Abstract
With new mutations of the COVID-19 or SARS-COV-2 virus being reported worldwide, one wishes to have access to a reliable, low-cost early diagnostic device that can be used at home. Biosensors able to detect these viruses have been sought out as point-of-care (POC) devices, which can be used in such situations and are hence gaining popularity. Recently, many research works are focused on this sector due to the pandemic. The electrochemical biosensors are a subcategory of biosensors based on the transducer used. The electrochemical means allow for more low-cost, efficient, and portable POC products. Different designs for a POC biosensor have been developed to detect different biological analytes of importance over the years. This review gathers information about the important designs of electrochemical biosensor devices currently being used and advances possible for COVID-19 virus detection. The information provided can be used for further design developments in the field of electrochemical biosensors that can be used to detect such viruses.
Riya Titus and Mukti Mandal are equal contribution.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
10.1 Introduction
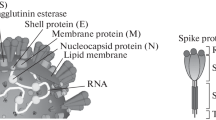
The year 2019 marked the start of a pandemic that is still evoking fear worldwide [55]. The SARS-COV-2 virus, a member of the B lineage of the genus Betacoronavirus (β-CoV), is more similar to the SARS-COV than the MERS virus. The various variants that are arising due to its mutation are causing havoc and concern among the public. SARS-CoV-2 is a virus of ~3 kb length that is single-stranded and possesses a positive RNA genome [56]. A virus is an intracellular parasite that uses host cellular mechanisms to replicate itself for its survival. A genomic material (DNA or RNA), a protein capsid for protecting the nucleic acid, and often an envelope for the capsid made of lipid, make up the essential components of the virus structure [54].
From the nineteenth to the twentieth, the turn of the century witnessed the emergence of formal studies on viruses. In 1933, the first Influenza virus (Flu) was isolated in a laboratory. The years 1918–1919 saw the Spanish Flu pandemic caused by a respiratory virus [25]. The respiratory tract of humans has physical barriers composed of epithelial cells and mucus and the alveolar macrophages in the lungs that protect us from most respiratory infections. The viruses usually attach to the receptors of the target-cell surface and penetrate the cytoplasm of the cell. The uncoated viral nucleic acid gets replicated and, upon assembly and maturation, becomes the infectious virus that leads to the visible symptoms. RNA viruses tend to mutate faster, such as respiratory syncytial virus (RSV), coronavirus (MERS and SARS-CoV), and Flu. These respiratory viruses cause concern in the public health sector because such mutations can lead to them being more virulent and potent [47]. SARS-CoV belongs to the subfamily coronavirinae of the family coronaviridae of the order Nidovirales. There are four genera within coronavirinae which are the alpha-, beta-, gamma-, and delta coronaviruses. Though widespread among mammals and causes mild respiratory or enteric infections, they were not a cause of much concern until the outbreak of severe acute respiratory syndrome (SARS) in 2002. The Middle East witnessed another coronavirus outbreak in 2014, caused by Middle East respiratory syndrome coronavirus (MERS-CoV) [41]. The SARS-CoV-2 or COVID-19 pandemic, which started toward the end of 2019, was declared a pandemic at the beginning of 2020. This time, it was caused by the coronavirus 2 (SARS-CoV-2) [50]. In viruses like SARS-CoV-2 and SARS-CoV, spike glycoprotein enters the host by binding to the angiotensin-converting enzyme 2 (ACE2) receptor found in several human body organs [38]. The whole genome analysis showed that β-CoVs encode several non-structural (2/3rd of SARS- CoV-2’s RNA) and mainly four structural proteins (spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N) (1/3rd of the genome) [37]. ORF1ab or ORF8 regions and the E, N, S, and RdRP genes of SARS-CoV-2 are the often genomic target regions of primers and probes. The main proteins (antigens) that trigger an antibody response in humans are S and N [44].
Cross-reactivity is a problem that can occur during diagnosis. This occurs when the test shows false positivity for any other species of coronavirus in humans. This happens due to the antibodies created against the proteins of one type of virus occasionally binding weakly to the proteins of another closely related virus, causing cross-reaction [9]. We can detect the viral infection either by directly detecting the virus or by the reactions or immune responses that occur in the body during the viral infection and detecting the products thus released during those reactions. Methods used for virus detection can be classified broadly as immunological assays, amplification techniques, and biosensors [42]. Immunological assays may include enzyme-linked immunosorbent assay (ELISA) and lateral flow immuno assay (LFIA), which can provide relatively fast on-site results, but some disadvantages exist like a high false-positive rate (5–11%) and relatively low sensitivity. These serological methods also face difficulties in detection due to the infection differences like being symptomatic or asymptomatic, immune response differences, and time duration of viral infection from patient to patient [3]. Amplification techniques include polymerase chain reaction (PCR), nanopore-targeted sequencing (NTS) [53], and recently the use of CRISPR technologies. Though they provide a relatively accurate result, some disadvantages exist. This is a slow process involving many steps for processing. The high mutation rate of RNA sometimes renders these processes inaccurate in detecting the mutated virus’s target genome [3]. The gold standard for immunoassays is radioimmunoassay (RIA), and the most commonly used immunoassay technique includes the enzyme-linked immunosorbent assay (ELISA) test [34]. The biosensors have proved themselves as a potential device to complement the ELISA tests in detecting several infections and diseases [17]. Especially now, upon the rise in COVID cases, different biosensors have been developed to detect the same.
10.1.1 Biosensors
Biosensors have mainly two parts: Bioreceptor and transducer. A biosensor is a device that can be used to detect biological analytes. The fear of the virus has driven the public to stay at their homes for safety. Thus, a device available for point-of-care (POC) applications like a biosensor catered to detecting SARS-COV-2 provided an option for the health sector to keep the COVID cases in check. Biosensors can be classified based on the analyte or reactions they can detect. They can be immunosensors (antigen–antibody interaction), enzymatic biosensors (enzyme-target analyte interaction) [18], DNA biosensors (hybridization), and whole cell biosensors as per this classification. Biosensors can also be of different types based on the transducers they use, such as electrochemical, optical, piezoelectric, calorimetric, or scanning probe microscopies. Optical biosensors use the principles of absorbance, fluorescence, chemiluminescence, or refractive index. Similarly, piezoelectric, calorimetric, and scanning probe microscope biosensors use the principles of affinity interactions, thermal characteristics, and atomic-level forces [30], respectively. Meanwhile, electrochemical biosensors use amperometric, potentiometric, conductometric, and impedimetric principles. If a biosensor uses an electrode system to convert any biological recognition event into an electrical signal, the thus obtained device is called an electrochemical biosensor [2]. Thus, electrochemical biosensing allows for comparing the change in the values of current, voltage, resistance, or capacitance due to any identifiable biological or chemical change. Commonly, these biosensors use three types of electrodes, namely working (electrode where the concerned reaction takes place), e.g., carbon, platinum, gold, etc., reference (the electrode whose potential remains constant against throughout the reaction), e.g., silver/silver chloride, saturated calomel electrodes, etc., and auxiliary electrodes or current-carrying electrodes (electrodes for making voltammetric and impedimetric measurements), e.g., inert solid electrodes like Pt, graphite, etc. These have low detection levels, inexpensive running costs, and simple instrumentation, making it easier for miniaturization. Based on the biological recognition elements and working principles also the electrochemical biosensors can be classified as enzymatic electrochemical biosensors, bioaffinity-based electrochemical biosensors, microbial biosensors, and nanobiosensors [2, 15].
10.1.2 Designs and Principles
There have been attempts to make simpler designs of these biosensors, and thus, microfluidics seemed a promising field as they are also based on total analysis system (TAS) [20]. Similarly, lab-on-chip designs apply microfluidics principles and integrate several laboratory steps into a single chip or circuit. Methods like lithography, laser machining, and xurography have been popular in recent times and modified to manufacture microfluidic devices such as lab-on-chips [26]. Electrochemical biosensor technologies and lab-on-chip principles, if appropriately implemented in a device, can reduce the cost, effort and increase the reliability of the detection kit thus developed [14]. As a result, much research has been initiated during these recent years to develop a biosensor design that will detect COVID-19 at high accuracy and precision with greater sensitivity and specificity [21]. There are different types of electrochemical biosensing techniques. Some of them are cyclic voltammetry, potentiometry, amperometry [22], electrochemical impedance spectroscopy [16], and conductivity. Cyclic voltammetry consists of changing different voltages and measuring the respective current values. Equipment used to detect phenolic antioxidants in chocolate food items uses this principle. Potentiometry measures the intrinsic voltage generated by the working electrode that is sensitive to the analyte versus the reference electrode insensitive to the analyte (Example: pH meter). Amperometry includes the applying of voltage and measuring the current value (Example: Glucose sensor). Electrochemical impedance spectroscopy is a non-destructive technique used to detect the time response of chemical systems at various voltage frequencies using low-amplitude alternating current (AC). An example of a system based on this principle is a microfluidic-based electrochemical biochip for diffusion-restricted DNA hybridization detection [4, 19], and conductivity is used for measuring salinity. Another type of popular principle used is that of Bio-FET. The (bio)molecules that bind to the substrate cause a surface potential change that leads to the device being gated. It is a type of field-effect transistor (FET) operating as an intrinsic amplifier. They can cause significant changes in the current when even small changes in the surface potential occur without any additional circuitry requirement. Here the design is such that a change in conductance occurs when biomolecules bind to the FET dielectric or gate electrode. As a result, it changes the dielectric material (FET gate dielectric), changing the underlying semiconductor material charge distributions.
An example of this is the label-free microfluidic integrated DNA FET on a printed circuit board by Xu et al. [58] to be used as a sensing FET and as an electrophoretic electrode to immobilize probe DNA at specific sites. Sometimes the principle of open-circuit potential (OCP) that uses net charge changes of an electrode surface and electrochemical impedance spectroscopy (EIS) is implemented together to create detection techniques that can detect MicroRNAs [28] and a microfluidic DC-biased AC electroosmotic vortex integrated DNA biosensing chip [19, 57]. Another principle we can add to an electrochemical biosensor to enhance its properties is carbon nanotubes. Carbon nanotubes show excellent conductivity, superior strength and sensitivity, remarkable physicochemical properties and chemical stability, and good biocompatibility. There are mainly two types of CNT based on the layers of walls: single-walled carbon nanotubes (SWCNT) and multi-walled carbon nanotubes (MWCNT). Carbon nanotube-based electrochemical biosensors are a type of nanobiosensor that can be broadly classified on the basis of the enzymes or biomolecules they detect. They are oxidase-based biosensors (e.g., glucose oxidase-based biosensors and cholesterol oxidase-based biosensors), dehydrogenase-based biosensors, DNA aptamer-based biosensors [36], CNT-based biosensors coated with antibodies for the detection of biomarkers, and other biosensors developed for biomedical applications based on carbon nanotubes [23].
Detection systems can be label-based systems or lab-free systems for biosensors. Label-based systems can be colorimetric, fluorescence, or optical fiber transducer-based. Colorimetric-based systems are fast, easy to use but have weak sensitivity. Fluorescence systems are of high efficiency, accuracy, fast labeling but costly. Optical fiber-based are controllable in sensing and sensitivity. Label-free systems can be electrical or conductance transducer-based or cantilever, or SPR based. Electrical and conductance transducer-based are of low cost, easy to use, and compatible with bioMEMS. Cantilever and SPR systems are too sensitive and can be affected by the environment.
Different materials can be used for designing a microfluidic biosensor. These materials can be inorganic, polymers, paper microfluidic chip, and hydrogels. Inorganic materials include silicon microfluidic chips, glass microfluidic chips, and ceramic microfluidic chips. Polymer materials include elastomeric materials like PDMS microfluidic chips and thermoset polyester (TPE) microfluidic chips. Polymer materials also include thermoplastic polymers like polystyrene (PS) microfluidic chips, polycarbonate (PC) microfluidic chips, polymethyl methacrylate (PMMA) microfluidic chips, polyethylene glycol diacrylate (PEGDA) microfluidic chip, microfluidic chip made of teflons like perfluorinated compounds (PFEP/PFA/PFPE), and polyurethane (PU) microfluidic chips. Composite materials such as cyclic olefin copolymer (COC) microfluidic chips and paper/polymer hybrid microfluidic chips can also be used [51].
When considering a suitable recognition element for detecting a virus, the nucleic acid-based biorecognition elements are more advantageous to use than antibodies due to their more stable nature. Designing any diagnostic device involves the proper selection of the biorecognition molecule/antigen that will finally be detected either qualitatively or quantitatively or both qualitatively and quantitatively by the device. In detecting the COVID-19 virus, we need a biorecognition molecule that one can detect easily and has the chance of least mutation so that we may be able to detect the virus even if specific mutations occur while spreading worldwide, making the diagnostic test a reliable one. It was found in the SARS-CoV-2 samples taken from India that ORF1ab is the most mutated region. After ORF1ab, the most mutated region was the N-gene, then the S gene, and ORF8 [43]. Thus, the ideal antigen/biorecognition molecule that can be taken for the diagnostic test can be either the spike protein or ORF8.
10.2 Some Designs of Electrochemical Biosensors for SARS-CoV-2 Detection
10.2.1 Electrochemical—Amperometry
The making of this sensor involved cutting a 1.5 × 1.5 cm Ti sheet from a G1 grade Ti sheet. Any formed oxide layer was removed by polishing with 600 grit polishing paper. To avoid any exposure to the electrolyte composed of n 96.5 mL (CH2OH)2, 3 mL DI H2O, and 0.505 g NH4F, the unpolished side was masked with Kapton tape. With the Ti foil (working electrode) kept 3 cm apart from a platinum foil (counter electrode) in a standard two-electrode configuration in a teflon beaker, the electrochemical anodization was carried out. Rinsing with DI H2O and baking in an oven for 4 h at 120 °C followed anodization. The sample was annealed after the tape had been removed from the baked sample. Thus, TiO2 nanotubes (TNTs) were synthesized, and then, using an incipient wetting method (a wet ion-exchange process), it was functionalized with cobalt. To detect the S-RBD Protein of SARS-CoV-2, amperometry was performed at a bias voltage of −0.8 V. It can detect the S-RBD protein in a 14–1400 nM concentration range in a very short time of ~30 s. In addition to this sensor’s evident rapid diagnostic application, it can also be modified with appropriate metallic elements to functionalize TNTs to detect other respiratory viruses. Figure 10.1 represents this electrochemical biosensor [52].

Reprinted (adapted) with permission from [52] from the Special Issue Detection and Diagnosis of the New Coronavirus under Open Access. Copyright © 2020 Sensors
An illustration of the working of the cobalt-functionalized TNT-based SARS-CoV-2 detecting platform.
10.2.2 Electrochemical—Paper-Based Amperometry
This design is a simple, rapid, quantitative, easy to implement, selective, and low-cost paper-based gold nanoparticle-mediated electrochemical sensor chip. A simple two-step procedure was used to fabricate the chip that does not require any amplification of genes as a step. First, a graphene suspension was coated on filter paper, forming a conductive film providing high carrier mobility. Then as the next step, a gold electrode was deposited with a predefined design. The special feature of this design is that four different types of highly specific antisense oligonucleotides (ssDNA) are used to cap the gold nanoparticles, which are used to target two domains of the viral nucleocapsid phosphoprotein (N-gene). This allows for a multimodal approach that helps to increase the reliability, sensitivity, and feasibility of the test even if some mutations occur in the viral gene. This type of electrochemical biosensor design is represented in Fig. 10.2 [1].

Reprinted (adapted) with permission from [1]. Copyright © 2020 American Chemical Society
A diagrammatic representation of a graphene-ssDNA-AuNP platform from sample collection, extraction, and getting added to the platform to incubation and recording of the digital electrochemical output.
10.2.3 Electrochemical—Differential Pulse Voltammetry (DPV)
The design used here is that of a super-sandwich-type electrochemical biosensor which makes use of a capture probe (CP), label probe (LP), auxiliary probe (AP), and target sequence. Nucleic acid amplification and reverse transcription are not needed in this method. For detection, the use of two premixes is required. Premix A was prepared by immobilizing CPs labeled with Thiol on the Au@Fe3O4 nanoparticle surfaces forming CP/Au@Fe3O4 nanocomposites. Then, Premix B was prepared. For this, Toluidine Blue (TB) was enriched for the SARS-CoV-2 detection by functionalizing graphene (SCX8-RGO) using p-sulfocalix[8]arene (SCX8). Au@SCX8-TB-RGO-LP biconjugate was formed by immobilizing the host–guest complexes (SCX8-TB) on RGO. Thus, producing the sandwich structure (CP-Target-LP). Long concatemers were formed due to the introduction of AP. The detection step involves the extraction of the target viral RNA (ORF1ab), then incubating it with Premix A for 1 h, and then next incubating this with Premix B for 2 h. Then detection occurs on the treated screen printing carbon electrode (SPCE), which is linked to a smartphone to show the results based upon the differential pulse voltammetry (DPV). This biosensor is reported not to show any cross-reactivity. This type of electrochemical biosensor design is shown in Fig. 10.3 [59].

Reprinted from [59], Copyright 2021, with permission from Elsevier
A diagram representing SARS-CoV-2 detection using a type of super-sandwich-type electrochemical biosensor and its detection with the help of smartphones.
Printed circuit board (PCB) technology was used to make electrodes for a low-cost electrochemical DNA biosensor, each costing just USD $0.55 (i.e., approximately INR ₹40) was used to detect COVID-19 virus N protein from wastewater samples using portable PCR instruments like miniPCR, without the requirement for qPCR reagents. Methylene Blue-DNA complex adsorption principles were used to increase the differential pulse voltammetry measurements. The PCB electrodes can also be reused after wiping them with lint-free wipes soaked with isopropyl alcohol. These electrodes were designed using Autodesk EAGLE software, and the standard ENIG plating process was used to form the gold electrodes on the PCB. Underneath the gold lies the nickel and copper layers of the electrode, respectively. PalmSens EmStat3 Blue potentiostat was used to perform voltammetry. PSTrace software was used to obtain peak values for DPV peak current and cathodic peak current in cyclic voltammetry voltammograms. The information thus obtained was used for the preparation of the required graphs. This type of electrochemical biosensor detection is represented in Fig. 10.4 [31].

Reprinted from [31], Copyright (2021), with permission from Elsevier
A schematic illustration of the workflow for detecting SARS-CoV-2 using the PCB-based electrochemical biosensor and the coding genes of major proteins used in this study.
10.2.4 Electrochemical—Electrochemical Impedance-Based Sensing (EIS)
Another research team hypothesized that the binding kinetics between the anti-SARS-CoV-2 antibody and the SARS-CoV-2 spike protein receptor-binding domain (RBD) mainly governed the detection mechanism. They introduced a design that uses a 16-well-plate xCELLigence system (RTCA S16) with integrated electrodes from ACEA Biosciences. This system design majorly allows for the non-invasive EIS detection of cell proliferation, morphology change, and attachment quality. Each well consists of an array of specially designed interdigitated electrodes fused to polyethylene terephthalate. Single-frequency measurements (10 kHz) are acquired every few seconds by each well, and the plate interface independently sees these. The interface attached to the well-plate is connected to the laptop via a USB. The timing measurements of independently addressable wells are controlled using a software; this type of electrochemical biosensor design is shown in Fig. 10.5 [45].

Reprinted from [45], Copyright (2021), with permission from Elsevier
96-well platform of ACEA Bioscience with the schematic layout of its electrode (a, b) and images of the electrode (c) and its magnified version (d), and illustration of electrochemical impedance-based sensing circuit (e) solution protein/antibody equivalent.
The 3D-printed COVID-19 test chip (3DcC) device has a working electrode (WE), a counterelectrode (CE), and a reference electrode whose base layers consist of coats of chromium and gold. Microdroplets containing gold nanoparticles were aerosol jet (AJ) printed on WE as micropillar arrays. The organic, non-polar solvent was used in the AJ printing technique, and this evaporated, leaving the dry nanoparticles and binders solidified. The printed structures were sintered to form the gold micropillars of the electrode. Shadow mask was used to coat thin silver or silver chloride layer on the RE. The replica-molding method was used to fabricate a PDMS microfluidic channel. For this, a poly(methyl methacrylate) (PMMA) mold and a poly(dimethylsiloxane) (PDMS), reverse mold was used. The micropillar array and the CE and RE electrodes on the glass slide were covered manually using the PDMS housing. There are also tubes out of the microfluidic channel to input the fluid into the channel. This followed the micropillar electrode array (3D-printed) functionalization using reduced graphene oxide (rGO) nanoflakes that enhance viral antigen bonding, increasing the antibody detection from the introduced fluid. The binding of the antibodies to the corresponding electrodes with the respective antigens increases the impedance due to the increasing thickness of the double-layer capacitance (Cdl) detected using the electrochemical impedance spectroscopic (EIS) measurements. This detection occurred in seconds, and regeneration of the electrodes for further use was also possible within 1 min in low pH chemistry using elution of antibody–antigen immunoaffinity. The results could be visualized on a smartphone-based platform, making it one of the best options for POC applications. Thus, the respective SARS-CoV-2 antibodies (S1 and receptor-binding-domain (RBD)) were detected. Figures 10.6a, b show the steps involved in the construction of these types of electrochemical biosensors [3].

Reprinted from [3], Copyright © 2020 Wiley–VCH GmbH, with Open Access permission from Advanced Materials, Wiley Online Library
An illustration of the process of manufacture of the 3D-printed COVID-19 test chip (3DcC) using aerosol jet nanoparticle 3D printing.
10.2.5 Electrochemical—Semiconductor Analyzer
This was designed as a graphene-based electrochemical biosensor using the principles of field-effect transistors (FETs). The wet transfer method was used to deposit the graphene layer onto a SiO2 substrate. Channels were constructed using photolithography and reactive ion etching. Thin-film deposition and lift-off methods were used to add the metal electrodes. The graphene layer is soaked with PBASE solution. The PBASE acts as a linker, especially a pyrene group of the compound that non-covalently attaches to graphene through pi-pi stacking. At the other end of the compound is the activated ester that reacts with the amines. Thus, the SARS-CoV-2 S protein antibody reacts with the linker to form a chemical bond. The sensing area dimensions were set as 100 × 100 μm2 (L × W). The device was also passivated with SU8-2010 so that the interferences during electrical measurements may be reduced. Also, the device showed no measurable MERS-CoV antigen cross-reactivity. Figure 10.7 shows the basic GFET design used for this type of detection method [48].

Reprinted (adapted) with permission from [48]. Copyright © 2020 American Chemical Society
A diagrammatic representation of the graphene FET sensor for detecting the SARS-CoV-2 spike antibody.
10.2.6 Electrochemical—Field-Effect Transistor (FET)
This device is an example of a FET nanobiosensor that uses a high-purity semiconducting (sc) single-walled carbon nanotube (SWCNT) for the detection of the SARS-CoV-2 spike protein and nucleocapsid protein. A specific antibody is used to functionalize the sc-SWCNT, enabling them to detect the two SARS-CoV-2 structural proteins from the nasopharyngeal swab samples. The high-purity sc-SWCNTs increase the sensitivity to detect the target analyte compared to unsorted SWCNT and graphene. Photolithography was performed to pattern the interdigitated gold electrodes on a Si/SiO2 substrate. Thus, the formation of microchannels of 10 μm took place, and dielectrophoresis (DEP) was used to deposit the sc-SWCNTs between the gold electrodes. Some treatments are done to activate the carboxylic groups and functionalize the SARS-CoV-2 antibody on SWCNTs. The devices were rinsed before and after soaking with a blocking buffer using nanopure water. This prepared the device for the FET measurements as nanopure water acted as the gating electrolyte. Detection of the SARS-CoV-2 antigens is done by studying its FET transfer characteristics in the liquid-gated FET device configuration. This type of electrochemical biosensor detection is shown in Fig. 10.8 [48].

Reprinted (adapted) with permission from [49]. Copyright © 2021 American Chemical Society
Diagram representing the structure of SARS-CoV-2 and schematic illustration of the basic working of the SWCNT FET biosensor.
10.2.7 Electrochemical—Square Wave Voltammetry (SWV)
The design of this electrochemical immunosensor for the coronavirus related to the MERS consisted of an array of gold nanoparticles modified carbon electrodes (DEP). It had eight working electrodes. Two control electrodes were used for comparative purposes. For the MERS-CoV antigen, four electrodes were used, while for human coronavirus (HCoV), two electrodes were used. This allowed for duplicate measurements and testing of cross-reactivity for each sample on the same chip. Due to the design having eight electrodes, multiplexed detection can be done for eight different CoVs by immobilizing their antigens on each of them. The spiked nasal samples give electrochemical measurements recorded as SWV using the ferrocyanide/ferricyanide as a probe [35].
10.2.8 Electrochemical—Magnetic Force-Assisted Immunoassay (MESIA)
Sampinute™ COVID-19 Antigen MIA is a product by Celltrion, USA Inc. It is a magnetic force-assisted electrochemical sandwich immunoassay (MESIA). This is used for detecting the SARS-CoV-2 receptor-binding domains (RBDs) spike proteins qualitatively present in the nasopharyngeal swab specimens either collected directly or through the means of a viral transport media. The product comes with 25 test cartridges. While in use, the dispersion of the sample into the sample inlets of the cartridges takes place via a pipette or a reagent tube. The sample then flows through the microfluidic channel. They form complexes with the anti-SARS-CoV-2 spike protein antibodies conjugated to magnetic nanoparticles (MNPs) if the sample contains SARS-CoV-2 spike proteins. Otherwise, no complexes are formed. In such an electrochemical sensor, the working electrode coated with anti-SARS-CoV-2 spike protein antibodies is encountered with these complexes, which also bind with the electrode. The principle of magnetism is used to actively control the antigen–antibody reactions, ensure that thorough mixing occurs between the MNPs and the antigens, and remove the unbound MNPs. The device makes use of a detection buffer, after which the electrochemical measurement step is done where an electric current is induced. This is because of the electrochemical oxidation and reduction of gold on the MNPs induced due to the voltage applied initially. The electric current quantity measured above a specific cut-off value indicates if the test result is positive (SARS-CoV-2 spike protein antigen present) or negative (SARS-CoV-2 spike protein antigen absent). The results are then displayed on screen by the Sampinute Analyzer device [7].
10.3 Comparison Table and Future Perspectives
From the designs mentioned above, we can see the common essential elements of a biosensor listed in Table 10.1. Some of the designs focused on solving the problems that existed before while they were used to detect other organisms other than SARS-CoV-2. The paper-based amperometry electrochemical biosensor uses the change in output voltage to address the issue related to signal amplification methodology and is working on how to integrate the technology with a portable mobile platform [1]. Some were novel technologies developed to overcome the shortcomings of PCR-based RNA assays. Ultrasensitive super-sandwich-type electrochemical sensors aim to develop high-throughput diagnostics through microfluidic-based cartridges in the future [59]. Future aims of the team that developed the printed circuit board electrodes include identifying optimal primers for electrochemical sensing and PCR amplification, assay integration with electrochemical sensing and onboard thermocycling, and also at consistent potentials to enhance the stability of the reference electrode for achieving redox peaks [32]. The electrochemical impedance-based sensing (EIS) using capacitive immunosensing assay established the possibility of detecting SARS-CoV-2 antibodies at clinically relevant concentrations utilizing a quantitative EIS method using widely accessible equipment. Such techniques might allow for more fast screening of patient samples, larger serological surveys to measure community anti-SARS-CoV-2 antibody levels, and possibly improved vaccination activity evaluation [45]. The biosensing technologies used will allow for early infection identification and isolation, potentially saving lives. Some of the test platforms like functionalized TiO2 nanotube-based [52] and the aerosol jet-printed 3D electrode biosensors [3] are general, which means they may be used to identify biomarkers for other diseases, including Zika, HIV, and Ebola. Finally, the platform will serve as a valuable tool for studying the infection and after recovery immune response dynamics. The sc-SWCNT FET detection method also allows for multiplex detection of viral antigens as well as antibodies that recognize these antigens [49]. Thus, we can see that new technologies and improvements made in either the fabrication or the working of these biosensors increased their utility and robustness. 2D materials like graphene and black phosphorus have also been utilized in biosensors for POC diagnostics [10]. Instead of the traditional and commonly used carbon-based screen-printed electrodes (SPCE) used in the super-sandwich-type electrochemical biosensor [59], screen-printed graphene electrodes (SPGE) can be synthesized and used. Reports suggest that SPGE have superior electrochemical properties than SPCE [10]. Similar studies are also being done on black phosphorous (BP), which shows excellent electrochemical properties due to their inherent redox properties. They have been used in an aptamer-functionalized and nanostructured label-free electrochemical biosensor [32]. BP-based biosensors also seem to show higher sensitivity and specificity in detecting IgG or IgM against the SARS-CoV-2 virus in blood samples compared to reduced graphene oxide (rGO) [10]. Also, it is seen that the detection sensitivity of the EIS method depends on various factors. Often there is an unaccounted change in impedance due to these factors. Thus, it has been found that the application of machine learning algorithms considering these factors can help predict the change in impedance more accurately [24].
10.4 Conclusion
Electrochemical biosensors used to detect several diseases have a huge potential due to their low cost, portable nature, easy instrumentation, and miniaturization (especially in making MEMS devices). Due to their electrochemical nature and signals that can be detected using a current/voltage change detector, they can be linked with other devices like smartphones to make customizable applications. These applications providing interactive and easy-to-use and detect platforms make diagnostic tests truly point of care (POC) in nature. Point-of-care tests should have the features according to the ASSURED guidelines. Thus, they have to be affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free (no complex equipment), and delivered to end-users [27]. Apart from electrochemical biosensors, chip-based, microfluidic-based [33], paper-based, or some material-based like film-based [8] and textile-based [40] biosensors or a combination of these can be developed for the detection of COVID-19 virus. The film-based biosensors can be used to detect pathogens from a crude sample. The textile-based biosensors can be cloth-based, thread-based, or fabric-based. These biosensors seem to provide ways to make the least complex ones that reduce cost, increase sensitivity, and provide a possible diagnostic method to detect the antibodies against the SARS-CoV-2 virus.
In the pandemic scenario, when we need regular testing and vaccination drives, releasing a massive amount of biomedical waste, one must focus on creating an ecological solution that causes the minimum amount of impact on the environment while providing the best utility. Thus, biodegradable alternatives for materials that we can use to make these devices must be considered. At last, the ultimate product we get must cause the least harm to the environment and be easy to dispose of. Simple bioinspired technologies or techniques can also be used in these test kits to make sample collection and processing easier. New innovative ideas are being used to develop these diagnostic kits, such as smartphone-based detection tests [39], using a collection pad on a lollipop stick to collect saliva samples [6], Carbon NanoTubes (CNT), and CRISPER-CAS technologies. Some of the current POC devices also incorporate CRISPR technologies for the diagnostic test process. Most of these tests, for sample preparation, made use of commercially available RNA extraction kits [5, 12] or made use of several pipetting steps for simplified lysis for the detection reaction [29, 39]. One notable achievement by Helena et al. (De Puig et al.) [11] involved developing the only diagnostic that can detect various variants of SARS-CoV-2 at nearly the CDC RT-qPCR standards. This device is called minimally instrumented SHERLOCK (miSHERLOCK) that uses biodegradable polylactic acid for 3D printing the device, thus minimizing cost and reducing the environmental footprint. It is also linked to an automated mobile app.
Asymptomatic cases of COVID-19 pose a threat to containing the spread of the virus since it does not show any typical symptoms and is hence difficult to recognize or detect if a person has SARS-CoV-2 infection. Also, during RT-PCR tests for COVID-19 [46], it was found that it failed to detect some cases of SARS-CoV-2 infection. All of these, coupled with the reckless behavior of the public, led to the subsequent waves of COVID-19 that several countries have faced till now. Chest X-ray proved to be a reliable method to detect the virus that infects the lungs early, even before significant symptoms are seen in the patients. AI-Based Intelligent COVID-19 detector Technology for Medical Assistance (ATMAN) is web-based software developed by CAIR, DRDO, and built using deep convolution neural network. This application can automatically pre-process the images from the X-ray test and classify the patient as normal, COVID-19, or pneumonia class. ATMAN is a tested and validated software approved for use. Thus, the use of AI is also proving itself as a valuable tool to enhance diagnostics [13].
Hence, we can see much potential for developments in the field of diagnostics in this world, where new diseases and organisms get reported while new technologies are also found to contain them. From this paper, one can get a rough idea of the developments in the field of electrochemical biosensors that have been developed to detect the SARS-CoV-2 virus. Some future aspects and advantages of these electrochemical biosensors are that the efficiency of the current systems can be increased through new materials and technologies that are being introduced. In addition to this, most of these biosensor designs mentioned before can also aid in the detection of other pathogens if they are modified accordingly.
References
Alafeef, M., Dighe, K., Moitra, P., Pan, D.: Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano 14, 17028–17045 (2020). https://doi.org/10.1021/ACSNANO.0C06392/SUPPL_FILE/NN0C06392_SI_001.PDF
Anik, U.: Electrochemical medical biosensors for POC applications. In: Medical Biosensors for Point of Care (POC) Applications, pp. 275–292. Elsevier Inc. (2017)
Ali, M.A., Hu, C., Jahan, S., et al.: Sensing of COVID-19 antibodies in seconds via aerosol jet nanoprinted reduced-graphene-oxide-coated 3D electrodes. Adv. Mater. 33, 2006647 (2021). https://doi.org/10.1002/ADMA.202006647
Ben-Yoav, H., Dykstra, P.H., Bentley, W.E., Ghodssi, R.: A microfluidic-based electrochemical biochip for label-free diffusion-restricted DNA hybridization analysis. Biosens. Bioelectron. 38, 114–120 (2012). https://doi.org/10.1016/J.BIOS.2012.05.009
Broughton, J.P., Deng, X., Yu, G., et al.: CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol 38(7), 38:870–874 (2020). https://doi.org/10.1038/s41587-020-0513-4
Carey, S.: Smartphone-based saliva test that can detect COVID-19 earns UF professor honors in national contest|UF Health, University of Florida Health. In: UF Health, University of Florida Health. https://ufhealth.org/news/2020/smartphone-based-saliva-test-can-detect-covid-19-earns-uf-professor-honors-national (2020). Accessed 30 Jan 2022
CELLTRION.: SAMPINUTETM. https://www.celltrion.com/en-us/kit/sampinute (2020). Accessed 30 Jan 2022
Ceylan Koydemir, H., Külah, H., Özgen, C.: Thin Film Biosensors. pp. 265–300 (2020). https://doi.org/10.1007/978-94-007-2592-8_8
Che, X.Y., Qiu, L.W., Liao, Z.Y., et al.: Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J. Infect. Dis. 191, 2033–2037 (2005). https://doi.org/10.1086/430355
Choi, J.R.: Development of point-of-care biosensors for COVID-19. Front. Chem. 8, 517 (2020). https://doi.org/10.3389/FCHEM.2020.00517/BIBTEX
De Puig, H., Lee, R.A., Najjar, D., et al.: (2021) Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci. Adv. 7. https://doi.org/10.1126/SCIADV.ABH2944/SUPPL_FILE/SCIADV.ABH2944_SM.PDF
Ding, X., Yin, K., Li, Z., et al.: Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 11(1) (2020). https://doi.org/10.1038/s41467-020-18575-6
DRDO.: AI Based Intelligent COVID-19 detector Technology for Medical Assistance (ATMAN). In: Defence Research and Development Organisation. https://www.drdo.gov.in/ai-based-intelligent-covid-19-detector-technology-medical-assistance-atman (2020). Accessed 30 Jan 2022
Dutta, G.: (2020) Nanobiosensor-based diagnostic System: transducers and surface materials. In: Nanobiomaterial Engineering: Concepts and Their Applications in Biomedicine and Diagnostics, pp. 1–13. https://doi.org/10.1007/978-981-32-9840-8_1
Dutta, G., Biswas, A., Chakrabarti, A. (eds.): Modern Techniques in Biosensors. p. 327 (2021). https://doi.org/10.1007/978-981-15-9612-4
Dutta, G., Jallow, A.A., Paul, D., Moschou, D.: Label-Free electrochemical detection of S. mutans exploiting commercially fabricated printed circuit board sensing electrodes. Micromachines 10 (2019). https://doi.org/10.3390/MI10090575
Dutta, G., Lillehoj, P.B.: Wash-free, label-free immunoassay for rapid electrochemical detection of PfHRP2 in whole blood samples. Sci. Rep. 8(1) (2018). https://doi.org/10.1038/s41598-018-35471-8
Dutta G, Park S, Singh A, et al (2015) Low-Interference Washing-Free Electrochemical Immunosensor Using Glycerol-3-phosphate Dehydrogenase as an Enzyme Label. Analytical Chemistry 87:3574–3578. https://doi.org/10.1021/AC504485A/SUPPL_FILE/AC504485A_SI_001.PDF
Dutta, G., Rainbow, J., Zupancic, U., et al.: Microfluidic devices for label-free DNA detection. Chemosensors 6, 43 (2018). https://doi.org/10.3390/CHEMOSENSORS6040043
Dutta, G., Regoutz, A., Moschou, D.: Electrochemical glucose biosensor with commercially fabricated printed circuit board sensing electrodes: the importance of electrode surface characteristics in sensor sensitivity. In: Electrochemical Glucose Biosensor with Commercially Fabricated Printed Circuit Board Sensing Electrodes: The Importance of Electrode Surface Characteristics in Sensor Sensitivity (2018)
Dutta, G., Regoutz, A., Moschou, D.: Enzyme-assisted glucose quantification for a painless Lab-on-PCB patch implementation. Biosens. Bioelectron. 167, 112484 (2020). https://doi.org/10.1016/J.BIOS.2020.112484
Dutta, G., Regoutz, A., Moschou, D.: Commercially fabricated printed circuit board sensing electrodes for biomarker electrochemical detection: the importance of electrode surface characteristics in sensor performance. Proceedings 2, 741 (2018). https://doi.org/10.3390/PROCEEDINGS2130741
Gupta, S., Murthy, C.N., Prabha, C.R.: Recent advances in carbon nanotube based electrochemical biosensors. Int. J. Biol. Macromol. 108, 687–703 (2018). https://doi.org/10.1016/J.IJBIOMAC.2017.12.038
Jafari, R.: Overcoming the Bottleneck in COVID-19 Detection: a machine-learning approach to improve accuracy of electrochemical impedance spectroscopy (EIS) detection sensitivity. Open Access J. Biomed. Sci. 3 (2020). https://doi.org/10.38125/OAJBS.000229
Javelle, E., Raoult, D.: COVID-19 pandemic more than a century after the Spanish flu. Lancet. Infect. Dis 21, e78 (2021). https://doi.org/10.1016/S1473-3099(20)30650-2
Jayamohan, H., Romanov, V., Li, H., et al.: Advances in microfluidics and lab-on-a-chip technologies. Mol. Diagn. (3), 197–217 (2017). https://doi.org/10.1016/B978-0-12-802971-8.00011-0
John, A.S., Price, C.P.: Existing and emerging technologies for point-of-care testing. Clin. Biochem. Rev. 35, 155 (2014)
Jolly, P., Wong, L.C.C., Miodek, A., et al.: A simple and highly sensitive electrochemical platform for detection of MicroRNAs. In: 2015 IEEE SENSORS—Proceedings (2015). https://doi.org/10.1109/ICSENS.2015.7370378
Joung, J., Ladha, A., Saito, M., et al.: Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 383, 1492–1494 (2020). https://doi.org/10.1056/NEJMC2026172/SUPPL_FILE/NEJMC2026172_DISCLOSURES.PDF
Jurado-Sánchez, B., Moreno-Guzmán, M., Perales-Rondon, J.V., Escarpa, A.: Nanobiosensors for food analysis. In: Handbook of Food Nanotechnology, pp. 415–457 (2020). https://doi.org/10.1016/B978-0-12-815866-1.00011-X
Kumar, M.S., Nandeshwar, R., Lad, S.B., et al.: Electrochemical sensing of SARS-CoV-2 amplicons with PCB electrodes. Sens. Actuators, B Chem. 343 (2021). https://doi.org/10.1016/j.snb.2021.130169
Kumar, V., Brent, J.R., Shorie, M., et al.: Nanostructured aptamer-functionalized black phosphorus sensing platform for label-free detection of myoglobin, a cardiovascular disease biomarker. ACS Appl. Mater. Interfaces. 8, 22860–22868 (2016). https://doi.org/10.1021/ACSAMI.6B06488/SUPPL_FILE/AM6B06488_SI_001.PDF
Lam, T., Devadhasan, J.P., Howse, R., Kim, J.: A chemically patterned microfluidic paper-based analytical device (C-µPAD) for point-of-care diagnostics. Sci. Rep. 7(1), 1–10 (2017). https://doi.org/10.1038/s41598-017-01343-w
Law, J.W.F., Mutalib, N.S.A., Chan, K.G., Lee, L.H.: Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 5, 770 (2014). https://doi.org/10.3389/FMICB.2014.00770/BIBTEX
Layqah, L.A., Eissa, S.: An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta 186, 1–10 (2019). https://doi.org/10.1007/S00604-019-3345-5/TABLES/1
Mandal, M., Dutta, N., Dutta, G.: Aptamer-based biosensors and their implications in COVID-19 diagnosis. Anal. Methods 13, 5400–5417 (2021). https://doi.org/10.1039/D1AY01519B
Monto, A.S., Cowling, B.J., Peiris, J.S.M.: Coronaviruses. In: Viral Infections of Humans, p. 199 (2014). https://doi.org/10.1007/978-1-4899-7448-8_10
Ni, W., Yang, X., Yang, D., et al.: Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care (London, England) 24 (2020). https://doi.org/10.1186/S13054-020-03120-0
Ning, B., Yu, T., Zhang, S., et al.: A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv. 7 (2021). https://doi.org/10.1126/SCIADV.ABE3703/SUPPL_FILE/ABE3703_SM.PDF
Parrilla, M., Cánovas, R., Jeerapan, I., et al.: A textile-based stretchable Multi-Ion potentiometric sensor. Adv. Healthcare Mater. 5, 996–1001 (2016). https://doi.org/10.1002/ADHM.201600092
Payne, S.: Family Coronaviridae. Viruses 149 (2017). https://doi.org/10.1016/B978-0-12-803109-4.00017-9
Phan, L.M.T., Tieu, M.V., Pham, T.T., Cho, S.: Clinical utility of biosensing platforms for confirmation of SARS-CoV-2 infection. Biosensors 11, 167 (2021). https://doi.org/10.3390/BIOS11060167
Raghav, S., Ghosh, A., Turuk, J., et al.: Analysis of Indian SARS-CoV-2 genomes reveals prevalence of D614G mutation in spike protein predicting an increase in interaction with TMPRSS2 and virus infectivity. Front. Microbiol. 11, 2847 (2020). https://doi.org/10.3389/FMICB.2020.594928/BIBTEX
Rahimi, A., Mirzazadeh, A., Tavakolpour, S.: Genetics and genomics of SARS-CoV-2: a review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics 113, 1221–1232 (2021). https://doi.org/10.1016/J.YGENO.2020.09.059
Rashed, M.Z., Kopechek, J.A., Priddy, M.C., et al.: Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. A R T I C L E I N F O
Ravi, N., Cortade, D.L., Ng, E., Wang, S.X.: Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 165, 112454 (2020). https://doi.org/10.1016/J.BIOS.2020.112454
Ribeiro, B.V., Cordeiro, T.A.R., e Freitas, G.R.O., et al.: Biosensors for the detection of respiratory viruses: a review. Talanta Open 2, 100007 (2020). https://doi.org/10.1016/J.TALO.2020.100007
Seo, G., Lee, G., Kim, M.J., et al.: Rapid detection of COVID-19 Causative Virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14, 5135–5142 (2020). https://doi.org/10.1021/ACSNANO.0C02823/SUPPL_FILE/NN0C02823_SI_001.PDF
Shao, W., Shurin, M.R., Wheeler, S.E., et al.: Rapid detection of SARS-CoV-2 antigens using high-purity semiconducting single-walled carbon nanotube-based field-effect transistors. ACS Appl. Mater. Interfaces. 13, 10321–10327 (2021). https://doi.org/10.1021/ACSAMI.0C22589/SUPPL_FILE/AM0C22589_SI_001.PDF
Singh, B., Datta, B., Ashish, A., Dutta, G.: A comprehensive review on current COVID-19 detection methods: From lab care to point of care diagnosis. Sens. Int. 2, 100119 (2021). https://doi.org/10.1016/J.SINTL.2021.100119
Team, E.: Materials for microfluidic chips fabrication: a review 2017. Elveflow (2021)
Vadlamani, B.S., Uppal, T., Verma, S.C., Misra, M.: Functionalized TiO2 nanotube-based electrochemical biosensor for rapid detection of SARS-CoV-2. Sensors 20, 5871 (2020). https://doi.org/10.3390/S20205871
Wang, M., Tong, Y., Li, Y., et al.: Nanopore targeted sequencing for the accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses. Small 16, 2002169 (2020). https://doi.org/10.1002/SMLL.202002169
Wessner, D.R.: Origin of Viruses. In: Nature Education. https://www.nature.com/scitable/topicpage/the-origins-of-viruses-14398218/ (2010). Accessed 30 Jan 2022
World Health Organization: Novel Coronavirus (2019-nCoV). https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf?sfvrsn=20a99c10_4 (2020). Accessed 30 Jan 2022
Wu, C.C., Huang, W.C., Hu, C.C.: An ultrasensitive label-free electrochemical impedimetric DNA biosensing chip integrated with a DC-biased AC electroosmotic vortex. Sens. Actuators B Chem. 209, 61–68 (2015). https://doi.org/10.1016/J.SNB.2014.11.078
Wu, F., Zhao, S., Yu, B., et al.: A new coronavirus associated with human respiratory disease in China. Nature 579(7798), 265–269 (2020). https://doi.org/10.1038/s41586-020-2008-3
Xu, G., Abbott, J., Qin, L., et al.: Electrophoretic and field-effect graphene for all-electrical DNA array technology. Nat. Commun. 5(1), 1–9 (2014). https://doi.org/10.1038/ncomms5866
Zhao, H., Liu, F., Xie, W., et al.: Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens. Actuators B Chem. 327, 128899 (2021). https://doi.org/10.1016/J.SNB.2020.128899
Acknowledgements
Authors gratefully acknowledge the Start-Up Research Grant (SRG) funded by Science & Engineering Research Board (SERB) (SRG/2020/000712), Department of Science and Technology (DST)(Government of India, Ministry of Science and Technology, (Technology Development and Transfer, TDP/BDTD/12/2021/General), Indo-German Science & Technology Centre (IGSTC) (IGSTC/Call 2019/NOMIS/22/2020-21/164), and Institute Scheme for Innovative Research and Development (ISIRD) (IIT/SRIC/ISIRD/2019–2020/17), Indian Institute of Technology Kharagpur (IIT Kharagpur), India, for the financial support.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Titus, R., Mandal, M., Dutta, G. (2023). Electrochemical Biosensor Designs Used for Detecting SARS-CoV-2 Virus: A Review. In: Dutta, G., Biswas, A. (eds) Next Generation Smart Nano-Bio-Devices. Smart Innovation, Systems and Technologies, vol 322. Springer, Singapore. https://doi.org/10.1007/978-981-19-7107-5_10
Download citation
DOI: https://doi.org/10.1007/978-981-19-7107-5_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-7106-8
Online ISBN: 978-981-19-7107-5
eBook Packages: EngineeringEngineering (R0)