, and a Young’s modulus of 3 GPa [11]. P3HP therefore stands out as a PHA member that holds great promise. No microorganism has been known to synthesize homopolymer P3HP so far. Thus, recombinants have been developed to produce P3HP. Andreessen et al. [28] first reported bacterial synthesis of P3HP using glycerol as carbon source in a two-step fed-batch fermentation. Wang et al. [77] modified the process by replacing the strict anaerobic glycerol dehydratase from Clostridium butyricum with the vitamin B12-dependent glycerol dehydratase DhaB123 from Klebsiella pneumonia. Zhou et al. [29] used 1,3-propanediol as a precursor to produce over 90% P3HP in E. coli cell dry weight (CDW). There were attempts to synthesize P3HP from an unrelated carbon source starting with acetyl-CoA [86, 87]. The related pathway involves carboxylation of acetyl-CoA to malonyl-CoA, reduction of malonyl-CoA to 3HP, its coupling to CoA, and their following polymerization. This recombinant pathway led to only 1.32 g/L CDW containing 0.98% P3HP [87].
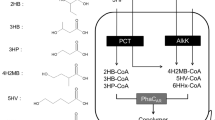
The authors’ lab reported that multiple genes from various sources were assembled into a new pathway for the production of P3HP from glucose as a sole carbon source, including gpd1 (glycerol-3-P dehydrogenase) and gpp2 (glycerol-3-P phosphatase) from Saccharomyces cerevisiae [88, 89], dhaB1-3 (glycerol dehydratase) and gdrAB (glycerol dehydratase reactivating factor) from Klebsiella pneumonia [90, 91], pduP (propionaldehyde dehydrogenase) from Salmonella typhimurium [92, 93], phaC (PHA synthase) from Ralstonia eutropha [26, 94], aldD (aldehyde dehydrogenase) and dhaT (1,3-propanediol dehydrogenase) from Pseudomonas putida KT2442 [79, 83], and pcs’ (propanoyl-CoA synthetase) from Chloroflexus aurantiacus [29]. When the plasmid containing the above multiple genes was transformed into E. coli, up to 18.4% P3HP homopolymer was produced from glucose (Fig. 6) [34].
Construction of P3HP and P(3HB-co-3HP) biosynthetic pathways from glucose as a sole carbon source in recombinant Escherichia coli [34]. Enzymes encoded by each gene are described below: gpd1 glycerol-3-P dehydrogenase (Saccharomyces cerevisiae), gpp2 glycerol-3-P phosphatase (Saccharomyces cerevisiae), dhaB1-3 glycerol dehydratase (Klebsiella pneumoniae), gdrAB glycerol dehydratase reactivating factors (Klebsiella pneumoniae), pduP propionaldehyde dehydrogenase (Salmonella typhimurium), phaC polyhydroxyalkanoate synthase (Ralstonia eutropha), aldD aldehyde dehydrogenase (Pseudomonas putida), dhaT 1,3-propanediol dehydrogenase (Pseudomonas putida), pcs’ propanoyl-CoA synthetase (Chloroflexus aurantiacus), phaA β-ketothiolase (Ralstonia eutropha), phaB NADPH-dependent acetoacetyl-CoA reductase (Ralstonia eutropha)
The expression of the two genes gpd1 and gpp2 allows dihydroxyacetone from glucose glycolysis to form glycerol-3-phosphate, which is further hydrolyzed to generate glycerol [95]. Glycerol is converted to 3-hydroxypropionaldehyde by glycerol dehydratase (DhaB1-3) from Klebsiella pneumonia, which is an important intermediate for P3HP, and 3-hydroxypropionaldehyde is converted to 3-hydroxypropionate (3HP) by aldehyde dehydrogenase (AldD) cloned from Pseudomonas putida KT2442. Propionyl-CoA synthetase (PCS’) from Chloroflexus aurantiacus should be able to change 3HP to 3HP-CoA. At the same time, 3-hydroxypropionaldehyde can also be directly turned into 3HP-CoA by propionaldehyde dehydrogenase (PduP) from Salmonella typhimurium. To increase the activity of glycerol dehydratase, gdrAB, a reactivation factor for glycerol dehydratase was inserted into the above-mentioned pathway. When gene pudP was used to replace aldD and dhaT, the resulting plasmid pDC02 became the only plasmid containing the entire pathway from glucose to P3HP. Recombinant E. coli Trans1-T1 (pDC02) produced over 18% P3HP in over 5 g/L CDW when grown in glucose LB medium whereas in the glucose mineral medium, 12% P3HP was accumulated in 3 g/L CDW. More P3HP accumulation from glucose is expected when the metabolic flux is further optimized [34].
When a P3HB synthesis pathway containing the P3HB synthesis operon phaCAB from Ralstonia eutropha was added to the P3HP synthesis pathway, the recombinant harboring the P3HB and P3HP pathways started to produce random copolymers of P3HB3HP from glucose as the sole carbon source. This study demonstrated that ultra-strong polyhydroxyalkanoates (PHA), mainly P3HP and P3HB3HP, can be synthesized from low cost glucose using synthetic biology approaches.
The two plasmids p15apCAB and pDC02, which harbor three genes and nine genes from different microorganisms responsible for P3HP and P3HB syntheses from glucose, respectively, can be regarded as bio-devices or bio-bricks that are assembled to perform their functions (Fig. 7). This study can serve as a typical synthetic biology example that uses bio-bricks or bio-devices to achieve biological functions. In this case, it was the synthetic biology for production of novel bio-polyesters. In total, 11 heterogeneous genes were cloned from other microorganisms and were assembled to become new pathways to meet our new demands.
Orders of gene arrangements on plasmids pDC02 and p15apCAB, respectively [34]
In the future, the two polyester synthesis pathways could be transformed into other microbial hosts after codon optimization to enhance P3HB3HP production by some industrial microbial hosts [34].
4.4 Engineering the β-Oxidation Pathway on the Chromosome for mcl PHA Synthesis
Many Pseudomonas spp. are able to utilize fatty acids via their β-oxidation to obtain both energy and substrates for cell growth. The β-oxidation pathway shortens the fatty acid chain lengths in each cycle by two carbon atoms, generating several PHA monomers of different lengths, which can result in the formation of random PHA copolymers (Fig. 8).
The weakened beta-oxidation cycle, reversed fatty acid beta-oxidation cycle and in situ fatty acid synthesis. Enzymes in β-oxidation cycle: FadD fatty acid-CoA ligase, FadE acyl-CoA dehydrogenase, FadBa S-enoyl-CoA hydratase, FadB 3-hydroxyacyl-CoA dehydrogenase, FadA acetyl-CoA acetyltransferase, PhaJ R-enoyl-CoA hydratase, PhaC PHA synthase, PhaG 3-hydroxyacyl-CoA-acyl carrier protein transferase. Genes in the reversed fatty acid β-oxidation cycle: yqeF/fadA thiolase, fadB hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase, ydiO enoyl-CoA reductase, ter trans-2-enoyl-CoA reductase, tesA/tesB/yciA thioesterase
Recently, the authors’ lab succeeded in engineering the β-oxidation pathway encoded on the chromosomes of Pseudomonas putida and Pseudomonas entomophiles, resulting in controllable PHA composition, including formation of PHA homopolymers and composition-adjustable random copolymers and block copolymers [35, 51, 60]. To avoid the changing of fatty acid substrate structures, chromosomal genes related to β-oxidation were selectively deleted to weaken β-oxidation in Pseudomonas spp., so that fatty acids can maintain their structures when used as PHA monomer precursors.
Mutant Pseudomonas putida KTQQ20, a derivative of P. putida KT2442, deleted key fatty acid degradation enzymes encoded by genes fadB, fadA, fadB2x, and fadAx, as well as PP2047 and PP2048 encoding 3-hydroxyacyl-CoA dehydrogenase and acyl-CoA dehydrogenase, respectively, combined with the deletion of phaG encoding 3-hydroxyacyl-CoA-acyl carrier protein transferase, becomes defective in fatty acid β-oxidation activity. The strain was now able to synthesize homopolymer poly(3-hydroxydecanoate) or PHD and P(3HD-co-84 mol% 3HDD) when grown on decanoic acid or dodecanoic acid, respectively [35]. When grown on mixtures of the sodium salt of hexanoate (C6) and decanoate (C10), it produced random copolymers of P(3HHx-co-3HD) with monomer compositions easily regulated by varying the C6:C10 ratio. P. putida KTQQ20 also produced diblock copolymer P3HHx-b-P(3HD-co-3HDD) when sodium salts of hexanoate (C6) and decanoate (C10) were fed to its culture one after another [60].
Pseudomonas entomophila strain L48, a strong fatty acid utilizer, was also investigated for microbial production of mcl PHA. A total of 70.2% of P. entomophila genes have orthologs with the P. putida genome, of which >96% are found in synteny. The β-oxidation activity of P. entomophila was weakened by deleting similar genes on its chromosome as in P. putida. The resulting P. entomophila LAC26 accumulated over 90 wt% PHA consisting of 99 mol% 3HDD. Homopolymers of C6–C14 were all accumulated, respectively, when an equal chain length of a fatty acid was fed to the mutant for related PHA homopolymer production [51].
4.5 Pathways for scl and mcl PHA Copolymers
P. putida KTOYO6 is a fatty acid β-oxidation impaired mutant in which genes of 3-ketoacyl-CoA thiolase (fadA) and 3-hydroxyacyl-CoA dehydrogenase (fadB) were deleted to a maximum level to improve fatty acid utilization for PHA synthesis [53]. When its mcl PHA synthase (C6–C14) was replaced by a less specific synthase operon phaPCJ Ac which could synthesize both scl and mcl monomers (C3–C7) from Aeromonas caviae, recombinant P. putida KTOYO6ΔC (phaPCJ Ac ) was able to produce a diblock copolymer of PHB-b-PHVHHp by controlling the sequential feeding time of sodium butyrate and sodium heptanoate. When cultivated on mixtures of sodium salts of butyrate (C4) and hexanoate (C6), random copolymers of P(3HB-co-3HHx) were accumulated with monomer contents adjustable by C4:C6 ratios [55].
5 Functional PHA
When cultures of engineered strains, such as P. putida KTQQ20 or P. entomophila LAC23, were fed with fatty acids containing functional groups such as double or triple bonds, epoxy, carbonyl, cyano, phenyl and halogen group, respectively [46], the resulting PHA contains the functional groups on the side chains, allowing further chemical modifications (grafting) on the side chains.
Homopolymers with 100 mol% content of aromatic moieties, random copolymers, or a blend of both have been produced [47]. Hydrophilic PHA bearing alkoxy, acetoxy, or hydroxyl groups are also of great interest, as they show enhanced solubility and biocompatibility [46].
The β-oxidation weakened P. entomophila LAC23 was found able to accumulate PHA containing phenyl groups on the side chains. When cultured in 5-phenylvaleric acid, only homopolymer poly(3-hydroxy-5-phenylvalerate) was synthesized. Copolyesters of 3-hydroxy-5-phenylvalerate (3HPhV) and 3-hydroxydodecanoate (3HDD) were also successfully produced by P. entomophila LAC23 when grown on mixtures of phenylvaleric acid and dodecanoic acid. Compositions of 3HPhV in P(3HPhV-co-3HDD) were controllable, ranging from 3% to 32%, depending on dodecanoic acid:5-phenylvaleric acid ratios [47]. Although the production of PHA with functional groups is still facing high costs and low productivity, the toxicity of substrates also affect the growth of microorganisms, and PHA with functional groups needs be produced from unrelated carbon sources in future studies.
6 Engineering the Bacterial PHA Synthesis Using CRISPRi
Clustered regularly interspaced short palindromic repeats interference (CRISPRi) is a powerful technology used to regulate eukaryotic genomes [96]. CRISPRi has also been reported to control PHA biosynthesis pathway flux and to adjust PHA composition. First, an E. coli strain was engineered by introducing a pathway for the production of P3HB4HB from glucose [18]. The native gene sad, encoding succinate semi-aldehyde dehydrogenase, was regulated by CRISPRi using five specially designed single guide RNAs (sgRNAs) for controlling carbon flux toward 4-hydroxybutyrate (4HB) biosynthesis in E. coli. The system allowed formation of P3HB4HB consisting of 1–9 mol% 4HB. Additionally, succinate, generated by succinyl-CoA synthetase and succinate dehydrogenase (respectively encoded by genes sucC, sucD, sdhA, and sdhB) was channeled preferentially to the 4HB precursor using selected sgRNAs such as sucC2, sucD2, sdhB2, and sdhA1 via CRISPRi. The resulting 4HB content in P3HB4HB could be adjusted from 1.4 mol% to 18.4 mol% depending on the expression levels of down-regulated genes (Fig. 9). The results show that CRISPRi is a feasible approach to simultaneously manipulate multiple genes and control metabolic flux in E. coli [97].
CRISPRi as a tool to control P3HB4HB biosynthesis pathway flux and to adjust 3HB/4HB composition [97]. Engineered pathways for P3HB4HB synthesis by recombinant Escherichia coli. The CRISPRi system was used to repress gene transcription initiation and elongation in the related pathways. To obtain P3HB4HB consisting of various 4HB ratios, several genes can be manipulated simultaneously, including following genes: phaA beta-ketothiolase, phaB NADPH-dependent acetoacetyl-CoA reductase, phaC PHA synthase, sucD succinate semi-aldehyde dehydrogenase, 4hbD 4-hydroxybutyrate dehydrogenase, orfZ 4-hydroxybutyrate CoA transferase
7 Engineering the Bacterial Shapes for Enhanced Polyhydroxyalkanoates Accumulation
Most bacteria have a small size ranging from 0.5 μm to 2 μm, preventing the bacterial cells from accumulating large amounts of inclusion bodies intracellularly, even though the bacteria are able to grow very fast. To overcome the size limitation, it is important to make bacterial cells larger. That is to say, a larger intracellular space is needed for more inclusion body accumulation. Various approaches were taken to increase the bacterial cell sizes, including deletion on actin-like protein gene mreB, weak expression of mreB in mreB deletion mutant, and weak expression of mreB in mreB deletion mutant under inducible expression of sulA, the inhibitor of division ring protein gene ftsZ. All of the methods resulted in different levels of increases in bacterial sizes and PHB granules accumulation [98].
MreB, the actin-like bacterial cytoskeletons, which also affects bacterial morphology, was considered a suitable engineering target for expanding the cell volumes [99]. When mreB was deleted, E. coli changed from rods to spherical shapes, and some cells even increased their sizes to diameters of around 10 μm. More PHB granules were accumulated in the large E. coli JM109SG (ΔmreB) cells. However, E. coli JM109SG (ΔmreB) also appeared to be fragile and a fraction of cells ruptured during the growth stage. This phenomenon showed that MreB may provide critical support for maintaining the cell shape. Ectopic expression of MreB in a wild-type bacterium was found to interfere with normal MreB cytoskeleton formation, resulting in a larger cell size compared with that of a wild type. To increase the cell size further, the mreB gene was compensated by constitutively expressing mreB in a weaker manner in MreB deleted E. coli JM109SG together with an arabinose-inducible sulA gene encoding an inhibitory protein for the formation of the cell division ring (FtsZ ring), the overexpression of which leads to elongated cells. Remarkably, an increase of over 100% PHB accumulation was observed in recombinant E. coli overexpressing mreB in an mreB deletion mutant under inducible expression of gene ftsZ inhibiting protein SulA (Fig. 10). The molecular mechanism of enlarged bacterial size was found to be directly related to the weakened cytoskeleton, which was the result of broken skeleton helix [98]. The larger E. coli cells make it possible to produce more PHA.
Electron microscopy studies on morphology and PHB production by E. coli JM109SG (ΔmreB) overexpressing mreB [98]. (a) Schematic of PHB accumulation in E. coli JM109SG (ΔmreB) overexpressing mreB. Scale bar: 0.5 μm; (b) Growth and PHB accumulation by recombinants harboring pBHR68 cultivated in minimal medium at 30°C for 10 h followed by addition of 20 g/L glucose and another 40 h of growth. Error bars: s.d. (n = 3). E. coli JM109SG (pTK/pBHR68) (c) and E. coli JM109SG(ΔmreB/pTK-mreB/pBHR68) (d) were grown in LB medium at 30°C for 10 h followed by addition of 20 g/L glucose and another 40 h of growth. TEM on sections of cells of control E. coli JM109SG (pTK/pBHR68) (e) and E. coli JM109SG (ΔmreB/pTK-mreB/pBHR68) (f) cultivated in the LB medium at 30°C for 10 h, followed by addition of 20 g/L glucose and another 40 h of growth. Scale bar: 5 μm
8 Conclusion
The application of PHA as a low-cost biodegradable plastic has been hampered by its higher production cost and the difficulty to control precisely their structures and properties. Global efforts have been made to develop technology for lowering the PHA production cost. With the successful construction of β-oxidation weakened Pseudomonas spp. as PHA production platforms, it is possible to control the formation of homopolymers and random- and block copolymers including monomer structures and ratios, and this allows us to obtain PHA with consistent properties. At the same time, it is possible to introduce various functional groups into the PHA side chains in a quantitative way, which provides more opportunities for side-chain grafting. Functional PHA together with endless possibilities for grafting have provided us with limitless ways of making new PHA, possibly with some high value-added functionalities. With the development of synthetic biology, it also becomes possible to construct unnatural pathways to produce novel PHA with strong value-added properties. It is widely held that within 5 or 10 years, many novel properties including environmental responsiveness, shape memory ability, controllable biodegradability, and mechanical ultra-strength will be developed from the diverse PHA materials. Thus, with the development of synthetic biology, we open a new PHA golden era.
References
Brandl H, Gross RA, Lenz RW, Fuller RC (1990) Plastics from bacteria and for bacteria: poly(beta-hydroxyalkanoates) as natural, biocompatible, and biodegradable polyesters. Adv Biochem Eng Biotechnol 41:77–93
Chen GQ (2009) A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev 38:2434–2446
Poirier Y, Nawrath C, Somerville C (1995) Production of polyhydroxyalkanoates, a family of biodegradable plastics and elastomers, in bacteria and plants. Biotechnology 13:142–150
Chen GQ, Wu Q (2005) The application of polyhydroxyalkanoates as tissue engineering materials. Biomaterials 26:6565–6578
Gao X, Chen JC, Wu Q, Chen GQ (2011) Polyhydroxyalkanoates as a source of chemicals, polymers, and biofuels. Curr Opin Biotechnol 22:768–774
Volova TG, Gladyshev MI, Trusova MY, Zhila NO (2006) Degradation of polyhydroxyalkanoates and the composition of microbial destructors under natural conditions. Microbiology 75:593–598
Steinbüchel A, Lütke-Eversloh T (2003) Metabolic engineering and pathway construction for biotechnological production of relevant polyhydroxyalkanoates in microorganisms. Biochem Eng J 16:81–96
Steinbüchel A, Valentin HE (1995) Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol Lett 128:219–228
Meng DC, Shen R, Yao H, Chen JC, Wu Q, Chen GQ (2014) Engineering the diversity of polyesters. Curr Opin Biotechnol 29:24–33
Pederson EN, McChalicher CW, Srienc F (2006) Bacterial synthesis of PHA block copolymers. Biomacromolecules 7:1904–1911
Meng DC, Shi ZY, Wu LP, Zhou Q, Wu Q, Chen JC, Chen GQ (2012) Production and characterization of poly(3-hydroxypropionate-co-4-hydroxybutyrate) with fully controllable structures by recombinant Escherichia coli containing an engineered pathway. Metab Eng 14:317–324
Doi Y (1989) Production, properties, and biodegradation of microbial copolyesters of 3-hydroxybutyrate and 4-hydroxybutyrate. Abstr Pap Am Chem S 197:45-Btec
Lemoigne M (1926) Products of dehydration and of polymerization of β-hydroxybutyric acid. Bull Soc Chem Biol 8:770–782
Savenkova L, Gercberga Z, Nikolaeva V, Dzene A, Bibers I, Kalnin M (2000) Mechanical properties and biodegradation characteristics of PHB-based films. Process Biochem 35:573–579
Fiedler S, Steinbuchel A, Rehm BH (2002) The role of the fatty acid beta-oxidation multienzyme complex from Pseudomonas oleovorans in polyhydroxyalkanoate biosynthesis: molecular characterization of the fadBA operon from P. oleovorans and of the enoyl-CoA hydratase genes phaJ from P. oleovorans and Pseudomonas putida. Arch Microbiol 178:149–160
Timm A, Steinbüchel A (1990) Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl Environ Microbiol 56:3360–3367
Chanprateep S (2010) Current trends in biodegradable polyhydroxyalkanoates. J Biosci Bioeng 110:621–632
Li ZJ, Shi ZY, Jian J, Guo YY, Wu Q, Chen GQ (2010) Production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) from unrelated carbon sources by metabolically engineered Escherichia coli. Metab Eng 12:352–359
Chen GQ, Patel MK (2012) Plastics derived from biological sources: present and future: a technical and environmental review. Chem Rev 112:2082–2099
Tyo KE, Kocharin K, Nielsen J (2010) Toward design-based engineering of industrial microbes. Curr Opin Microbiol 13:255–262
Kichise T, Fukui T, Yoshida Y, Doi Y (1999) Biosynthesis of polyhydroxyalkanoates (PHA) by recombinant Ralstonia eutropha and effects of PHA synthase activity on in vivo PHA biosynthesis. Int J Biol Macromol 25:69–77
Lee SY, Wong HH, Choi JI, Lee SH, Lee SC, Han CS (2000) Production of medium-chain-length polyhydroxyalkanoates by high-cell-density cultivation of Pseudomonas putida under phosphorus limitation. Biotechnol Bioeng 68:466–470
Tan IKP, Kumar KS, Theanmalar M, Gan SN, Gordon B (1997) Saponified palm kernel oil and its major free fatty acids as carbon substrates for the production of polyhydroxyalkanoates in Pseudomonas putida PGA1. Appl Microbiol Biotechnol 47:207–211
Hyakutake M, Saito Y, Tomizawa S, Mizuno K, Tsuge T (2011) Polyhydroxyalkanoate (PHA) synthesis by class IV PHA synthases employing Ralstonia eutropha PHB-4 as host strain. Biosci Biotechnol Biochem 75:1615–1617
Rehm BHA, Steinbüchel A (1999) Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int J Biol Macromol 25:3–19
Yuan W, Jia Y, Tian J, Snell KD, Muh U, Sinskey AJ, Lambalot RH, Walsh CT, Stubbe J (2001) Class I and III polyhydroxyalkanoate synthases from Ralstonia eutropha and Allochromatium vinosum: characterization and substrate specificity studies. Arch Biochem Biophys 394:87–98
Sudesh K, Abe H, Doi Y (2000) Synthesis, structure and properties of polyhydroxyalkanoates: biological polyesters. Prog Polym Sci 25:1503–1555
Andreessen B, Lange AB, Robenek H, Steinbüchel A (2010) Conversion of glycerol to poly(3-hydroxypropionate) in recombinant Escherichia coli. Appl Environ Microbiol 76:622–626
Zhou Q, Shi ZY, Meng DC, Wu Q, Chen JC, Chen GQ (2011) Production of 3-hydroxypropionate homopolymer and poly(3-hydroxypropionate-co-4-hydroxybutyrate) copolymer by recombinant Escherichia coli. Metab Eng 13:777–785
Slater SC, Voige WH, Dennis DE (1988) Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-beta-hydroxybutyrate biosynthetic pathway. J Bacteriol 170:4431–4436
Steinbüchel A, Valentin H, Schönebaum A (1994) Application of recombinant gene technology for production of polyhydroxyalkanoic acids: biosynthesis of poly(4-hydroxybutyric acid) homopolyester. J Environ Polym Degrad 2:67–74
Zhou XY, Yuan XX, Shi ZY, Meng DC, Jiang WJ, Wu LP, Chen JC, Chen GQ (2012) Hyperproduction of poly(4-hydroxybutyrate) from glucose by recombinant Escherichia coli. Microb Cell Fact 11:54
Steinbüchel A, Debzi E-M, Marchessault R, Timm A (1993) Synthesis and production of poly(3-hydroxyvaleric acid) homopolyester by Chromobacterium violaceum. Appl Microbiol Biotechnol 39:443–449
Meng DC, Wang Y, Wu LP, Shen R, Chen JC, Wu Q, Chen GQ (2015) Production of poly(3-hydroxypropionate) and poly(3-hydroxybutyrate-co-3-hydroxypropionate) from glucose by engineering Escherichia coli. Metab Eng 29:189–195
Liu Q, Luo G, Zhou XR, Chen GQ (2011) Biosynthesis of poly(3-hydroxydecanoate) and 3-hydroxydodecanoate dominating polyhydroxyalkanoates by beta-oxidation pathway inhibited Pseudomonas putida. Metab Eng 13:11–17
Chen JY, Song G, Chen GQ (2006) A lower specificity PhaC2 synthase from Pseudomonas stutzeri catalyses the production of copolyesters consisting of short-chain-length and medium-chain-length 3-hydroxyalkanoates. Anton Leeuw Int J G 89:157–167
Loo CY, Lee WH, Tsuge T, Doi Y, Sudesh K (2005) Biosynthesis and characterization of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from palm oil products in a Wautersia eutropha mutant. Biotechnol Lett 27:1405–1410
Gao X, Yuan XX, Shi ZY, Guo YY, Shen XW, Chen JC, Wu Q, Chen GQ (2012) Production of copolyesters of 3-hydroxybutyrate and medium-chain-length 3-hydroxyalkanoates by E. coli containing an optimized PHA synthase gene. Microb Cell Fact 11:130
Huisman GW, de Leeuw O, Eggink G, Witholt B (1989) Synthesis of poly-3-hydroxyalkanoates is a common feature of fluorescent pseudomonads. Appl Environ Microbiol 55:1949–1954
Klinke S, Ren Q, Witholt B, Kessler B (1999) Production of medium-chain-length poly(3-hydroxyalkanoates) from gluconate by recombinant Escherichia coli. Appl Environ Microbiol 65:540–548
Verwoert II, Verbree EC, van der Linden KH, Nijkamp HJ, Stuitje AR (1992) Cloning, nucleotide sequence, and expression of the Escherichia coli fabD gene, encoding malonyl coenzyme A-acyl carrier protein transacylase. J Bacteriol 174:2851–2857
Jung YK, Kim TY, Park SJ, Lee SY (2010) Metabolic engineering of Escherichia coli for the production of polylactic acid and its copolymers. Biotechnol Bioeng 105:161–171
Zou XH, Chen GQ (2007) Metabolic engineering for microbial production and applications of copolyesters consisting of 3-hydroxybutyrate and medium-chain-length 3-hydroxyalkanoates. Macromol Biosci 7:174–182
Wallen LL, Rohwedde WK (1974) Poly-beta-hydroxyalkanoate from activated-sludge. Environ Sci Technol 8:576–579
Li SJ, Cai LW, Wu LP, Zeng GD, Chen JC, Wu Q, Chen GQ (2014) Microbial synthesis of functional homo-, random, and block polyhydroxyalkanoates by beta-oxidation deleted Pseudomonas entomophila. Biomacromolecules 15:2310–2319
Olivera E, Arcos M, Naharro G, Luengo J (2010) Unusual PHA biosynthesis. In: Chen GG-Q (ed) Plastics from bacteria. Microbiol Monographs, vol 14. Springer, Berlin, Heidelberg, pp. 133–186
Shen R, Cai L, Meng D, Wu L, Guo K, Dong G, Liu L, Chen J, Wu Q, Chen G (2014) Benzene containing polyhydroxyalkanoates homo- and copolymers synthesized by genome edited Pseudomonas entomophila. Sci China Life Sci 57:4–10
Abraham GA, Gallardo A, San Roman J, Olivera ER, Jodra R, Garcia B, Minambres B, Garcia JL, Luengo JM (2001) Microbial synthesis of poly(beta-hydroxyalkanoates) bearing phenyl groups from Pseudomonas putida: chemical structure and characterization. Biomacromolecules 2:562–567
Slater S, Houmiel KL, Tran M, Mitsky TA, Taylor NB, Padgette SR, Gruys KJ (1998) Multiple beta-ketothiolases mediate poly(beta-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J Bacteriol 180:1979–1987
Wang HH, Li XT, Chen GQ (2009) Production and characterization of homopolymer polyhydroxyheptanoate (P3HHp) by a fadBA knockout mutant Pseudomonas putida KTOY06 derived from P. putida KT2442. Process Biochem 44:106–111
Chung AL, Jin HL, Huang LJ, Ye HM, Chen JC, Wu Q, Chen GQ (2011) Biosynthesis and characterization of poly(3-hydroxydodecanoate) by beta-oxidation inhibited mutant of Pseudomonas entomophila L48. Biomacromolecules 12:3559–3566
Jagoda A, Ketikidis P, Zinn M, Meier W, Kita-Tokarczyk K (2011) Interactions of biodegradable poly([R]-3-hydroxy-10-undecenoate) with 1,2-dioleoyl-sn-glycero-3-phosphocholine lipid: a monolayer study. Langmuir 27:10878–10885
Ouyang SP, Luo RC, Chen SS, Liu Q, Chung A, Wu Q, Chen GQ (2007) Production of polyhydroxyalkanoates with high 3-hydroxydodecanoate monomer content by fadB and fadA knockout mutant of Pseudomonas putida KT2442. Biomacromolecules 8:2504–2511
Lütke-Eversloh T, Bergander K, Luftmann H, Steinbüchel A (2001) Biosynthesis of poly(3-hydroxybutyrate-co-3-mercaptobutyrate) as a sulfur analogue to poly(3-hydroxybutyrate) (PHB). Biomacromolecules 2:1061–1065
Li SY, Dong CL, Wang SY, Ye HM, Chen GQ (2011) Microbial production of polyhydroxyalkanoate block copolymer by recombinant Pseudomonas putida. Appl Microbiol Biotechnol 90:659–669
Hu D, Chung AL, Wu LP, Zhang X, Wu Q, Chen JC, Chen GQ (2011) Biosynthesis and characterization of polyhydroxyalkanoate block copolymer P3HB-b-P4HB. Biomacromolecules 12:3166–3173
Tripathi L, Wu LP, Chen J, Chen GQ (2012) Synthesis of diblock copolymer poly-3-hydroxybutyrate-block-poly-3-hydroxyhexanoate [PHB-b-PHHx] by a beta-oxidation weakened Pseudomonas putida KT2442. Microb Cell Fact 11:44
Wang Q, Yang P, Xian M, Liu H, Cao Y, Yang Y, Zhao G (2013) Production of block copolymer poly(3-hydroxybutyrate)-block-poly(3-hydroxypropionate) with adjustable structure from an inexpensive carbon source. ACS Macro Lett 2:996–1000
Tripathi L, Wu LP, Meng D, Chen J, Chen GQ (2013) Biosynthesis and characterization of diblock copolymer of p(3-hydroxypropionate)-block-p(4-hydroxybutyrate) from recombinant Escherichia coli. Biomacromolecules 14:862–870
Tripathi L, Wu LP, Meng DC, Chen JC, Wu Q, Chen GQ (2013) Pseudomonas putida KT2442 as a platform for the biosynthesis of polyhydroxyalkanoates with adjustable monomer contents and compositions. Bioresour Technol 142:225–231
Hazer B (1994) Preparation of polystyrene poly(β-hydroxy nonanoate) graft copolymers. Polym Bull 33:431–438
Cakmakli B, Hazer B, Borcakli M (2001) Poly(styrene peroxide) and poly(methyl methacrylate peroxide) for grafting on unsaturated bacterial polyesters. Macromol Biosci 1:348–354
Zhang J, Kasuya K, Takemura A, Isogai A, Iwata T (2013) Properties and enzymatic degradation of poly(acrylic acid) grafted polyhydroxyalkanoate films by plasma-initiated polymerization. Polym Degrad Stab 98:1458–1464
Hu SG, Jou CH, Yang MC (2003) Antibacterial and biodegradable properties of polyhydroxyalkanoates grafted with chitosan and chitooligosaccharides via ozone treatment. J Appl Polym Sci 88:2797–2803
Yalpani M, Marchessault RH, Morin FG, Monasterios CJ (1991) Synthesis of poly(3-hydroxyalkanoate) (PHA) conjugates: PHA-carbohydrate and PHA-synthetic polymer conjugates. Macromolecules 24:6046–6049
Domenek S, Langlois V, Renard E (2007) Bacterial polyesters grafted with poly(ethylene glycol): behaviour in aqueous media. Polym Degrad Stab 92:1384–1392
Kim HW, Chung CW, Hwang SJ, Rhee YH (2005) Drug release from and hydrolytic degradation of a poly(ethylene glycol) grafted poly(3-hydroxyoctanoate). Int J Biol Macromol 36:84–89
Renard E, Tanguy PY, Samain E, Guerin P (2003) Synthesis of novel graft polyhydroxyalkanoates. Macromol Symp 197:11–18
Chung MG, Kim HW, Kim BR, Kim YB, Rhee YH (2012) Biocompatibility and antimicrobial activity of poly(3-hydroxyoctanoate) grafted with vinylimidazole. Int J Biol Macromol 50:310–316
Kim HW, Chung MG, Kim YB, Rhee YH (2008) Graft copolymerization of glycerol 1,3-diglycerolate diacrylate onto poly(3-hydroxyoctanoate) to improve physical properties and biocompatibility. Int J Biol Macromol 43:307–313
Le Fer G, Babinot J, Versace D-L, Langlois V, Renard E (2012) An efficient thiol-ene chemistry for the preparation of amphiphilic PHA-based graft copolymers. Macromol Rapid Commun 33:2041–2045
Ishida K, Hortensius R, Luo X, Mather PT (2012) Soft bacterial polyester-based shape memory nanocomposites featuring reconfigurable nanostructure. J Polym Sci B Polym Phys 50:387–393
Park J, Park JG, Choi WM, Ha CS, Cho WJ (1999) Synthesis and photo- and biodegradabilities of poly[(hydroxybutyrate-co-hydroxyvalerate)-g-phenyl vinyl ketone]. J Appl Polym Sci 74:1432–1439
Lee HS, Lee TY (1997) Graft polymerization of acrylamide onto poly(hydroxybutyrate-co-hydroxyvalerate) films. Polymer 38:4505–4511
Shen XW, Yang Y, Jian J, Wu Q, Chen GQ (2009) Production and characterization of homopolymer poly(3-hydroxyvalerate) (PHV) accumulated by wild type and recombinant Aeromonas hydrophila strain 4AK4. Bioresour Technol 100:4296–4299
Park SJ, Lee TW, Lim SC, Kim TW, Lee H, Kim MK, Lee SH, Song BK, Lee SY (2012) Biosynthesis of polyhydroxyalkanoates containing 2-hydroxybutyrate from unrelated carbon source by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol 93:273–283
Wang Q, Yang P, Liu CS, Xue YC, Xian M, Zhao G (2013) Biosynthesis of poly(3-hydroxypropionate) from glycerol by recombinant Escherichia coli. Bioresour Technol 131:548–551
Andreessen B, Steinbüchel A (2010) Biosynthesis and biodegradation of 3-hydroxypropionate-containing polyesters. Appl Environ Microbiol 76:4919–4925
Zhang L, Shi ZY, Wu Q, Chen GQ (2009) Microbial production of 4-hydroxybutyrate, poly-4-hydroxybutyrate, and poly(3-hydroxybutyrate-co-4-hydroxybutyrate) by recombinant microorganisms. Appl Microbiol Biotechnol 84:909–916
Hein S, Sohling B, Gottschalk G, Steinbuchel A (1997) Biosynthesis of poly(4-hydroxybutyric acid) by recombinant strains of Escherichia coli. FEMS Microbiol Lett 153:411–418
Sohling B, Gottschalk G (1996) Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J Bacteriol 178:871–880
Alber BE, Fuchs G (2002) Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J Biol Chem 277:12137–12143
Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, dos Santos VAPM, Fouts DE, Gill SR, Pop M, Holmes M, Brinkac L, Beanan M, DeBoy RT, Daugherty S, Kolonay J, Madupu R, Nelson W, White O, Peterson J, Khouri H, Hance I, Lee PC, Holtzapple E, Scanlan D, Tran K, Moazzez A, Utterback T, Rizzo M, Lee K, Kosack D, Moestl D, Wedler H, Lauber J, Stjepandic D, Hoheisel J, Straetz M, Heim S, Kiewitz C, Eisen J, Timmis KN, Dusterhoft A, Tummler B, Fraser CM (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4:799–808
Celinska E (2010) Debottlenecking the 1,3-propanediol pathway by metabolic engineering. Biotechnol Adv 28:519–530
Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R, Estadilla J, Teisan S, Schreyer HB, Andrae S, Yang TH, Lee SY, Burk MJ, Van Dien S (2011) Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol 7:445–452
Alber B, Olinger M, Rieder A, Kockelkorn D, Jobst B, Hugler M, Fuchs G (2006) Malonyl-coenzyme A reductase in the modified 3-hydroxypropionate cycle for autotrophic carbon fixation in archaeal Metallosphaera and Sulfolobus spp. J Bacteriol 188:8551–8559
Wang Q, Liu CS, Xian M, Zhang YG, Zhao G (2012) Biosynthetic pathway for poly(3-hydroxypropionate) in recombinant Escherichia coli. J Microbiol 50:693–697
Albertyn J, Vantonder A, Prior BA (1992) Purification and characterization of glycerol-3-phosphate dehydrogenase of Saccharomyces cerevisiae. FEBS Lett 308:130–132
Norbeck J, Pahlman AK, Akhtar N, Blomberg A, Adler L (1996) Purification and characterization of two isoenzymes of DL-glycerol-3-phosphatase from Saccharomyces cerevisiae – identification of the corresponding GPP1 and GPP2 genes and evidence for osmotic regulation of Gpp2p expression by the osmosensing mitogen-activated protein kinase signal transduction pathway. J Biol Chem 271:13875–13881
Forage RG, Foster MA (1982) Glycerol fermentation in Klebsiella pneumoniae – functions of the coenzyme-B12-dependent glycerol and diol dehydratases. J Bacteriol 149:413–419
Liang QF, Zhang HJ, Li SN, Qi QS (2011) Construction of stress-induced metabolic pathway from glucose to 1,3-propanediol in Escherichia coli. Appl Microbiol Biotechnol 89:57–62
Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC (1999) The propanediol utilization (pdu) operon of Salmonella enterica serovar typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B-12-dependent 1,2-propanediol degradation. J Bacteriol 181:5967–5975
Leal NA, Havemann GD, Bobik TA (2003) PduP is a coenzyme-a-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch Microbiol 180:353–361
Hiramitsu M, Doi Y (1993) Microbial synthesis and characterization of poly(3-hydroxybutyrate-co-3-hydroxypropionate). Polymer 34:4782–4786
Salles IM, Forchhammer N, Croux C, Girbal L, Soucaille P (2007) Evolution of a Saccharomyces cerevisiae metabolic pathway in Escherichia coli. Metab Eng 9:152–159
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183
Lv L, Ren YL, Chen JC, Wu Q, Chen GQ (2015) Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: controllable P(3HB-co-4HB) biosynthesis. Metab Eng 29:160–168
Jiang XR, Wang H, Shen R, Chen GQ (2015) Engineering the bacterial shapes for enhanced inclusion bodies accumulation. Metab Eng 29:227–237
van den Ent F, Amos LA, Lowe J (2001) Prokaryotic origin of the actin cytoskeleton. Nature 413:39–44
Acknowledgements
This research was financially supported by 973 Basic Research Fund (Grant No. 2012CB725201), National High Tech 863 Grants (Grant No. 2012AA023102), and a Grant from National Natural Science Foundation of China (Grant No. 31270146).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Meng, DC., Chen, GQ. (2017). Synthetic Biology of Polyhydroxyalkanoates (PHA). In: Zhao, H., Zeng, AP. (eds) Synthetic Biology – Metabolic Engineering. Advances in Biochemical Engineering/Biotechnology, vol 162. Springer, Cham. https://doi.org/10.1007/10_2017_3
Download citation
DOI: https://doi.org/10.1007/10_2017_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-55317-7
Online ISBN: 978-3-319-55318-4
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)