Abstract
The present work introduces a novel segmentation approach for detection of brain tumor in presence of surrounding obscured tissues. In this view, kernel-based fuzzy clustering algorithm is employed to capture the clear boundary of the tumors. Proposed method also considers two significant features of brain MRI for segmentation; one is regional entropy and the other regional brightness. The most important issue of fuzzy clustering algorithm is the selection of optimal number of clusters prior to the clustering. This work determines the optimal cluster number by introducing the concept of cluster validity indices. Employing five different cluster validity indices, the optimal cluster number is obtained for both of the features. Then, these two features are integrated using principal component analysis method. Following this, shape characteristics of the segmented tumors are extracted for grading the benignancy/malignancy of the tumors. Finally, the superiority of the proposed segmentation approach is compared with similar research works in this field and its efficiency is studied in terms of the classification indices.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Cancer is a life-threatening disease, and one of the most frightening among them is the brain cancer. The survival rate for people with brain cancer decreases with age (American Society of Clinical Oncology (ASCO) 2020). More than a million cases of brain tumor are diagnosed per year in India. A popular and effective technique, Magnetic Resonance Imaging (MRI) provides exquisite detail of the brain, to detect the prognosis of tumor, but sometimes the presence of surrounding soft tissues obscures the tumor outline. In this view, image segmentation is one of the most crucial steps in tumor analysis. There are several popular image segmentation techniques applied in medical imaging but a single approach is not applicable for all types of brain MRI. Hence, there is a need for more advanced and automated approach, which would mostly eliminate the inconveniences present in the conventional techniques and would provide better result for diagnosis of brain tumor.
Based on these, present work introduces a novel cluster validity index-based fuzzy c-means (FCM) clustering algorithm for segmentation of brain MRI. FCM is one of the most popular and widely used algorithms due to its robustness in presence of ambiguity and impreciseness. Two significant regional features of MRI; local entropy and brightness captured by appropriate kernel are utilized as the data of FCM model which accurately detects the prognosis of brain tumors. To combat the problem of manual selection of cluster numbers in FCM algorithm, this work employs five cluster validity indices for prediction of appropriate cluster numbers. Following this segmentation approach, discrimination of benignancy/malignancy of tumors also produces encouraging results.
The proposed method is discussed in Sect. 2. Section 3 provides the experimental results using the proposed detection model and finally Sect. 4, Sect. 5 draws some discussion and conclusion of this work.
2 Proposed Methodology
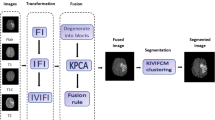
To resolve high degrees of inhomogeneity present in brain MRI, a novel cluster validity index-based FCM algorithm has been proposed using two significant spatial characteristics of MRI. The framework of the proposed approach, consisting of different blocks is shown in Fig. 1. To abide by the principles of medical ethics, multimodal brain MRIs (T1-weighted, T2-weighted, Gad, and PD) have been used from benchmarked databases from ‘The Whole Brain Atlas-Harvard Medical School’ (Johnson and Becker 2003) and ‘The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS)’ (Lastname et al. 2015).
In the following sections, the different blocks of the proposed methodology have been further discussed in detail.
2.1 Significant Feature Selection
From several research works, it has been observed that statistical features like local entropy, brightness, homogeneity, relative variances have been individually used to resolve the impreciseness in edge detection (Despotovic et al. 2015). On the contrary, Das et al. (2019) studied that, the combination of local entropy and local brightness is an effective pair to model the impreciseness present in mammograms. Sometimes, in brain MRI, the presence of overlapped soft tissues flattens the brightness of tumor and hence leads to over/under segmentation problems. As local entropy of a region estimates the information carried by it, surrounding soft tissues occupy different grade of entropy compared to the tumors. Following this, the present study shows that the combinations of local entropy (E) and brightness (B) efficiently resolve the uncertainties present in the process of tumor segmentation.
Regional entropy (E) and brightness (B) are mathematically expressed by the following two equations
where X = {x1, x2, …, xi,…, xn} denotes the set of image pixels and p(xi) is the probability of occurrence of pixel intensity I(xi) inside the kernel.
A kernel of size 3 × 3 (n = 9) has been moved across the entire image to fetch different characteristics of E and B, which in turn are used as the data for the proposed approach.
2.2 Cluster Number Selection
A conventional clustering algorithm Fuzzy c-means (FCM) has a great advantage of being more flexible than hard clustering techniques and provides better result in case of overlapped imprecise data. In spite of being advantageous, the most important challenge in FCM algorithm is to predict the optimal number of clusters (Gueorguieva et al. 2017). In this view, present work addresses optimal cluster number selection in terms of various cluster validity indices, prior to the segmentation using FCM algorithm.
Cluster Validity Indices
Cluster validation approach evaluates the obtained results and finds the best partitioning of the given data (Gueorguieva et al. 2017). The two important criteria for selection of the cluster number are compactness and separation of the underlying data which has been clearly shown in Fig. 2.
Compactness refers to the closeness of the members in each cluster, measured by the variance of cluster members and it should be minimized for better clustering. Separation refers to the distance between two different clusters, the inter-cluster distance (Wang and Zhang 2007). In the present work, five effective cluster validation indices have been employed for evaluating the cluster numbers (Wang and Zhang 2007; Bataineh et al. 2011; Pakhira et al. 2004). The validation indices and their mathematical formulations are described below
-
(i)
Partition Coefficient (PC): It measures the overlapping between the clusters and its value lies in the range of [1/KC, 1], where KC is the number of clusters (Capitaine and Frelicot 2011). Closer the value of PC to unity, more crisp the clustering would be (Gueorguieva et al. 2017).
$${\text{PC}} = \frac{1}{n}\mathop \sum \limits_{i = 1}^{{K_{{\text{c}}} }} \mathop \sum \limits_{j = 1}^{n} \left( {\mu_{ij}^{2} } \right)$$(3)where n is the size of the data matrix, \(\mu_{ij}\) is the membership value of jth data point in ith cluster.
-
(ii)
Classification Entropy (CE): CE measures the fuzziness of a given cluster (Capitaine and Frelicot 2011). Hence, CE estimates the effectiveness of partitioning and its value lies between the intervals [0, logKc]. Lower value of CE reflects better partitioning of dataset Z.
$${\text{CE}} = - \frac{1}{n}\mathop \sum \limits_{i = 1}^{{K_{{\text{c}}} }} \mathop \sum \limits_{j = 1}^{n} \mu_{ij} \log \mu_{ij}$$(4) -
(iii)
Separation Coefficient (SC): SC computes the ratio of sum of the separation and compactness of clusters (Gueorguieva et al. 2017). Minimum value of SC indicates the better separation between the clusters.
$$SC = \mathop \sum \limits_{{i = 1}}^{{K_{{\text{c}}} }} \frac{{\mathop \sum \nolimits_{{j = 1}}^{n} ~\left( {\mu _{{ij}} } \right)^{x} \left\| {z_{j} - c_{i} ^{2} } \right\|}}{{n_{i} ~~~\mathop \sum \nolimits_{k}^{{K_{c} }} \left\| {c_{k} - c_{i} ^{2} } \right\|~}}$$(5)where x is the fuzzifier index taken as 2, \(\left\| {z_{j} - c_{i} } \right\|\) is the Euclidean distance for jth data point and ith cluster.
-
(iv)
Separation Index (SI): SI utilizes the minimum distance of separation for the partition validity (Gueorguieva et al. 2017). Lower value of SI indicates better separation.
$${\text{SI}} = ~\frac{{\mathop \sum \nolimits_{{i = 1}}^{{K_{{\text{c}}} }} ~\mathop \sum \nolimits_{{j = 1~}}^{n} \left( {\mu _{{ij}} } \right)^{2} \left\| {z_{j} - c_{i} } \right\|^{2} }}{{n~\min _{{ik}} \left\| {c_{k} - c_{i} } \right\|^{2} }}$$(6) -
(v)
Xie-Beni's Validation (XB): XB estimates the ratio of total variation within the given clusters and the separation between clusters and its smaller value indicates that the clusters are compact as well as well separated (Xie and Beni 1991).
$${\text{XB}} = ~\frac{{\mathop \sum \nolimits_{{i = 1}}^{{K_{{\text{c}}} }} ~\mathop \sum \nolimits_{{j = 1~}}^{n} \left( {\mu _{{ij}} } \right)^{x} \left\| {z_{j} - c_{i} } \right\|^{2} }}{{n~\min _{{ik}} \left\| {z_{j} - c_{i} } \right\|^{2} }}$$(7)
The optimal number of clusters has been selected by the following selection rule—“For optimal cluster number, the majority of cluster validity indices (PC, CE, SC, SI, XB) must satisfy their respective criteria”.
2.3 FCM Algorithm
FCM is an objective function-based algorithm in which the membership values are assigned to each of the data point (E/B) corresponding to a cluster depending on the Euclidean distance between the data point and the cluster center (Bezdek et al. 1984; Kang et al. 2009; Adhikari et al. 2015). Higher degrees of membership are assigned to those data points which are close to the cluster center, and hence, the objective function gets minimized accordingly. The objective function of the FCM algorithm has been shown below
where U = [µij] denotes the matrix of membership values, Z is the given ‘q’ dimensional data of ‘n’ objects, Di is the local norm inducing matrix which is used as an optimization variable in U = [µij], Kc denotes the center of the clusters, \(d_{ik}^{2}\) is the Euclidean metrics which depends on the corresponding Euclidean distance \(d_{ij}^{2}\) and m denotes the fuzzifier index usually taken as 2 for best results.
The degree of membership value of given dataset Z in the cluster Kc, satisfies the equation below,
where \(\mu_{ij } = \frac{1}{{\mathop \sum \nolimits_{t = 1}^{{K_{{\text{c}}} }} \left( {\frac{{d_{ij} }}{{d_{it} }}} \right)^{{\frac{2}{m - 1}}} }}\).
Also, the equations for the cluster center (\(c_{i}\)) and the Euclidean metric (\(d_{ik}^{2}\)) has been stated below-
where Kc denotes the cluster number, dij2 is the Euclidean distance between jth data point and the ith cluster center.
2.4 Feature Combination Using Principal Component Analysis
To measure the contributions of each of the statistical features that is, regional entropy and brightness, a popular and extensively used computational technique known as principal component analysis (PCA) has been employed in this work. PCA is an useful approach to find out the principal component of the datasets for both of the features and henceforth, determines the respective weight factors for them (Mukherjee and Das 2020). It works on the principle of computing the covariance matrix created from the datasets of both of the regional features. Hence, it makes the segmentation approach automated and robust.
2.5 Shape-Based Feature Extraction
Several research works have been conducted, to extract information of the region of interest (ROI). The aim of this work is to determine the better feature extraction technique, in case of a particular scenario. From several studies, it has been observed that the malignant tumors consist of uneven shape irregularities in comparison with benign tumors as shown in Fig. 3. Hence, the characterization of tumors in terms of its shape has been the main focus to capture these shape irregularities.
In this view, the proposed method addresses a combination of some conventional shape metrics with a radius vector (r) based shape descriptor (Kurtosis) to study the tumor characteristics of brain MRI. The radius vector-based feature is insensitive to image orientation and alignments (Kobayashi et al. 2008). The mathematical formulations are as follows
where N is the total number of contour pixels and mean \(m = \frac{1}{N}\mathop \sum \nolimits_{n = 1}^{N} \left[ {r\left( n \right)} \right]\)
where W, L, A, P, CA denotes width, length, area, perimeter and convex area of the tumor, respectively.
Based on these extracted feature characteristics, the segmented tumors have been categorized into malignant/benign groups.
2.6 Classification
In this work, a conventional k nearest neighbor (KNN) classifier has been chosen, to classify the brain tumors into malignant/benign classes. KNN is a supervised classification algorithm which classifies a data point based on its neighboring data points. From known training datasets, KNN classifies the test data based on a similarity measure. The parameter ‘k’ in KNN algorithm refers to the number of nearest neighbors which are determined based on some distance parameters (Zhang et al. 2018). In this work, Euclidean distance measures between the known and the unknown data points have been considered. The performance of the KNN classifier on the proposed FCM model has been evaluated based on some statistical measures and the performance indices such as sensitivity, specificity and accuracy have been estimated by the following mathematical formulations
Sensitivity: It estimates how correctly the classifier can predict the malignant tumors.
Specificity: It estimates how correctly the classifier can predict the benign tumors.
Accuracy: It estimates the total correctly predicted malignant and benign tumors.
where (TP): number of previously known malignant tumors correctly identified as malignant; (TN): number of previously known benign tumors correctly identified as benign; (FP): number of previously known benign tumors incorrectly identified as malignant; (FN): number of previously known malignant tumors incorrectly identified as benign.
Further, this classifier has been validated using a popular technique, k-fold cross-validation. Specifically, fivefold cross-validation approach has been employed here which estimates the performance of the KNN classifier. The entire dataset is divided into k-subsets, such that every time each of the k subset is considered as test set and the remaining (k − 1) subsets as training sets, to validate the performance. The average estimation of total k number of trials provides the total effectiveness of the model.
3 Experimental Results
The dataset in the present work consists of 50 randomly chosen brain MRIs (31 benign and 19 malignant samples) specified by expert radiologists, taken from the benchmarked sources as mentioned in Sect. 2. To detect brain tumor using the proposed segmentation approach, following steps have been executed.
3.1 Cluster Number Selection Procedure
The following Tables 1 and 2 present the dataset of five validity indices computed by varying the cluster numbers (KC) from 3 to 9 with respect to the features; regional entropy and regional brightness, respectively. By thorough analysis of the datasets and following the selection rule—“For optimal cluster number, the majority of cluster validity indices (PC, CE, SC, SI, XB) must satisfy their respective criteria”, the cluster selection procedure has been carried out.
By analyzing from Tables 1 and 2, it can be observed that KC = 9 and KC = 3 has been selected as the optimal cluster numbers, following the above-mentioned selection rule for both the features regional entropy (E) and regional brightness (B), respectively.
3.2 Data of the Proposed Segmentation Approach
The segmentation results of brain MRIs with various illumination and contrast, obtained after successful execution of the proposed approach, are shown in Fig. 4.
By varying the nearest neighbor ‘k’ of the KNN classifier, it has been found empirically that for k = 7, the maximum average accuracy of 96.0%; sensitivity of 96.42% and specificity of 95.45% have been obtained.
4 Discussion
The efficiency of the proposed segmentation approach is estimated in terms of classification accuracy. Table 3, shows a brief comparison of overall classification accuracy, of the proposed approach with other related research works. It is found that the present work shows superior/comparable performances with respect to recent studies.
5 Conclusion
The present work has addressed an automated, robust and efficient segmentation technique based on five effective cluster validity indices to estimate the optimal cluster numbers in an automated manner. The combination of two significant regional features makes the proposed segmentation method more efficient. Moreover, following the detection procedure, the shape describing feature set makes the classification easier and the results also show the superiority of the proposed approach with other related studies. As the cluster number selection procedure takes a significant time frame hence, further investigation of more sophisticated segmentation model may lead to more robust and cost-effective diagnostic system for real-time use.
References
Adhikari SK, Sing JK, Basu DK, Nasipuri M (2015) Conditional spatial fuzzy C-means clustering algorithm for segmentation of MRI images. Appl Soft Comput 34:758–769
American Society of Clinical Oncology (ASCO) (2020) Brain tumor: statistics. https://www.cancer.net/cancer-types/brain-tumor/statistics. Last accessed Apr 2020
Amin J, Sharif M, Gul N, Yasmin M, Shad SA (2020) Brain tumor classification based on DWT fusion of MRI sequences using convolutional neural network. Pattern Recogn Lett 129:115–122
Bataineh KM, Naji M, Saqer M (2011) A comparison study between various fuzzy clustering algorithms. Jordan J Mech Industr Eng 5(4):335–343
Bezdek JC, Ehrlich R, Full W (1984) FCM: the fuzzy c-means clustering algorithm. Comput Geosci 10(2–3):191–203
Capitaine HL, Frelicot C (2011) A cluster-validity index combining an overlap measure and a separation measure based on fuzzy-aggregation operators. IEEE Trans Fuzzy Syst 19(3):580–588
Das P, Das A (2019) A fast and automated segmentation method for detection of masses using folded kernel based fuzzy c-means clustering algorithm. Appl Soft Comput 85(105775)
Despotovic I, Goossens B, Philips W (2015) MRI segmentation of the human brain: challenges, methods and applications. Comput Math Methods Med 23(450341)
Gueorguieva N, Valova I, Georgiev G (2017) M&MFCM: fuzzy C-means clustering with Mahalanobis and Minkowski distance metrics. Procedia Comput Sci 114:224–233
Johnson KA, Becker JA (2003) The whole brain atlas (Media). Lippincott Williams and Wilkins. https://www.med.harvard.edu/AANLIB/
Jothi G, Inbarani HH (2016) Hybrid tolerance rough set-firefly based supervised feature selection for MRI brain tumor image classification. Appl Soft Comput 46:639–651
Kang J, Min L, Luan Q, Li X, Liu J (2009) Novel modified fuzzy c-means algorithm with applications. Dig Signal Process 19(2):309–319
Kobayashi T, Otsu N (2008) Image feature extraction using gradient local auto-correlations. In: Forsyth D, Torr P, Zisserman A (eds) Computer vision-ECCV 2008. Lecture notes in computer science, vol 5302. Springer, Berlin, pp 346–358
Menze et al (2015) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imag 34(10)
Mukherjee S, Das A (2020) Effective fusion technique using FCM based segmentation approach to analyze Alzheimer’s disease. In: Pattnaik P, Mohanty S, Mohanty S (eds) Smart healthcare analytics in Iot enabled environment. Intelligent Systems Reference Library, vol 178. Springer, Cham, pp 91–107
Nabizadeh N, Kubat M (2015) Brain tumors detection and segmentation in MR images: Gabor wavelet versus statistical features. Comput Electr Eng 45:286–301
Pakhira MK, Bandyopadhyay S, Maulik U (2004) Validity index for crisp and fuzzy clusters. Pattern Recogn 37(3):487–501
Sachdeva J, Kumar V, Gupta I, Khandelwal N, Ahuja CK (2016) A package-SFERCB-“Segmentation, feature extraction, reduction and classification analysis by both SVM and ANN for brain tumors.” Appl Soft Comput 47:151–167
Wang W, Zhang Y (2007) On fuzzy cluster validity indices. Fuzzy Sets Syst 158(19):2095–2117
Xie XL, Beni G (1991) A validity measure for fuzzy clustering. IEEE Trans Pattern Anal Mach Intel 13:841–847
Zhang S, Li X, Zong M, Zhu X, Wang R (2018) Efficient kNN classification with different numbers of nearest neighbors. IEEE Trans Neural Netw Learn Syst 29(5):1774–1785
Acknowledgements
The present work is partially supported by the Center of Excellence (CoE) in Systems Biology and Biomedical Engineering, University of Calcutta, funded by the World Bank, MHRD India. Authors would also like to thank Dr. S. K. Sharma of EKO X-ray and Imaging Institute, Kolkata for providing the valuable comments on the subjective evaluation.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Das, K., Das, A. (2021). Segmentation of Brain Tumor Using Cluster Validity Index-Based Fuzzy C-Means Algorithm. In: Mukherjee, M., Mandal, J., Bhattacharyya, S., Huck, C., Biswas, S. (eds) Advances in Medical Physics and Healthcare Engineering. Lecture Notes in Bioengineering. Springer, Singapore. https://doi.org/10.1007/978-981-33-6915-3_6
Download citation
DOI: https://doi.org/10.1007/978-981-33-6915-3_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-33-6914-6
Online ISBN: 978-981-33-6915-3
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)