Abstract
This chapter addresses the adverse effect of heat stress on plant growth, genes associated with heat stress tolerance and adaptive strategies that can be used to create heat-tolerant plants. CRISPR/Cas9 seems a promising approach regarding stress tolerance. The modified versions of CRISPR/Cas9 like CRISPRi, CRISPRa, base editing and CRISPR multiplexing offers more and more specificity and advanced editing options and minimizes the off-target effect. The versatility of CRISPR/Cas9 brings a new revolution in the field of plant science to alleviate abiotic stress like heat stress.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
13.1 Introduction
In view of the ever-growing population, agricultural biotechnology has offered tremendous potential to overcome conventional methods of crop improvement, crop protection, quality management and improving other agronomical traits against several stresses. In recent year, biotic and abiotic stresses are being the limiting factors that significantly affect crop yield and quality. There is a need to enhance food production with nutritional qualities that can possibly help to combat malnutrition in developing countries. This can be achieved by improving upon qualitative and quantitative traits of crop plants by adopting new analytical tools and technologies. Plant researcher are accountable to discover the frontiers of the natural biosphere and their fundamental mechanisms like genome editing which may improve lifestyle of human being.
Global agricultural production is facing unprecedented challenges due to climate changes. The world population by 2050 will reach 9.6 billion, aftermath the demand for staple food will have increased by 60%. Meanwhile, the swiftness of increasing yield caused by the revolution has been steadily declining. Unfavourable global climate change is foreseen to further limit plant production, thus cultivars with enhanced resilience to drastic environmental situations and with increased yield and improved quality need to be generated. However, the strategies used traditionally for crop breeding are very laborious, complicated and time-consuming methods of transferring desired traits into a superior cultivar. To tackle abiotic stress, different combinations of chemicals are used that may have adverse effects on human health and the environment, which may also develop chemical resistance in insects and weeds. Henceforth more-effective and time-saving plant breeding methods are required. Thus, breeding climate-smart crops that can tolerate various abiotic stresses such as heat stress, drought and salinity would be a sustainable approach to cope with such challenges. The most eco-friendly approach to cope with the challenge of abiotic stress is breeding tolerant cultivars. The use of recent advances in genome editing technologies assure new opportunities for improvement of crop by employing precision editing for target traits (Driedonks et al. 2016).
13.2 What Is Genome Editing and Why It Is Needed?
The recent few years have witnessed enormous excitement with the discovery of genome editing. Genome editing is a collection of advanced molecular biology techniques which allows precision, efficient and targeted modifications at genomic loci. The different approaches to editing involve use of site-specific nucleases (ZFN, TALEN, CRISPR/Cas), which create double-strand breaks (DSB) in DNA. All these genome editing techniques use a sequence-specific nuclease that allows to identify the target DNA sequence, once the target DNA sequence is identified a double-stranded break (DSB) is created. After the creation of a double-stranded break, the endogenous repair systems fix them by one of the two approaches that are Non-Homologous End Joining (NHEJ) and Homologous Direct Repair (HDR). In case of NHEJ, there is insertion or deletions of nucleotide causing gene knockouts while in case of HDR there is reconstruction of the cleaved DNA with the use of template DNA analogue to the break site sequence. However, out of the three nucleases, CRISPR/Cas9 attracted the maximum attention for developing several plant and animal products with desired genetic modifications through genome editing. Soon an alternative for Cas9 in the form of Cpf1 became available which paves the way to a superior system in the form of CRISPR/Cpf1, and has several advantages over CRISPR/Cas. ZFN/TALEN/CRISPR-mediated genome editing has been an approach that is preferred over transgenics, as no foreign gene is being introduced, and only an existing gene is altered or edited, using cell own machinery. Therefore, it has been largely debated point that products of genome editing technologies like CRISPR/Cas9 should not be subjected to the regulatory system, which is typically used in case of genetically modified organisms (GMOs). At least in some countries, this has made commercialization of genome-edited products easier. As an example, a strain of ‘mushroom’ with white buttons, which cannot turn brown (when stored) was developed using CRISPR and commercialized in the USA without being subjected to regulatory systems that are commonly applied to GMOs. In this, a gene PPO (polyphenol oxidase) which is responsible for browning was altered which results in reduction of PPO quantity by 30%. A mutant waxy corn that gave higher yield under drought conditions has also been developed through genome editing by DuPont, the same genome-edited waxy corn was also approved in the USA for commercial cultivation and may become available to the farmers for commercial cultivation within the next few years. CRISPR/Cas9 creates an opportunity to enhance productivity by creating genetic variability for breeding purpose, supply of disease-free and healthy planting material, improvement in different stress tolerance etc. CRISPR approach has been proved to give a number of options to provide resistance against different biotic and abiotic stresses and is used to create tolerant crops. This technique has been quite successfully used for creating resistant crop plants. Amidst all the issues raised against genetically modified crops, it is imperative to highlight the scientific principles involved so as to make full use of a technology that might solve the problem of food shortage.
CRISPR/Cas9 is a natural inspiration, one of the most powerful tools available which is derived from natural products. Originally, it is bacterial adaptive immune system used in their defence mechanism. In last few years, CRISPR/Cas9 captivated researchers globally by creating several plant and animal products with desirable genetic modifications through genome editing. The studies first recognized Cas9 as a large multifunctional protein which have two putative nuclease domains namely HNH and RuvC that are responsible for introducing DSBs into invading phages (Jinek et al. 2014) and plasmid enables in vivo targeting of temperate phages and plasmids in bacteria (Garneau et al. 2010). This defence system inserts or deletes bases, which turn DNA code into knockout situation of the targeted gene.
The popular CRISPR/Cas9 system for genome editing makes use of Cas9 having endonuclease activity for creating a double-strand break (DSB) at the target site of the target DNA strand. The target site is recognized with the help of a single-guide RNA (sgRNA), which is programmable and is designed using the target sequence that is intended to be edited. The sgRNA consists of a scaffold sequence that is of ⁓20 bp length and facilitates DNA binding to Cas9. CRISPRs thus have two main components such as nuclease Cas9 acting as a molecular scalpel and synthetic single-guide RNA (sgRNA). sgRNA is a complex of crRNA and trRNA in which trRNA is required for maturation of pre-crRNA to crRNA. Thus, sgRNA works in sense of a GPS system that guides the Cas9 to the exact target site of cleavage. Together, sgRNA–Cas9 complex creates site-specific double-strand break. Along with these components for genome editing PAM sequence is needed, in the absence of PAM, genome editing may not take place. The sgRNA is used to design in such a manner that it should lie upstream of a protospacer adjacent motif, 5′-NGG-3′ (PAM sequence). Cas9 and SgRNA together attach to a specific stretch of DNA bases due to complementary base pairing between one of the target strand and crRNA, endonuclease activity of Cas9 causes a cut in the double helix. Once DSB is formed the cellular repair mechanism tries to fix loss by rejoining the cut DNA ends, either by NHEJ or HDR. NHEJ competes with the preferred HDR-dependent genome editing, and creates a high frequency of indels and off-site alterations during genome editing. But HDR works poorly unless cells are dividing, which means this strategy does not function in cells such as brain and muscle cells that no longer copy themselves.
13.3 Food Security
Climate change causes substantial risks to food production and global food security. There is an adverse impact on agricultural production due to extreme weather and the impact of extreme weather is likely to become more frequent in upcoming years which provides additional challenges to farmers to increase productivity for increasing population within the less available area of land. Climate change brings a Cascade of risks from physical impacts to ecosystems, agroecosystems, agriculture production, food chains, income and trade with economic and social impacts on livelihoods and food security and nutrition (FAO 2015). Food security exists when all people, at all times, have physical and economic access to sufficient, safe and nutritious food that meets their dietary needs and food preferences for an active and healthy life (World Food Summit 1996).
The above definition gives four dimensions of food security: availability of food, accessibility (economically and physically), utilization (the way it is used and assimilated by the human body) and stability of these three dimensions.
The key to increase crop productivity despite climate change is the ability to adapt to alternative climates, enabling expansion of cultivation and yield resilience using modern genome editing tools and technology. The relationship between climate-related events and vulnerability to food insecurity worldwide in developing and least-developed countries is shown in Fig. 13.1.
The relationship between climate-related events and vulnerability to food insecurity worldwide in developing and least-developed countries. The vulnerability to food insecurity at present day is shown on the map at the bottom of the poster. Future projections are shown for a range of scenarios of different future global greenhouse gas emissions and adaptation levels. This shows that with both adaptation and mitigation, it is possible to successfully tackle the impact of climate change on future food insecurity. Source: United Nations World Food Programme 2015, Food Insecurity and Climate Change Map
13.4 Engineered Crops Through Advanced Plant Breeding Approach
We know that in crop improvement programme, there are varieties of approaches used in plant breeding which has done an excellent job in the last 100 years for improvement of crop. Plant breeding arises with domestication and after that selection. In this, we were utilizing only those variable genotypes which are present in nature but to fulfil hunger of increasing population we need more variability to create a high-yielding resistant variety of crops. Then we came to hybridization breeding, in this we believe natural hybridization. We were using only those genotypes which are naturally compatible for hybridization. But that was not enough to fulfil increasing hunger. The researchers started studying artificial hybridization or wide hybridization but as we know that breeding approaches are time consuming. Then to overcome time barrier scientists moved towards mutation breeding. The results were really appreciating because a lot of variabilities we were getting using mutation breeding. But the drawback of mutation breeding method is sometimes mutation cannot be heritable or undesirable mutations can take place. Then to increase the efficiency scientists came to transgenic breeding. The beauty of transgenic breeding is that the desirable clone gene can transfer from any source to the host regardless of origin. But there are drawbacks to transgenic breeding also. Products of transgenic breeding called GMOs have to go through a lot of regulatory processes, in most countries GMOs are banned to prevent unpredictable risks to environment and food safety.
Therefore, the era of genome editing emerged massively. Genome editing is a type of engineering plants in which DNA is inserted, deleted or modified or replaced within plant genome. Spotlight came on genome editing in 2005, ZNF: Zinck Finger Nucleases. In 2010 TALEN: transcription activator-like effector nucleases and in 2013 CRISPR/Cas9 came. CRISPR/Cas9 is the most widely used genome editing tool to date. But some drawbacks are there in CRISPR/Cas9 too, to overcome these drawbacks of CRISPR the improved technology that is Base Editing has emerged in 2016.
Ethical and biosafety issues involving genetically modified organisms (GMOs) have been the subject of discussion for the last almost three decades. Consequently, GMOs have faced stringent regulations globally, thus restricting commercialization of many products that were developed. However, recently it has been argued that the products of gene editing following CRISPR technology should not be subjected to the same regulatory restrictions, which are used for GMOs. This argument has been accepted in America, so that the products of gene editing by CRISPR in America no longer require to undergo the regulations that are required for GMOs. Consequently, the issue has been examined globally, particularly in America (USA, Canada, Argentina) and Europe. In the USA, these products no longer require regulatory clearance like GMOs, but in Europe, the highest court recently decided that the gene-edited crops should be subject to the same stringent regulations that are used for GMOs. It is not surprising, since Europe has always been conservative in dealing with the subject of the release of GMOs. One would expect that the products of base editing (discussed in this chapter) would be treated like any other mutant product of conventional breeding, since only a single base is altered (as in base substitution mutants).
The cultivars with enhanced resilience to adverse environments and with increased yields and improved quality need to be generated. However, the traditional strategies used for crop breeding are laborious, time consuming and complicated, thus more effective and time-saving breeding methods are required. With the rapid progress in sequencing technologies, genomic information on an ever-increasing number of plant species is becoming available, and genome editing systems are offering the chance to edit genes with precision and creating new opportunities for crop improvement. Different strategies to generate improved crop varieties are shown in Fig. 13.2.
13.4.1 CRISPR-Mediated Genome Editing: The Evolution of Site-Specific Nucleases
Since the discovery of double-stranded DNA helix, the technologies for manipulating DNA have enabled advances in biology. Among these introducing site-specific modifications in the genomes of cells and organisms remain elusive. Recently, the site-directed Zinc Finger Nucleases (ZFNs) and TAL effector nucleases (TALENs) using the principles of DNA protein recognition were developed.
Difficulties in protein design, synthesis and validation have become a barrier to widespread adoption for routine use of these engineered nucleases. We have seen that field of biology is going through transformative phase with the arrival of facile genome engineering in animals and plants with the use of RNA-programmable CRISPR-Cas9. The CRISPR/CAS9 technology is originated from type II CRISPR-Cas system, which provides an adaptive immune system to bacteria against viruses and plasmids. Cas9 is CRISPR-associated protein having endonuclease activity. Cas 9 uses a guide sequence within an RNA duplex, tracrRNA: crRNA, which forms complementary base pairing with the target DNA sequence, enabling Cas9 to introduce a double-stranded break. Single-guide RNA (sgRNA) has two critical features: one is the sequence at the 5′ end—determines the target DNA site by complementary base pairing and the second is the sequence at 3′ side which binds to Cas9 (Doudna and Charpentier 2014).
CRISPR was first described in 1987 by Japanese researchers as a series of short direct repeats interspaced with short sequences in the genome of Escherichia coli (Ishino et al. 1987). Later CRISPRs were detected in numerous bacteria and archaea and predictions were made on the role of CRISPRs in DNA repair or gene regulation (Makarova et al. 2002). In the year 2005, observations were made that many spacer sequences within the CRISPRs are derived from plasmid and viral origin. Along with the findings that CRISPR loci are transcribed and with the observation that Cas (CRISPR-associated) genes encode proteins having putative nuclease and helicase domains, it was proposed that CRISPR-Cas is an adaptive defence system that might use antisense RNA as memory signatures of past invasions (Makarova et al. 2006). The experiment conducted in 2007 in which the infection of lactic acid bacterium Streptococcus thermophilus with lytic phages provided the first evidence of CRISPR/Cas-mediated adaptive immunity (Barrangou et al. 2007). The finding of CRISPR/Cas-mediated adaptive immune system led an idea that CRISPR/Cas systems which exist naturally in cultured bacteria used in dairy industry can be utilized for immunization against phages this is the first successful application of CRISPR/Cas for biotechnological purposes (Barrangou and Horvath 2012). In the year 2008, it was noted that mature CRISPR RNAs (crRNAs) acts as guide in a complex with Cas proteins to interfere with virus proliferation in E. coli (Brouns et al. 2008). In the same year, the DNA targeting activity of the CRISPR/Cas system was found in Staphylococcus epidermidis (Marraffini and Sontheimer 2008). Functional CRISPR/Cas loci consist of a CRISPR array which are identical repeats intercalated between invader DNA targeting spacers which later encode the crRNA components and also an operon of Cas genes encoding the Cas protein components. Naturally, viruses can be matched to their bacterial or archaeal hosts by detecting CRISPR spacers (Andersson and Banfield 2008). Different studies depicted that viruses are evolving constantly to overcome CRISPR-mediated attenuation.
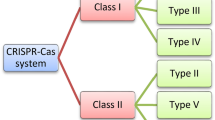
CRISPR/Cas immune response is usually divided into three distinct groups, i.e. Adaptation, Expression and Interference. CRISPR/CAS systems are extremely diverse and are classified into 2 classes, 6 types, and 19 subtypes (Makaroya et al. 2015). Despite this diversity, there is a common feature in all systems and that is a CRISPR locus with alternative repeats and spacer structures and a set of associated Cas genes. There are two proteins that are present almost in all CRISPR/Cas systems and they are Cas 1 and Cas 2. Cas 1 and Cas 2 are functions in the adaptation phase.
There are two CRISPR/Cas classes which are divided into three types each. Class 1 consist of type I, III and IV, and Class 2 consist of types II, V and VI. Each type of CRISPR system has distinct architectures of the effector modules that include unique signature proteins. Each type is further divided into multiple subtypes. There is a difference in the mechanism of pre-crRNA processing in Class 1 and Class 2 CRISPR/Cas systems. In case of Class 1 system, the maturation of crRNA is catalyzed by a dedicated complex of multiple Cas proteins which was first identified in subtype I-E and designated cascade (CRISPR-associated complex for antiviral defence). Binding takes place between cascade complex and pre-crRNA and additional Cas protein, Cas 6 is recruited, which have nuclease activity and is responsible directly for processing. In case of type II systems, an external bacterial enzyme, RNAse III catalyzed the prototype of Class 2, with the help of an additional RNA species, the trans-acting CRISPR RNA (trRNA), encoded within the CRISPR/CAS locus. trRNAs were also found in subtype V-B systems, but the cleavage, in this case, remains uncharacterized. In case of types V and VI, incompletely characterized nuclease activity of the same large effector protein that is involved in target cleavage catalyzed the pre-crRNA processing (Koonin and Makarova 2019).
13.5 Strategies to Design Abiotic Stress-Tolerant Plants with CRISPR Technologies
Nowadays, CRISPR/Cas9 is used in developing abiotic stress-tolerant plants. It is possible to target the abiotic stress tolerance mechanisms by CRISPR/Cas9 system as it allows CRISPRi (CRISPR interference) and CRISPRa (CRISPR activation) of genes (Zafar et al. 2020). So, CRISPR/CAS9 can be used for activating tolerant genes as well as suppressing sensitivity genes. CRISPR multiplexing and base editing can also be implemented to design abiotic stress-tolerant plants (Fig. 13.3).
CRISPRi
The unique property of Cas9 to bind DNA at sites recognized by the sgRNA sequence and the PAM permits its application beyond permanent modifications of DNA. A catalytically inactivated version of Cas9 called as dCAS9 can be utilized for targeted gene regulation on a genome-wide scale. This strategy is well known as CRISPR interference (CRISPRi). CRISPRi functions by directly blocking transcription and thus causing gene silencing or suppressing the expression of a particular gene. Thus, the CRISPRi system can also be used to knockdown various gene expressions. In CRISPRi some effector domain such as KRAB/SID functions as repressor which binds to dCAS9. CRISPRi is one of the promising platform for modulating gene expression in a broad range of host cells. Presence of complex host factor is not required for CRISPRi instead of that it only depends on dCAS9 protein and guide RNAs and due to this reason, CRISPRi is flexible and highly designable. sgRNA-guided targeting is specific due to specificity dictated by its sequence identity and is not affected by the presence of other sgRNAs this enables regulation of multiple genes simultaneously by CRISPRi. The silencing by CRISPRi is very specific with no detectable off-target effects. CRISPRi can be used efficiently to suppress the susceptible gene for different abiotic stress (Qi et al. 2013).
CRISPRa
In CRISPR/Cas9-based transcriptional activation, i.e. CRISPRa the catalytically inactivated version of Cas9 called as dCAS9 is used which is genetically fused to an activator domain such as VP16/VP64. Dead Cas9 (dCas9) also called as nuclease null Cas9 fuses with an effector domain which is an activator and allows users to precisely direct a given functional activity to any random locus within the genome (Chavez et al. 2012).
Base Editing
Base editing offers precision-targeted nucleotide editing without requiring double-stranded break or donor DNA template and does not rely on HDR. Due to CRISPR repair mechanism, Liu’s and co-workers in 2016 made changes in CRISPR’s tool kit by modifying Cas enzyme known as base editors, they fused sgRNA with a dead Cas9 (dCas9). Now disable dead Cas9 (dCas9) is unable to cut whole double helix but still unzip at appropriate spot. Cas9 to dCas9 (Asp10Ala, His840Ala) repair mechanism is Base excision Repair (BER) that undo the change which means reverting U to the original G base due to U glycolase enzyme due to this lacuna. The disabled Cas9 with nickase activity together with cytosine/adenine deaminases for the event of four generations of cytosine base editors (BE1–BE4) for C → U conversion and a minimum of seven generations of adenine base editors (ABE1– ABE7) for A → I conversion. These base editors exhibited improved efficiency and reduced frequency of deletions among the products (Gupta 2019). The base editing technology will bring precision to gene editing technology for crop improvement. Further dCas9 is modified into nCas9, the nCas9 also known as NICKASE snips the unedited strand which gooses the cell DNA Mismatch Repair (MMR) mechanism and converts C:G to U:G to UA to T:A (Yan et al. 2018; Gupta 2019).
13.6 Heat Stress: Impact on Crop Production
Heat stress causes adverse effects on the performance of yield of the crop. High-temperature shocks at the reproductive phase can cause a drastic reduction in yield of cereals in temperate regions. There is an adverse effect on the quality of final produce like oil, starch and protein in cereals and oilseed crops due to heat stress. Reduction in grain weight and total number of grain was reported due to elevated temperatures. There is a significant decline in the yield of rice due to temperature stress as it causes a reduction in different rice growth and yield traits. Researchers have reported that tillering stage in rice is very sensitive to elevated temperature. The grain weight of rice is not affected under stress-free environment but in contrast at high night temperature leads to significant reduction in yield of rice per unit area. Drastic yield reduction due to heat stress is reported in case of common bean (Phaseolus vulgaris L.) and peanut (Arachis hypogea L.). Drastic effects of heat stress on the yield performance of tomato (Solanum lycopersicum) are reported as it affects meiosis, fertilization and growth of fertilized embryo (Fahad et al. 2017).
Thus, heat stress affects adversely crops and there is a significant reduction in growth and yield of several important crops. The extent of damage due to heat stress depends on crop stage and severity of stress. Basically, it is found that the reproductive stage is more sensitive to the stresses and causes a drastic reduction in the yield.
13.6.1 Plant Response to Heat Stress and Adaptive Strategies
The Global agricultural production is facing challenges and in future will have to face unescapable demand due to unpredictable environments, specially heat stress. Heat stress is among one of the major abiotic factors that affect plant growth, development and yield. Rise in temperature persistently above optimal for plant growth can induce heat stress and which results in low yield. At some threshold, the effect of heat stress may be lethal. When plants encounter heat stress, the percentage of seed germination, photosynthetic efficiency and yield declines. Under heat stress, during the reproductive growth period, the function of tapetal cells is lost and therefore the anther is dysplastic. In general, heat stress have a negative effect on crop physiology which causes decrease in rate of photosynthesis and increase in the rate of respiration and ultimately affects the plant growth and yield. Plant root system affects negatively due to heat stress which in turn causes adverse effects on nutrient and water uptake and its transfer to various plant parts leading to disrupted pollination, flowering, root development and growth stages. Seed size and quality also have negative impact due to heat stress (Janni et al. 2020).
The excessive emission of greenhouse gases leads to rise in global temperature and it is predicted to be responsible for reducing food grain yield which threats global food security. The negative impact of abiotic stresses is much severe in regions like Africa and South Asia as these regions are already experiencing food insufficiency. Therefore, making climate smart crops is a need of hour to tackle food insecurity and this can be achieved with use of advanced genome editing approach (Zafar et al. 2020).
13.6.2 Strategies for Heat Stress Management
The different molecular-biotechnological approaches are used for the development of heat stress tolerance in plants. Along with biochemical and physiological mechanisms, molecular approaches are advancing to understand the concept of heat tolerance. Plant acquired stress tolerance by modulating multiple genes and by co-ordinating the expression of gene in different pathways. In general, heat stress triggers the upregulation of several heat-inducible genes, commonly referred to as ‘heat shock genes’ (HSGs), these are master players in heat stress tolerance. HSGs encode HSPs and these products are significantly necessary for plant’s survival under fatal high temperatures. The high temperature induces most of those proteins' constitutive expression to protect intracellular proteins from being denaturation hence, preserve their stability with high performance through protein folding, thus act as chaperones. Though plant produces HSPs in certain developmental stages the expression of these proteins is restricted such as in embryogenesis, seed germination and fruit maturation hence it can be the reason for heavy losses to plants in heat stress. Plants give response to heat stress by enhanced expression of heat-shock protein (HSP), other stress-related proteins and production of reactive oxygen species. There are various mechanisms that plant implement to cope with the heat stress some of them are maintenance of membrane stability, scavenging of ROS, production of antioxidants, accumulation and adjustment of compatible solutes, induction of mitogen-activated protein kinase (MAPK) and calcium-dependent kinase (CDPK) cascade, chaperon signalling and transcriptional activation. These are the different mechanism that enables the plants to thrive under heat stress (Wahid et al. 2007).
Thermotolerance counter to heat stress is accomplished in plants transferred with heat-shock regulatory proteins. In most of the plants, HSFs are expressed constitutively; in ordinary conditions, these HSPs proteins exist as a monomer bound to one of the HSP70 within the cytoplasm. Once the plant has sensed a heat stress, nucleus activity for tolerance starts, the HSP70 dissociates from cytoplasmic monomeric HSFs then it enters into the nucleus and forms a trimer that can bind with the HSEs. Upon binding of heat-shock factor, it recruits other transcriptional components, resulting in natural phenomenon within minutes in increased temperature. Since all HSGs contain HSE conserved sequences, overexpression of HSF gene intern turned on most HSGs and consequently provides protection against heat stress. Although this basic system is universal to eukaryotic cells, it is highly complicated in plants. Unlike animals and yeasts, which may have four or fewer HSFs, plants are shown to possess multiple copies of these genes: tomato features a minimum of 17 and Arabidopsis has 21 different HSF genes. These genes are classified into three groups (classes A, B and C), which are discriminated by features of their flexible linkers and oligomerization domains. Many of the HSFs are heat inducible, suggesting that the precise HSF involved in transcription of a selected gene may vary relying on the timing and intensity of the strain. Generally, overexpression of plant HSFs can increase plant’s thermotolerance, but gene knockouts of individual HSFs tested thus far have had little effect on survival at HT. Thus, plants appear to possess a stimulating ability to finely control the expression of heat-induced genes through the HSF system. Some studies also support that there is an immediate correlation between the HSP level within the cell and respective stress tolerance.
13.6.3 Genes Associated with Heat Stress Tolerance
In light of global warming production of plants that are tolerant to heat stress is of immense importance. Plant cells show response to heat stress by the use of genetic machinery present in themselves for survival and reproduction. By altering expression of heat shock protein genes/ factors high temperature tolerance in transgenic plants had been largely achieved. Overexpression of transcription factors such as DREB2A, bZIP28 and WRKY proteins have the potential to impart heat stress tolerance (Table 13.1). Several transcription factors other than HSFs, DREBs and WRKYs have been reported their significant role in high temperature tolerance. Nuclear transcription factor X-box binding 1 gene promotes acquired high-temperature tolerance.
13.6.4 CRISPR-Mediated Approach to Enhance Heat Stress Tolerance
The CRISPR/Cas 9 have the potential to edit gene essential for the development of heat-tolerant crops. Tomato seems an ideal model for testing editing by CRISPR/Cas9 due to its ability to undergo efficient transformation for achieving quality improvements (Pan et al. 2016). CRIPSPR/Cas-mediated editing of the slagamous-like 6 (SLAGL6) gene gives parthenocarpic fruits additional heat tolerance (Klap et al. 2017). With the use of CRISPR/Cas editing tool mutation of the thermosensitive genic male-sterile maize plants had been done (Li et al. 2017). BZR1 overexpressing and CRISPR-bzr1-mutant tomato lines depicted the involvement of BZR1 in thermo-tolerance due to regulation of the Feronia (Fer) homologs (Yin et al. 2018). Due to CRISPR-bzr 1-mutant, there is impaired production of H2O2 in apoplast, reduction in induction of Respiratory Burst Oxidative Homolog 1 (RBOH1), and heat tolerance, in contrast, its overexpression enhances H2O2 production and recovery of thermo-tolerance. The rationale steps that are involved in CRISPR are depicted in Fig. 13.4.
13.7 Limitations and Future Prospects of CRISPR
13.7.1 Limitations
In actual program of editing, it has been noticed that to get desirable product, a variety of products need to be exercised selection process vigorously, which generates only a frequency of not more than 5% of the desirable one. Being such a magical tool for genome editing CRISPR-Cas9 introduces random insertions, translocations, deletions and unwanted base to base conversions, off-target editing which is one of the major limitations associated with this tool.
Off-target Effects
Off-target effect is the most debated criticism in case of CRISPR editing tool. There is a risk of accidentally mutating non-target genes of target organism which can lead to unintended effects on ecosystem. There may be chances of activation of unwanted genes such as disease susceptibility genes due to mutation at unexpected sites. Several strategies have been adopted to reduce off-target risks from Cas9 by optimizing the sgRNA or proper design of sgRNA. Truncated sgRNA use is found effective to reduce the undesired mutations at some of the off-target sites without the sacrifice of on-target genome editing efficiencies (Ding et al. 2016). Improving specificity by minimizing the off-target effects of CRISPR-Cas system have been achieved by adopting the strategies like Cas9 nickase, Cas9n and dCas9, along with careful design and gRNA truncated at 50 ends (trugRNAs) (Osakabe et al. 2016).
13.7.2 Future Prospects
With the use of CRISPR/Cas9 technique precise editing of target gene is possible. Utilization of CRISPR/Cas 9 technology widens the scope of crop improvement for different aspects through genetic manipulation. Site-specific cleavage or site-directed mutagenesis seems to be a boon for crop improvement as it provides more specificity and efficiency. CRISPRs have potential to act on exons or coding sequences and thus can create null alleles, it can also act on regulatory sequences and ORFs which thus leads to enhanced expression. CRISPRs have potential ability to create single or multiple mutations either in homologous or non-homologous regions. Hiring these potential applications of CRISPR researchers have focused on improvement of biotic and abiotic stress tolerance. One of the other important advantages is non-transfer of transgenes to the next generation as they can be excluded with the use of process of segregation leading to the production of transgene-free plants which can be used in further study. The use of CRISPRi, CRISPRa, base editing and CRISPR multiplexing offers more and more specificity and advanced editing options and minimizes the off-target effect. Thus, the versatile technology CRISPR/Cas 9 promises to bring a new revolution in the field of plant science to alleviate abiotic and biotic stresses.
References
Andersson AF, Banfield JF (2008) Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047–1050
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Barrangou R, Horvath P (2012) CRISPR: new horizons in phage resistance and strain identification. Annu Rev Food Sci Technol 3:143–162
Brouns SJJ, Jore MN, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, Oost JVD (2008) Small CRISPR RNAs guide antiviral defense in pro karyotes. Science 321:960–964
Chavez A, Tuttle M, Pruitt BM, Campen BE, Chari R, Ovanesyan DT, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, Housden BE, Perrimon N, Collins JJ, Church G (2012) Comparison of Cas9 activators in multiple species. Nat Methods 13:563–567
Ding Y, Li H, Chen LL, Xie K (2016) Recent advances in genome editing using CRISPR/Cas9. Front Plant Sci 7(703):1–12
Doudna JA, Charpentier E (2014) Genome editing: the new frontier of genome engineering with CRISPR-Cas9. Science 346(6213):1258096
Driedonks N, Rieu I, Vriezen WH (2016) Breeding for plant heat tolerance at vegetative and reproductive stages. Plant Reproduction 29:67–79
Fahad S, Bajwa AA, Nazir U, Anjum SA, Farooq A, Zohaib A, Sadia S, Nasim W, Adkins S, Saud S, Ihsan MZ, Alharby H, Wu C, Wang D, Huang J (2017) Crop production under drought and heat stress: plant responses and management options. Front Plant Sci 8:1147
FAO (2015) Climate change and food security: risks and responses
Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magada AH, Moineau S (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71
Gupta PK (2019) Beyond CRISPR: single base editors for human health and crop improvement. Curr Sci 116:396–357
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169:5429–5433
Janni M, Gulli M, Maestri E, Marmiroli M, Valliyodan B, Nguyen HT, Marmiroli N (2020) Molecular and genetic bases of heat stress responses in crop plants and breeding for increased resilience and productivity. J Exp Bot 71(13):3780–3802
Jinek M, Jiang F, David WT, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Lavarone AT, Charpentier E, Nagales E, Doudna JA (2014) Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343(6176):1247997
Klap C, Yeshayahou E, Bolger AM, Arazi T, Gupta SK, Shabtai S, Usadel B, Salts Y, Barg R (2017) Tomato facultative parthenocarpy results from SLAGAMOUS-LIKE 6 loss of function. Plant Biotechnol J 15(5):634–647
Koonin EV, Makarova KS (2019) Origins and evolution of CRISPR-Cas systems. Phil Trans R Soc B 374:20180087
Li J, Zhang H, Si X, Tian Y, Chen K, Liu J, Chen H, Gao C (2017) Generation of thermosensitve male-sterile maize by targeted knockout of the ZmTMS5 gene. J Genet Genomics 44(9):465–468
Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV (2002) A DNA repair system specific for thermophilic archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res 30:482–496
Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV (2006) A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1:7
Makaroya KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garret RA, Oost JV, Backofen R, Koonin EV (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736
Marraffini LA, Sontheimer EJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845
Osakabe Y, Watanabe T, Sugano SS, Ueta R, Ishihara R, Shinozaki K, Osakabe K (2016) Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci Rep 6(26685):1–12
Pan C, Ye L, Qin L, Liu X, He Y, Wang J, Chen L, Gang L (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep 6:24765
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152(5):1173–1183
Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61:199–223
World Food Summit (1996) Rome Declaration on World Food Security. FAO, Rome
Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, Yang B, Zhou X, Zhou H (2018) Highly efficient a•T to G•C base editing by Cas9nguided tRNA adenosine deaminase in rice. Mol Plant 11:631–634
Yin Y, Qin K, Song X, Zhang Q, Zhou Y, Xia X, Yu J (2018) BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signalling in tomato. Plant Cell Physiol 59(11):2239–2254
Zafar SA, Zaidi SS, Gaba Y, Pareek SLS, Dhankher OP, Li X, Mansoor S, Pareek A (2020) Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J Exp Bot 71(20):470–479
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Mote, G., Jadhav, P., Magar, S., Thakur, P., Moharil, M., Biradar, R. (2022). CRISPR/Cas-Based Genome Editing to Enhance Heat Stress Tolerance in Crop Plants. In: Kumar, R.R., Praveen, S., Rai, G.K. (eds) Thermotolerance in Crop Plants. Springer, Singapore. https://doi.org/10.1007/978-981-19-3800-9_13
Download citation
DOI: https://doi.org/10.1007/978-981-19-3800-9_13
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-3799-6
Online ISBN: 978-981-19-3800-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)