Abstract
Tumor aggressiveness is encouraged by hypoxia, which also lowers patient survival. Numerous individuals with hypoxic tumors had poor outcomes, which shows that additional factors may affect how the tumors react to hypoxia. Hypoxia is frequently found in solid tumors and is known to influence aggressive tumor activity, chemotherapy resistance, and radiation resistance, all of which lead to a bad prognosis for the cancer patient. PIWI-interacting RNAs (piRNAs) control how tumor cells react to hypoxia, but little is known about how piRNAs function in hypoxic tumors. PiRNAs are brand-new, tiny, noncoding RNA molecules with 24 and 31 nucleotides length. They frequently interact with proteins from the PIWI protein family to regulate the epigenetic regulation of gene expression, which is essential for understanding cancer genetics. Numerous studies have demonstrated that abnormal piRNA expression is a hallmark of various tumor forms, although their precise tumorigenic roles are still unknown. Patients' cancer type-specific piRNA signatures differ from one another. The malignancy renal cell carcinoma, defined by constitutive activation of hypoxia-related signaling brought on by a common mutation or deletion of the von Hippel–Lindau factor, is found to have highly uniform piRNA profiles across patients (VHL). According to prior reports, piRNAs and PIWI proteins may be crucial in cancer development, prognosis, and management. However, it has yet to be determined how these compounds might be relevant in therapeutic settings. The utilization of piRNAs and PIWI proteins as cancer treatments and diagnostic and prognostic biomarkers is also considered prospective options. In this chapter, we cover recent research on the biogenetic mechanisms, roles, and emerging roles of piRNAs in hypoxia cancer, offering fresh perspectives on the possible uses of piRNAs and PIWI proteins in the detection and clinical management of hypoxic cancer.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
8.1 Introduction
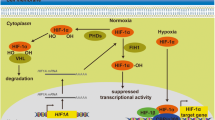
Several solid tumors form areas that are hypoxic or poorly oxygenated. Hypoxia-inducible factor-1 (HIF-1) and HIF-2, heterodimeric transcription factors, activate more than 90 genes involved in anaerobic glycolysis, pH control, angiogenesis, cellular migration, and metastasis (Giatromanolaki et al. 2001; Forsythe et al. 1996). There are several ways to identify hypoxic tumors, and patients with hypoxic tumors typically have worse outcomes than patients with normoxic tumors (Wilson and Hay 2011; Simi et al. 2006; Hung et al. 2009). Patients with hypoxic tumors usually have an aggressive tumor phenotype and decreased patient survival. In addition, hypoxic tumor cells increase the risk of metastatic disease and reduce the effectiveness of radiation therapy and most forms of chemotherapy. However, there is a range of poor outcomes in patients with hypoxic tumors, and identifying those patients who would have the worst effect is a significant unmet need in the field of oncology. The impact of hypoxia on RNA expression is extensive. PiRNAs are a subclass of small noncoding RNAs (sncRNAs) that have recently been realized to be important in cancer biology. However, their effects on other small noncoding RNAs (sncRNAs), such as PIWI-interacting RNAs (piRNAs), are unknown. In humans, more than 30,000 piRNAs have been identified (Ostheimer et al. 2014; Ku and Lin 2014). Their ability to direct related chromatin-silencing machinery to transposon-encoding DNA regions in the genome (Huang et al. 2013a) best describes them.
A subset of piRNAs may also be able to control protein-coding genes through DNA methylation, which, if the regulatory targets are cancer-relevant, may impact the development of cancer (Watanabe et al. 2011; Jacobs et al. 2016). Humans have four different PIWI protein isoforms, including PIWIL1 (HIWI), PIWIL2 (HILI), PIWIL3, and PIWIL4 (HIWI2); rodents have three different isoforms, including PIWIL1 (MIWI) PIWIL2 (MILI), and PIWIL4 (MIWI2); and Drosophila has three different isoforms, including PIWI, Aub, and PiRNAs that are essential in controlling gene expression and acting as transposon silencers (Dharap et al. 2011; Yan et al. 2011; Rizzo et al. 2014; Liu et al. 2018). piRNA expressions that are abnormally high have been found in various illnesses, particularly neoplasms. PiRNAs are intriguing new therapeutic targets for onco-medicine and promising biomarkers for early diagnosis. Germline cells have high levels of piRNA expression, which controls genomic stability by identifying DNA target sequences and aids in the recruitment of the required machinery to cause transposable element epigenetic silencing (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006).
Recent data suggest that they are expressed, functionally active, and connected to epigenetic pathways of cancer development in somatic tissue (Esteller 2008; Rouget et al. 2010; Esteller 2011; Fu et al. 2014; Ha et al. 2014a; Barckmann et al. 2015; Gebert et al. 2015; Moyano and Stefani 2015; Ng et al. 2016a). Nevertheless, little is known about piRNA expression mechanisms in somatic cells or solid malignancies. Patients with renal cell carcinoma (RCC) have indicated that the solid tumor microenvironment may have a role in controlling piRNA expression and perhaps contributing to the diverse piRNA expression seen between patients, according to Martinez and his colleagues (2015) (Martinez et al. 2015a). Since von Hippel–Lindau factor (VHL) loss-of-function mutations frequently occur in RCC tumors, constitutive, oxygen-independent stabilization of HIF-1(Maxwell et al. 1999) and HIF-mediated upregulation of hypoxia-associated gene products, Maxwell and his team (1999) discovered that piRNA expression was remarkably consistent between RCC tumors.
8.2 Biogenesis of piRNAs and Generation of Mature piRNAs
Genetic areas called piRNA clusters, which can be classified as uni-strand or dual-strand clusters, where a significant majority of piRNA precursors are created. Dual-strand clusters produce precursors that map to both genomic strands, unlike uni-strand collections, which make precursors that only map to one strand. Additionally, some piRNA precursors may be originated from individual transposons or the 30 UTR of protein-coding genes (Robine et al. 2009; Saito et al. 2009). Uni-strand cluster transcription is comparable to canonical mRNA transcription. The transcription-associated histone 3 lysine 4 demethylation (H3K4me2) mark is present at promoters in uni-strand clusters. Additionally, piRNA precursors are 30 terminated, and 50 methyl-guanosine capped (Mohn et al. 2014; Lim and Kai 2015). On the other hand, dual-strand clusters lack distinct RNA polymerase II (RNA Pol II) promoter signatures, such as H3K4me3 and RNA Pol II peaks, and non-polyadenylated piRNA precursors are generated (Le Thomas et al. 2013; Chen et al. 2016).
Several proteins, including RNA Pol II, the Rhino-Cutoff-Deadlock complex (RDC complex), Moonshiner, TATA-box binding protein-related factor (TRF2), and the three prime repair exonuclease, are involved in the transcription of dual-strand clusters into piRNA precursors (TREX). piRNA precursors are moved out of the nucleus after transcription is complete. The RNA helicase Armitage first resolves the secondary structures (Armi). After deserialization, the mitochondria-associated endonuclease Zucchini cleaves piRNA precursors to produce pre-piRNAs with a 50 monophosphate (Zuc). Then, a 30–50 exonuclease called Nibbler trims (Nbr) the pre-piRNAs at the 30 ends after loading them onto PIWI proteins. Primary piRNAs are created through a process known as primary piRNA biogenesis, known as primary piRNAs. The production of piRNAs is boosted by the participation of Ago3 and Aub proteins and is primed by initial piRNAs.
8.3 piRNAs: Novel Functions in Cancer Expression and Selectively Deregulation by Hypoxic Tumors
Martinez and his group (2015) used our previously published bespoke small RNA sequencing analysis pipeline (Martinez et al. 2015a) to assess the expression levels of 23,440 human piRNAs on a per-tumor basis. PiRNAs were only included in analyses if they had a median expression of 10 (reads per kilobase million) RPKM in at least one of the groups (hypoxic and/or normoxic) and a minimum of twofold change in median RPKM expression values. It was done to identify the most significant changes in piRNA expression between hypoxic and nonhypoxic groups. Comparisons can be made using the nonparametric Mann–Whitney U test, with the Benjamini–Hochberg technique being used to compensate for multiple testing's false discovery rates (Lee and Lee 2018).
8.4 piRNAs Maintain Genomic Integrity by Silencing Transposable Elements
Transposable elements (TEs) can cause genetic variety and instability, making them prime candidates for increased cancer-causing potential in humans (Cordaux and Batzer 2009; Chenais 1835; Suntsova et al. 2015). The establishment of a repressive chromatin state can be caused by the position of the PIWI-piRNA complex in the nucleus. It has been discovered that the ectopic expression of piRNAs increases heterochromatin-1 (HP1) and H3K9me3, which, when bound, results in a heterochromatin state (Suntsova et al. 2015). Restoring tumor suppressor p53 and associated piRNAs may be a promising approach for treating cancer (Levine et al. 2016) in light of the carcinogenic impact of LINE-1 and other repetitive elements.
8.5 piRNAs Contribute to Tumorigenesis Through Regulation of DNA Methylation
Another epigenetic process with a functional connection to piRNAs is DNA methylation. Recent research suggests that piRNAs and associated PIWI proteins can encourage retrotransposons' de novo DNA methylation(Aravin et al. 2007; Kuramochi-Miyagawa et al. 2008; Kojima-Kita et al. 2016). Additionally, piRNAs can control DNA methylation on non-transposon loci as well as in the context of transposons. It is noteworthy that the mutation rs1326306 G > T related to piR-021285 discovered by Fu, and his colleagues (2015) was found to be substantially connected with breast cancer (Fu et al. 2015). They discovered methylation variations in several genes involved in cancer development by comparing the genome-wide methylation profiles in MCF7 cells transfected with either the wild type or mutant piR-021285. The mechanisms behind its oncogenic action also revealed that the suppression of piR-823 resulted in a notable decrease in the production of DNMT3A and DNMT3B, which decreased global DNA methylation and caused the methylationsilencedp16INK4A tumor suppressor gene to become active once again.
8.6 Post-Transcriptional Regulation of Gene Expression by piRNAs
DNA methylation and chromatin silencing are just a few of the things piRNAs can do. Numerous studies have shown that piRNAs can suppress the expression of their target genes like miRNAs (Ha et al. 2014b; Martinez et al. 2015b; Zhang et al. 2015). In contrast to CDSs and 5′-UTRs, expressed piRNAs have been concentrated in the human testis' 3′-untranslated regions (UTRs). The mRNAs targeted by piRNA in the case of MIWI mutant mice were overexpressed due to their inability to be cleaved in the absence of MIWI. Indeed, several studies have shown that piRNAs can work as a surveillance system to destroy retrotransposon-associated mRNAs to avoid the ubiquitous dissemination of transposon sequences (Watanabe et al. 2006).
8.7 piRNAs Have Tumorigenic or Suppressive Roles in Cancer Development
Research shows that piRNAs have a role in cancer development as either tumor suppressors or oncogenes. Additionally, they might aid in the growth and spread of cancer cells. For instance, in patients with non-small cell lung carcinoma, the upregulation of piR-651 was discovered to be related to cancer progression (NSCLC). Notably, piR-651 overexpression dramatically increased tumor development and metastasis in the A549 lung cancer cell line. Additionally, piR-651 overexpression causes cell cycle arrest by upregulating the production of CDK4 and cyclin D1. It implies that piR-651 may function as an oncogene in this cancer (Li et al. 2016; Yao et al. 2016). Another well-known oncogenic piRNA, PiR-Hep1, aids in the invasion, migration, and proliferation of liver cancer cells (Law et al. 2013).
8.8 piRNAs in the Maintenance of Cancer Stemness and Chemoresistance
The dogma in the piRNA field is being challenged by a growing body of recent studies that show the PIWI pathway is also connected to the preservation of cancer stemness. Zhang and his colleagues (2013) extracted CD44 (+)/CD24 (-) tumor cells (cancer stem cells [CSC]) from 1086 clinical specimens for breast cancer and found that these cells expressed more PIWIL2 and piR-932 than control cells did (Zhang et al. 2013). Furthermore, the authors hypothesized that by encouraging abnormal methylation of the Latxin gene the piR-932 PIWIL2 complex might support the persistence of breast cancer stem cells. One of the defining traits of cancer stem cells is chemoresistance. Greater tumorigenicity, tumor sphere formation, and increased chemoresistance in vivo were all caused by the overexpression of HIWI (PIWI proteins in humans) in cervical cancer cells, together with the activation of many stem-associated genes (Liu et al. 2014a). Targeting the PIWI pathway may be a promising technique for chemotherapy and other clinical applications of cancer since PIWI proteins and their associated piRNAs play a vital role in maintaining the stem cell characteristics of cancer cells (Table 8.1).
8.9 The Role of piRNAs in Hypoxic Cancer
In human malignancies, the roles of PIWI proteins and piRNAs have begun to be revealed (Liu 2016). There is mounting evidence that many tumor cells express PIWI proteins in mice and humans, including PIWIL2-like proteins, HIWI, and PIWIL2 (Esteller 2011; Siddiqi and Matushansky 2012). Additionally, piRNAs were found in these cells (Esteller 2011). Human cancers such as gastric, bladder, breast, colorectal, and lung cancer have been documented to express piRNAs aberrantly. These data suggest that the piRNA pathway may be involved in the emergence of cancer. Although the possible involvement of piRNAs in cancer is still being explored, little is known about the functional role that particular piRNAs play in human cancer. These findings underline how critical it is to comprehend the precise function of the piRNA pathway during carcinogenesis and present brand-new therapeutic options. Table 8.2 and Fig. 8.1 show piRNA expression concerning the various cancer types.
8.10 Gastric Cancer
Gastric cancer is the second-largest contributor to cancer-related fatalities worldwide (Pan et al. 2013; Shomali et al. 2017) and the fifth most common cancer worldwide (Torre et al. 2012; Jiang et al. 2017). There was no correlation between the expression levels of piR-823 and its clinical–pathological characteristics in gastric cancer tissue (Cheng et al. 2012a). Due to piR-651's overexpression as an oncogene in gastric cancer, the TNM (tumor node metastasis) stage showed a positive connection. Additionally, a piR-651 inhibitor could reduce cell development in the G2/phase, proving that piRNAs are essential for carcinogenesis (Cheng et al. 2011). Additionally, it was noted that piR-651 and piR-823 had reduced levels of CTCs (circulating tumor cells) in the peripheral blood of patients with gastric cancer compared to normal controls (Cui et al. 2011).
However, piR-823 and piR-651 were more sensitive than commonly used biomarkers for gastric cancer, such as CA19-9 (carbohydrate antigen 19-9) and CEA (serum carcinoembryonic antigen). It is because piRNAs are short fragments that are less likely to be degraded, and levels of piR-651 and piR-823 in blood samples are generally stable. Additionally, these piRNAs can pass through cell membranes and are, therefore, easily detected. These results imply that piRNAs may represent novel therapeutic targets for treating gastric cancer (Li et al. 2014).
8.11 Bladder Cancer
The most prevalent malignancy of the urinary system and the ninth most common cancer overall are bladder cancers (Park et al. 2014; Ploeg et al. 2009). The researchers profiled three pairs of bladder cancer samples and their surrounding normal tissues using the ArrayAtarHG19 piRNA array, which is for human piRNAs. They discovered that the crucial piRNA piRABC (also known as DQ594040) was downregulated in bladder cancer (Chu et al. 2015). PiRABC demonstrated extremely high differential expression levels between healthy tissues and bladder cancer. Tumor Necrosis Factor Superfamily Member 4 (TNFSF4) and piRABC may interact, leading researchers to speculate that piRABC may encourage bladder cancer cell death by upregulating TNFSF4 (Pardini and Naccarati 2018).
8.12 Breast Cancer
Deep sequencing was done on four matched nontumor tissues and four breast cancer tissues to identify differentially expressed piRNAs. Four piRNAs (piR-20365, piR-20582, piR-20485, and piR-4987) were later upregulated in 50 breast cancer samples by RT-PCR. Patients' clinical pathology characteristics were noted, including lymph node status, tumor size, estrogen receptor (ER) status, and Her2 status. Additionally, lymph node metastasis correlated favorably with piR-4987 upregulation (Huang et al. 2013b).
A study demonstrated that the piR-932/PIWIL2 complex might favorably influence the process of breast cancer stem cells, which promotes EMT, by encouraging the methylation of Latex (epithelial–mesenchymal transition). PIWIL2 and piR-932 have been proposed as potential targets for preventing the spread of breast cancer (Zhang et al. 2013). Similar findings were made by another study, which discovered that piR-021285 plays a role in methylation at several known breast cancer-related genes, specifically attenuated 5′ untranslated regions (UTR)/first exon methylation at the pro-invasive ARHGAP11A gene and invasiveness in an in vitro cell line model (Han et al. 2017).
8.13 Lung Cancer
Lung cancer is the most common cause of cancer-related death worldwide and is broadly classified as non-small cell lung cancer (about 85% of cases) and small cell lung cancer (about 15% of cases) (Blandin Knight et al. 2017). Furthermore, they demonstrated that piR-L-163 could regulate phosphorylated ERM and play a crucial part in protein activation by directly binding to and doing so (Mei et al. 2015). Targeting piR-L-138 could be a possible technique to overcome chemoresistance in patients with lung squamous cell carcinoma (LSCC) as the researchers discovered that it was elevated by cisplatin (CDDP)-based chemotherapy both in vivo and in vitro (McClatchey and Fehon 2009). The top-downregulated piR-L-163 in NSCLC cells prevented cell migration and invasion by maintaining the activity of phosphorylated ezrin-radixin-moesin (p-ERM), which links transmembrane proteins like EBP50 and filamentous actin (F-actin). In a different investigation, piR-55490 was discovered to inhibit lung cancer cell proliferation by interacting with the 30 UTR of the mTOR mRNA (Peng et al. 2016). In NSCLC A549 and HCC827 cell lines, elevated piR-651 increased cell proliferation, migration, and invasion while suppressing cell death (Zhang et al. 2018). However, further research is required to understand its mechanism. Additionally, compared to primary cancer cells, metastatic lung adenocarcinoma cells drastically decreased the expression of piR-57125 (Daugaard et al. 2017).
8.14 Liver Cancer
According to predictions, liver cancer will rank as the sixth most frequently diagnosed cancer worldwide and the fourth most prevalent cause of cancer mortality. According to research by Law et al., piR-Hep1 is elevated in nearly half of HCC tumors (46.6%) compared to the neighboring nontumor liver. Cirrhotic nodules (CNs), low-grade dysplastic nodules (LGDNs), high-grade dysplastic nodules (HGDNs), early hepatocellular carcinoma (eHCC), and advanced HCC (pHCC) are the different stages of the development of liver cancer (Ng et al. 2016b). Numerous piRNAs served as markers for each stage.
PiR-LLi-24894 was only expressed in CNs, while piR-LLi-30552, hsa-piR-020498, and hsa-piR-013306 were largely expressed in HGDNs, eHCCs, and pHCCs, and were exclusively accumulated in HCC. PiR-823 also encouraged the formation of extracellular matrix components such as collagen type I alpha 1 and a-smooth muscle actin (a-SMA), leading to cirrhosis (Tang et al. 2018). By triggering the signal transducer and activator of transcription 3 (Stat3)/Bcl-xL signaling pathway, piR-Hep1 may have a role in reducing cell death(Law et al. 2013).
8.15 Stomach Cancer
The third most common cancer-related cause of death worldwide and the fifth most frequently diagnosed malignancy (Bray et al. 2018), gastric cancer is still a severe disease. PiR-823 was found to be markedly downregulated in gastric cancer tissues. PiR-823 mimics reduced cell proliferation, and a xenograft nude mice model demonstrated that piR-823 prevented tumor growth (Cheng et al. 2012b). Additionally, peripheral blood showed decreased expression of piR-651 and piR-823 in patients with gastric cancer. There have been reports of an increase in piR-32105, piR-58099, and piR-59056 in gastric cancer tissues (Cui et al. 2011). It is commonly recognized that surgery or endoscopic treatment can cure stomach cancer in its early stages, but the prognosis for advanced gastric cancer is uncertain. While immunotherapy, like anti-PD-1/PD-L1 therapy, is a potential treatment for cancer, patients with advanced gastric cancer are not exceptionally responsive to immunotherapy alone or when paired with chemotherapy in first-line treatment.
8.16 Colorectal Cancer
The second most frequent disease in women and men worldwide is colorectal cancer (Law et al. 2013; Tang et al. 2018). According to several experts, PIWI has been linked to the onset of colorectal cancer (Weng et al. 2018; Mai et al. 2018a). According to Weng and his team's (2018) hypothesis, piRNA-823 was one of the piRNAs that aided in developing colorectal cancer. These genes included the activating transcription factor 3 (ATF3), the BTG anti-proliferation factor 3 (BTG1), the dual specificity phosphatase 1 (DUSP1), the fas cell surface death receptor (FAS), the nuclear factor kB (NF-kB) inhibitor an (NFkBIa), the uridine phosphorylase 1 (UPP1), the sestrin 2 (SESN2), and the tumor protein p53 inducible nuclear Additionally, piR-54265 hampered treatment, and individuals with more serum piR-54265 levels responded to chemotherapy noticeably less (Mai et al. 2018a). By encouraging its phosphorylation at Ser326, piR-823 has been shown to stimulate the production of heat shock transcription factor 1 (HSF1) and promote colorectal carcinogenesis (Yin et al. 2017). Additionally downregulated in CRC patients were piR-5937, piR-001311, piR-004153, piR-017723, piR-017724, piR-020365, piR-28876, piR-32105, piR-58099, and piR-59056 (Vychytilova-Faltejskova et al. 2018; Qu et al. 2019; Ng et al. 2016c).
8.17 PIWIs May Be Used for Cancer Diagnosis and Prognosis
Most of the existing research on PIWIs and carcinogenesis and cancer progression is derived from reports of clinical–pathological findings. PIWIs may be employed as biomarkers for clinical diagnosis and prognosis relating to poor outcomes, according to these studies. However, examining all four human PIWI homologs, most of this work concentrated on PIWIL1 and PIWIL2. According to research, HIWI expression gradually rises in normal gastric tissues, atrophic gastritis, intestinal metaplasia, and gastric malignancies, indicating that HIWI may have a role in the emergence of gastric cancer (Liu et al. 2006). HIWI was demonstrated to promote chemoresistance in cervical cancer and was proposed as a cancer stem cell marker (Liu et al. 2014b). Elevated HIWI mRNA and protein levels in pancreatic ductal adenocarcinoma have no overall effect on patient survival.
However, patients who had aberrant HIWI mRNA expression had a substantially higher probability of dying from a malignancy (Grochola et al. 2008). HIWI expression significantly increased with the progression of tumor grades in gliomas, demonstrating the relationship between higher positive HIWI and worse outcomes (Sun et al. 2010). In contrast, PIWIL1 has not been linked to the clinical– pathological characteristics of endometrioid cancer (Liu et al. 2010). Human testicular seminomas, prostate cancer, breast cancer, gastrointestinal cancer, ovarian cancer, and endometrial cancer were all shown to have increased PIWIl2 expression, as were mouse breast cancer, rhabdomyosarcoma, and medulloblastoma (Lee et al. 2010). It is interesting to note that human testicular non-seminoma cancers did not exhibit elevated PIWIl2 expression, and the equivalent PIWIL2 isoform is PL2L60A, not PL2L80A (Gainetdinov et al. 2014). PIWIl2 was primarily expressed in cancer stem cells, 81% of in situ carcinomas, and 90% of invasive carcinomas in breast cancer (Lee et al. 2010). Additionally, he and his coworkers (2010) noted that PIWIl2 was expressed at different stages of cervical cancer (He et al. 2010). EIF2C1 and PIWIL2 may serve as potential colon cancer biomarkers with early diagnostic value, according to another study that found that their elevated expressions were substantially related to the disease (Li et al. 2010). Curiously found that overexpression of PIWIl2 caused cisplatin resistance in human ovarian cancer cell lines, indicating that PIWIL2 was a marker for cisplatin resistance in cancer chemotherapy (Wang et al. 2011).
Clinical patients with breast cancer revealed significant expression levels of PIWIL2 and PIWIL4, but not PIWIL1 or PIWIL3 (Hashim et al. 2014). Elevated PIWIL1 and PIWIL2 expressions were associated with worse overall survival, and PIWIL1 was suggested to be an independent prognostic factor (Wang et al. 2012). In gastric cancer, the expression of PIWIL1-4 was significantly correlated with the T-stage, lymph node metastasis, and clinical TNM (cTNM). Stage III epithelial ovarian cancer patients' primary and metastatic tumors showed significantly increased PIWIL1-4 expression (Chen et al. 2014). Some piRNAs have been linked to the characteristics of malignancies so far, including human piR-Hep1, piR-823, piR-651, piR-4987, piR-20365, piR20485, piR-20582, and piRABC (Tan et al. 2015). By controlling de novo DNA methylation and angiogenesis in multiple myeloma, piR-823 was discovered to support carcinogenesis (Yan et al. 2015). By enhancing the methylation process of Lactein in breast cancer stem cells, it has been proposed that piR-932 binds PIWIL2 (Zhang et al. 2013). However, more research is needed to understand how piRNAs in association with PIWI proteins contribute to carcinogenesis, invasion, and metastasis.
8.18 piRNAs as Biomarkers in Cancer Potential Clinical Applications of piRNAs as Cancer Biomarkers
Determining the various expression patterns of piRNAs that are particular to different cancer types might enable the development of new cancer biomarkers, given the significance of piRNAs and their associated proteins in many cellular processes. The prognosis for cancer is improved by early detection and treatment. RNA sequencing has shown that human blood contains piRNAs in addition to miRNAs and other types of noncoding RNAs (Huang et al. 2013c; Freedman et al. 2016). Since piRNAs are similar in length to miRNAs, they can easily pass through cell membranes and enter the bloodstream (Mei et al. 2013). Additionally, they are exceedingly stable and resistant to destruction by ribonucleases in bodily fluids (Mitchell et al. 2008). Therefore, the piRNAs found in circulating tumor cells (CTCs) are intriguing novel complimentary tumor indicators for cancer.
It was observed that GC patients have lower levels of piR-651 and piR-823 in their peripheral blood than healthy controls. Liu and his team (2019) found that piR-651 and piR-823 are more sensitive compared with the favorable detection rates of serum carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) levels, suggesting that these piRNAs are more sensitive for gastric cancer screening than the frequently used biomarkers (Liu et al. 2019). Although piR-5937 and piR-28876 had an excellent diagnostic value even for individuals in clinical stage I, their expression in the serum of CRC patients dropped dramatically with the advanced clinical stage. Serum piR-54265 levels also serve as a therapeutic measure that predicts how well chemotherapy treats CRC patients. Patients with low serum piR-54265 levels exhibited a more excellent response to chemotherapy than those with high levels (Mai et al. 2018b). Compared to serum from healthy persons at diagnosis, piR-651, which is downregulated, originates from circulating rather than tumor cells. The patients' downregulation may result from variations in the peripheral blood populations linked to the existence of lymphoma (Cordeiro et al. 2016). Numerous studies have identified a piRNA signature in clear cell renal carcinoma (ccRCC) that may act as a prognostic indicator. Cordeiro et al. (2016) found three piRNAs (piR-30,924, piR-57,125, and piR-38,756) that were substantially linked with tumor recurrence and overall survival by employing piRNA microarray and subsequently validating candidate piRNAs in a larger cohort (Cordeiro et al. 2016).
8.19 New Therapeutic Approaches Using piRNAs
It is an exciting application to use synthesized piRNAs as weapons to inhibit the manufacture of cancer-related proteins by attaching to mRNAs since it may avoid the need for processing by enzymes like Dicer. In addition to being used as a post-translational strategy in combinatory therapy for various hypoxia cancers, PIWI antibodies may have a therapeutic effect on the proliferation of hypoxic cancer. Because the suspected regulators of mRNA PIWI expression, at least in the form of piRNAs, are already known, piRNA sequences should be employed for post-transcriptional silencing. At first impression, it could be preferable to prevent the development of a dangerous component rather than combat the undesirable consequences of a molecule already in operation. It is feasible to use specialized synthetic piRNAs designed to bind to PIWI proteins and exert genomic silence on PIWI genes at a transcriptional level, which differs from miRNAs and piRNA post-transcriptional mRNA inhibition. In a reverse way, this tactic is comparable to the "ping-pong" method of piRNA synthesis (Ross et al. 2014). For example, PIWI antibodies could be used to deliver drugs to hypoxic cancer cells, a delivery strategy already used with other antibodies. It would reduce the side effects of conventional cytotoxic medicines and perhaps improve the response to hypoxic cancer therapy.
8.20 Database for piRNAs and Functional Predictions
Lakshmi and his colleagues first created the piRNABank as a website resource on classified and clustered piRNAs in 2008 (Sai Lakshmi and Agrawal 2008). This database details the piRNAs found in humans, mouse, rats, and drosophila. The website (http://pirnabank.ibab.ac.in/) gathers all potential piRNA clusters and displays piRNAs and their related genomic elements, such as genes and repeat regions, on a genome-wide map. Chinese researchers Wang et al. and Zhang et al. have developed a more potent tool for piRNA functional studies called piRNA database-piRBase (Wang et al. 2018; Zhang et al. 2014). This database combined 264 datasets from 21 different taxa, and more than 173 million piRNAs were gathered. Additionally, it includes possible data on piRNA targets and piRNAs linked to diseases. Additionally, documented piRNA targets and epigenetic data are gathered. As a result, these databases (http://www.regulatoryrna.org/database/piRNA/) incorporate post-transcriptional regulation and epigenetic control data to facilitate the functional study of piRNA.
8.21 Future Directions in piRNA Research in Hypoxic Oncology
According to the studies we evaluated, PIWIs may be crucial in regulating the development of some cancer hallmarks in hypoxic cancer cells. Various pieces of evidence proved the prognostic and therapeutic potential of PIWIs. It is necessary to conduct additional research to explain this discrepancy. Further research is needed to determine whether PIWI scan is an essential marker for clinical diagnosis and prognosis because there is currently little clinical data available. There is undoubtedly a need for more information regarding the specific molecular mechanisms through which PIWIs contribute to carcinogenesis and the growth of hypoxic tumors. PiRNAs may be used as possible tumor biomarkers, a hot area of research, as several recent studies have revealed that several piRNAs are significantly expressed in blood samples. Additionally, several piRNAs' expression was linked to pathogenic factors or clinical outcomes. Regarding potential limitations, most investigations involved retrospective clinical material, indicating that prospective, multicenter clinical trials should confirm these findings. There is still much to learn about the roles of piRNAs and the proteins that interact with them in cancer, and this subject could provide a wealth of helpful information. Future usage of thorough and high-throughput methods could uncover more useful piRNA biomarkers and their interacting proteins unique to various types or stages of cancer. piRNAs and the proteins interacting with them will draw a lot of attention if their functions and mechanisms are investigated since they would be excellent biomarkers for diagnosis and novel targets for skillful therapeutic modulation.
References
Aravin A et al (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442:203–207. https://doi.org/10.1038/nature04916
Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316:744–747
Barckmann B et al (2015) AubergineiCLIP Reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep. https://doi.org/10.1016/j.celrep.2015.07.030
Blandin Knight S, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C (2017) Progress and prospects of early detection in lung cancer. Open Biol 7(9):170070
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424
Chalbatani GM, Dana H, Memari F, Gharagozlou E, Ashjaei S, Kheirandish P, Marmari V, Mahmoudzadeh H, Mozayani F, Maleki AR, Sadeghian E, Nia EZ, Miri SR, Nia NZ, Rezaeian O, Eskandary A, Razavi N, Shirkhoda M, Rouzbahani FN (2018) Biological function and molecular mechanism of piRNA in cancer. Pract Lab Med 13:e00113. https://doi.org/10.1016/j.plabm.2018.e00113. Erratum in: Pract Lab Med. 2020 Dec 02;22:e00194. PMID: 30705933; PMCID: PMC6349561
Chen C, Liu J, Xu G (2014) Overexpression of PIWI proteins in human stage III epithelial ovarian cancer with lymph node metastasis. Cancer Biomark 13:315–321
Chen YA, Stuwe E, Luo Y, Ninova M, Le Thomas A, Rozhavskaya E, Li S, Vempati S, Laver JD, Patel DJ et al (2016) Cutoff suppresses RNA polymerase II termination to ensure expression of piRNA precursors. Mol Cell 63:97–109
Chenais B (1835) Transposable elements and human cancer: a causal relationship? Biochim Biophys Acta 2013:28–35
Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J (2012a) piR-823, a novel noncoding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett 315:12–17
Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J (2012b) piR823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett 315:12–17
Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN (2011) piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta 412:1621–1625
Chu H, Hui G, Yuan L, Shi D, Wang Y et al (2015) Identification of novel piRNAs in bladder cancer. Cancer Lett 356:561–567
Cordaux R, Batzer MA (2009) The impact of retrotransposons on human genome evolution. Nat Rev Genet 10:691–703
Cordeiro A, Navarro A, Gaya A, Diaz-Beya M, Gonzalez-Farre B, Castellano JJ et al (2016) PiwiRNA-651 as marker of treatment response and survival in classical Hodgkin lymphoma. Oncotarget. 7(29):46002–46013
Cui L, Lou Y, Zhang X, Zhou H, Deng H, Song H, Yu X, Xiao B, Wang W, Guo J (2011) Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem 44:1050–1057
Daugaard I, Venø MT, Yan Y, Kjeldsen TE, Lamy P, Hager H, Kjems J, Hansen LL (2017) Small RNA sequencing reveals metastasis-related microRNAs in lung adenocarcinoma. Oncotarget 8:27047–27061
Dharap A, Nakka VP, Vemuganti R (2011) Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke 42:1105–1109
Esteller M (2008) Epigenetics in cancer. N Engl J Med 358:1148–1159. https://doi.org/10.1056/NEJMra072067
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12:861–874. https://doi.org/10.1038/nrg3074
Forsythe JA et al (1996) Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol 16:4604–4613
Freedman JE, Gerstein M, Mick E, Rozowsky J, Levy D, Kitchen R et al (2016) Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun. 7:11106
Fu A, Jacobs DI, Hoffman AE, Zheng T, Zhu Y (2015) PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis 36(10):1094–1102. https://doi.org/10.1093/carcin/bgv105. Epub 2015 Jul 25. PMID: 26210741; PMCID: PMC5006152
Fu A, Jacobs DI, Zhu Y (2014) Epigenome-wide analysis of piRNAs in gene-specifc DNA methylation. RNA Biol 11:1301–1312. https://doi.org/10.1080/15476286.2014.996091
Gainetdinov IV, Skvortsova YV, Stukacheva EA, Bychenko OS, Kondratieva SA, Zinovieva MV, Azhikina TL (2014) Expression profiles of PIWIL2 short isoforms differ in testicular germ cell tumors of various differentiation subtypes. PLoS One 9:e112528
Gebert D, Ketting RF, Zischler H, Rosenkranz D (2015) piRNAs from pig testis provide evidence for a conserved role of the Piwi pathway in post-transcriptional gene regulation in mammals. PLoS One 10:e0124860. https://doi.org/10.1371/journal.pone.0124860
Giatromanolaki A et al (2001) Relation of hypoxia inducible factor 1 alpha and 2 alpha in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br J Cancer 85:881–890. https://doi.org/10.1054/bjoc.2001.2018
Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specifc class of small RNAs binds mammalian Piwi proteins. Nature 442:199–202. https://doi.org/10.1038/nature04917
Grivna ST, Beyret E, Wang Z, Lin H (2006) A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20:1709–1714. https://doi.org/10.1101/gad.1434406
Grochola LF, Greither T, Taubert H, Moller P, Knippschild U, Udelnow A, Henne-Bruns D et al (2008) The stem cell-associated Hiwi gene in human adenocarcinoma of the pancreas: expression and risk of tumour-related death. Br J Cancer 99:1083–1088
Ha H, Song J, Wang S, Kapusta A, Feschotte C, Chen KC, Xing J (2014b) A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics 15:545
Ha H et al (2014a) A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics 15:545. https://doi.org/10.1186/1471-2164-15-545
Han YN, Li Y, Xia SQ, Zhang YY, Zheng JH, Li W (2017) PIWI proteins and PIWI-interacting RNA: emerging roles in cancer. Cell Physiol Biochem 3(1):1–20. (44)
Hashim A, Rizzo F, Marchese G, Ravo M, Tarallo R, Nassa G, Giurato G et al (2014) RNA sequencing identifies specific PIWI-interacting small noncoding RNA expression patterns in breast cancer. Oncotarget 5:9901–9910
He G, Chen L, Ye Y, Xiao Y, Hua K, Jarjoura D, Nakano T et al (2010) Piwil2 expressed in various stages of cervical neoplasia is a potential complementary marker for p16. Am J Transl Res 2:156–169
Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X (2013b) Altered expression of piRNAs and relation with clinicopathologic features of breast cancer. Clin Transl Oncol 15:563–568
Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H (2013a) A major epigenetic programming mechanism guided by piRNAs. Dev Cell 24:502–516
Huang XYT, Tschannen M et al (2013c) Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 14:319
Hung JJ et al (2009) Prognostic significance of hypoxia-inducible factor-1alpha, TWIST1 and Snail expression in resectable non-small cell lung cancer. Torax 64:1082–1089. https://doi.org/10.1136/thx.2009.115691
Jacobs DI, Qin Q, Lerro MC, Fu A, Dubrow R, Claus EB, DeWan AT, Wang G, Lin H, Zhu Y (2016) PIWI-interacting RNAs in gliomagenesis: evidence from Post-GWAS and functional analyses. Cancer Epidemiol Biomark Prev 25(7):1073–1080
Jiang D, Jiang L, Liu B, Huang H, Li W, Zhang T, Zu G, Zhang X (2017) Clinicopathological and prognostic significance of FoxM1 in gastric cancer: a metaanalysis. Int J Surg 48:38–44
Kojima-Kita K, Kuramochi-Miyagawa S, Nagamori I, Ogonuki N, Ogura A, Hasuwa H, Akazawa T, Inoue N, Nakano T (2016) MIWI2 as an effector of DNA methylation and gene silencing in embryonic male germ cells. Cell Rep 16:2819–2828
Ku H-Y, Lin H (2014) PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl Sci Rev:205–218
Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T (2008) DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22:908–917
Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, Lung RW, Chan AW, Chan TF, Wong N (2013) Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol 58:1165–1173
Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Tóth KF (2013) Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27:390–399
Lee JH, Jung C, Javadian-Elyaderani P, Schweyer S, Schutte D, Shoukier M, Karimi-Busheri F et al (2010) Pathways of proliferation and antiapoptosis driven in breast cancer stem cells by stem cell protein piwil2. Cancer Res 70:4569–4579
Lee S, Lee DK (2018) What is the proper way to apply the multiple comparison test? Korean J Anesthesiol 71(5):353–360. https://doi.org/10.4097/kja.d.18.00242. Epub 2018 Aug 28. Erratum in: Korean J Anesthesiol. 2020 Dec;73(6):572. PMID: 30157585; PMCID: PMC6193594
Levine AJ, Ting DT, Greenbaum BD (2016) P53 and the defenses against genome instability caused by transposons and repetitive elements. BioEssays 38:508–513
Li D, Luo Y, Gao Y, Yang Y, Wang Y, Xu Y, Tan S, Zhang Y, Duan J, Yang Y (2016) piR-651 promotes tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. Int J Mol Med 38:927–936
Li L, Yu C, Gao H, Li Y (2010) Argonaute proteins: potential biomarkers for human colon cancer. BMC Cancer 10:38
Li P-F, Chen S-C, Xia T et al (2014) Non-coding RNAs and gastric cancer. World J Gastroenterol 20(18):5411–5419
Lim RS, Kai T (2015) A piece of the pi(e): the diverse roles of animal piRNAs and their PIWI partners. Semin Cell Dev Biol 47-48:17–31
Liu L, Zhang J, Li A, Liu Z, He Z, Yuan X, Tuo S (2018) Prediction of cancer-associated piRNA-mRNA and piRNA-lncRNA interactions by integrated analysis of expression and sequence data. Tsinghua Sci Technol 23:115–125
Liu W, Gao Q, Chen K, Xue X, Li M, Chen Q, Zhu G, Gao Y (2014a) Hiwi facilitates chemoresistance as a cancer stem cell marker in cervical cancer. Oncol Rep 32:1853–1860
Liu W, Gao Q, Chen K, Xue X, Li M, Chen Q, Zhu G et al (2014b) Hiwi facilitates chemoresistance as a cancer stem cell marker in cervical cancer. Oncol Rep 32:1853–1860
Liu WK, Jiang XY, Zhang ZX (2010) Expression of PSCA, PIWIL1, and TBX2 in endometrial adenocarcinoma. Onkologie 33:241–245
Liu X, Sun Y, Guo J, Ma H, Li J, Dong B, Jin G et al (2006) Expression of hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int J Cancer 118:1922–1929
Liu Y (2016) MicroRNAs and PIWI-interacting RNAs in oncology. Oncol Lett 12(4):2289–2292
Liu Y, Dou M, Song X, Dong Y, Liu S, Liu H, Tao J, Li W, Yin X, Xu W (2019) The emerging role of the piRNA/piwi complex in cancer. Mol Cancer 18(1):123. https://doi.org/10.1186/s12943-019-1052-9. PMID: 31399034; PMCID: PMC6688334
Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai R, Li C, Li M, Zhou Y, Tan W et al (2018a) PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics 8:5213–5230
Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai R et al (2018b) PIWI-interacting RNA54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics 8(19):5213–5230
Martinez VD, Vucic EA, Thu KL, Hubaux R, Enfield KS, Pikor LA, Becker-Santos DD, Brown CJ, Lam S, Lam WL (2015b) Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep 5:10423
Martinez VD et al (2015a) Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep 5:10423. https://doi.org/10.1038/srep10423
Maxwell PH et al (1999) Tetumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271–275. https://doi.org/10.1038/20459
McClatchey AI, Fehon RG (2009) Merlin and the ERM proteins-regulators of receptor distribution and signaling at the cell cortex. Trends Cell Biol 19:198–206
Mei Y, Clark D, Mao L (2013) Novel dimensions of piRNAs in cancer. Cancer Lett 336(1):46–52
Mei Y, Wang Y, Kumari P et al (2015) A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells. Nat Commun 6:7316
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL et al (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 105(30):10513–10518
Mohn F, Sienski G, Handler D, Brennecke J (2014) The rhino-deadlockcutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157:1364–1379
Moyano M, Stefani G (2015) piRNA involvement in genome stability and human cancer. J HematolOncol 8:38. https://doi.org/10.1186/s13045-015-0133-5
Ng KW, Anderson C, Marshall EA, Minatel BC, Enfield KS, Saprunoff HL, Lam WL, Martinez VD (2016c) Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol. Cancer 15:5
Ng KW et al (2016a) Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol Cancer 15:5. https://doi.org/10.1186/s12943-016-0491-9
Ng KW, Anderson C, Marshall EA, Minatel BC, Enfield KSS, Saprunoff HL, Lam WL, Victor D, Martinez. (2016b) Piwiinteracting RNAs in cancer: emerging functions and clinical utility. Mol Cancer 15:5
Ostheimer C, Bache M, Guttler A, Reese T, Vordermark D (2014) Prognostic information of serial plasma osteopontin measurement in radiotherapy of non-small-cell lung cancer. BMC Cancer 14:858. https://doi.org/10.1186/1471-2407-14-858
Pan HW, Li SC, Tsai KW (2013) Tsai, MicroRNA dysregulation in gastric cancer. Curr Pharm Des 19(7):1273–1284
Pardini B, Naccarati A (2018) Altered piRNA profiles in bladder cancer: a new challenge in the next-generation sequencing era? J Genet Genomes 1:110
Park JC, Citrin DE, Agarwal PK, Apolo AB (2014) Multimodal management of muscle invasive bladder cancer. Curr Probl Cancer 38(3):80–108
Peng L, Song L, Liu C, Lv X, Li X, Jie J, Zhao D, Li D (2016) piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour Biol 37:2749–2756
Ploeg M, Aben KK, Kiemeney LA (2009) The present and future burden of urinary bladder cancer in the world. World J Urol 27:289–293
Qu A, Wang W, Yang Y, Zhang X, Dong Y, Zheng G, Wu Q, Zou M, Du L, Wang Y, Wang C (2019) A serum piRNA signature as promising non invasive diagnostic and prognostic biomarkers for colorectal cancer. Cancer Manag Res 11:3703–3720
Rizzo F, Hashim A, Marchese G, Ravo M, Tarallo R, Nassa G, Giurato G, Rinaldi A, Cordella A, Persico M et al (2014) Timed regulation of Pelement-induced wimpy testis-interacting RNA expression during rat liver regeneration. Hepatology 60:798–806
Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC (2009) A broadly conserved pathway generates 3’UTR-directed primary piRNAs. Curr Biol 19:2066–2076
Ross RJ, Weiner MM, Lin H (2014) PIWI proteins and PIWIinteracting RNAs in the soma. Nature 505(7483):353–359
Rouget C et al (2010) Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467:1128–1132. https://doi.org/10.1038/nature09465
Sai Lakshmi S, Agrawal S (2008) piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res 36:D173–D177
Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC (2009) A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461:1296–1299
Shomali N, Mansoori B, Mohammadi A, Shirafkan N, Ghasabi M, Baradaran B (2017) MiR-146a functions as a small silent player in gastric cancer. Biomed Pharmacother 96:238–245
Siddiqi S, Matushansky I (2012) Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem 113:373–380
Simi L et al (2006) Quantitative analysis of carbonic anhydrase IX mRNA in human non-small cell lung cancer. Lung Cancer 52:59–66. https://doi.org/10.1016/j.lungcan.2005.11.017
Sun G, Wang Y, Sun L, Luo H, Liu N, Fu Z, You Y (2010) Clinical significance of Hiwi gene expression in gliomas. Brain Res 1373:183–188
Suntsova M, Garazha A, Ivanova A, Kaminsky D, Zhavoronkov A, Buzdin A (2015) Molecular functions of human endogenous retroviruses in health and disease. Cell Mol Life Sci 72:3653–3675
Tan Y, Liu L, Liao M, Zhang C, Shuanggang H, Zou M, Gu M, Li X (2015) Emerging roles for PIWI proteins in cancer. Acta Biochim Biophys Sin 47(5):315–324
Tang X, Xie X, Wang X, Wang Y, Jiang X, Jiang H (2018) The combination of piR-823 and eukaryotic initiation factor 3 B (EIF3B) activates hepatic stellate cells via upregulating TGF-b1 in liver fibrogenesis. Med Sci Monit 24:9151–9165
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2012) Global cancer statistics, 2012. CA Cancer J Clin 65(2):87–108
Vychytilova-Faltejskova P, Stitkovcova K, Radova L, Sachlova M, Kosarova Z, Slaba K, Kala Z, Svoboda M, Kiss I, Vyzula R et al (2018) Circulating PIWIinteracting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol Biomarkers Prev 27:1019–1028
Wang J, Zhang P, Lu Y, Li Y, Zheng Y, Kan Y, Chen R, He S (2018) piRBase: a comprehensive database of piRNA sequences. Nucleic Acids Res. https://doi.org/10.1093/nar/gky1043
Wang QE, Han C, Milum K, Wani AA (2011) Stem cell protein Piwil2 modulates chromatin modifications upon cisplatin treatment. Mutat Res 708:59–68
Wang Y, Liu Y, Shen X, Zhang X, Chen X, Yang C, Gao H (2012) The PIWI protein acts as a predictive marker for human gastric cancer. Int J Clin Exp Pathol 5:315–325
Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H (2006) Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 20:1732–1743
Watanabe T, Tomizawa S-I, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S et al (2011) Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332:848–852
Weng W, Li H, Goel A (2019) Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochim Biophys Acta Rev Cancer 1871(1):160–169. https://doi.org/10.1016/j.bbcan.2018.12.005. Epub 2018 Dec 30. PMID: 30599187; PMCID: PMC6392428
Weng W, Liu N, Toiyama Y, Kusunoki M, Nagasaka T, Fujiwara T, Wei Q, Qin H, Lin H, Ma Y, Goel A (2018) Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol. Cancer 17:16
Wilson WR, Hay MP (2011) Targeting hypoxia in cancer therapy. Nat Rev Cancer 11:393–410. https://doi.org/10.1038/nrc3064
Wu X, Pan Y, Fang Y, Zhang J, Xie M, Yang F, Yu T, Ma P, Li W, Shu Y (2020) The biogenesis and functions of piRNAs in human diseases. Mol Ther Nucleic Acids 21:108–120. https://doi.org/10.1016/j.omtn.2020.05.023. Epub 2020 May 23. PMID: 32516734; PMCID: PMC7283962
Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L, Chen L et al (2015) piRNA823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 29:196–206
Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, Hu Y, Hu H, Li N, Chen W, Khaitovich P (2011) Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res 39:6596–6607
Yao J, Wang YW, Fang BB, Zhang SJ, Cheng BL (2016) piR-651 and its function in 95-D lung cancer cells. Biomed Rep 4:546–550
Yin J, Jiang XY, Qi W, Ji CG, Xie XL, Zhang DX, Cui ZJ, Wang CK, Bai Y, Wang J, Jiang HQ (2017) piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci 108:1746–1756
Zhang H, Ren Y, Xu H, Pang D, Duan C, Liu C (2013) The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg Oncol 22:217–223
Zhang P, Kang JY, Gou LT, Wang J, Xue Y, Skogerboe G, Dai P, Huang DW, Chen R, Fu XD, Liu MF, He S (2015) MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res 25:193–207
Zhang P, Si X, Skogerbo G, Wang J, Cui D, Li Y, Sun X, Liu L, Sun B, Chen R, He S, Huang DW (2014). piRBase: a web resource assisting piRNA functional study, Database (Oxford), 2014 (bau110)
Zhang SJ, Yao J, Shen BZ, Li GB, Kong SS, Bi DD, Pan SH, Cheng BL (2018) Role of piwi-interacting RNA-651 in the carcinogenesis of non-small cell lung cancer. Oncol Lett 15:940–946
Acknowledgments
None.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
Not applicable.
Funding
None.
Conflict of Interest
No.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Ray, S.K., Mukherjee, S. (2023). piRNA-Based Cancer Therapy in Hypoxic Tumor. In: Mukherjee, S., Kanwar, J.R. (eds) Hypoxia in Cancer: Significance and Impact on Cancer Therapy. Springer, Singapore. https://doi.org/10.1007/978-981-99-0313-9_8
Download citation
DOI: https://doi.org/10.1007/978-981-99-0313-9_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-0312-2
Online ISBN: 978-981-99-0313-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)