Abstract
Image artifact removal in computed tomography (CT) allows clinicians to make more accurate diagnoses. One method of artifact removal is iterative reconstruction. However, reconstructing large amounts of CT data using this method is tedious, which is why researchers have proposed using filtered back-projection paired with neural networks. The purpose of this paper is to compare the performances of various forms of training data for convolutional neural networks in three low level CT image processing tasks: sinogram completion, Poisson noise removal, and focal spot deblurring. Specifically, modified U-nets are trained with either CT sinogram data or reconstruction data for each of the tasks. Then, the predicted results of each model are compared in terms of image quality and viability in a clinical setting. The predictions show strong evidence of increased image quality when training models with reconstruction data, thus the reconstruction strategy possesses a clear edge in practicality over the sinogram strategy.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Computed tomography
- X-ray
- Sinogram completion
- Poisson noise removal
- Focal spot deconvolution
- Convolutional neural networks
1 Introduction
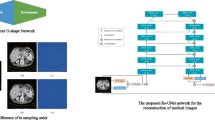
Computed Tomography is a commonly used medical imaging modality due to its advantages over other conventional imaging methods. These advantages include rapid image acquisition, larger amounts of data, and clear, detailed images of internal structures [1]. While CT possesses many advantages over other imaging modalities, a prominent issue that CT medical professionals struggle to deal with are image artifacts, which refer to misrepresentations of the scanned objects. Deep learning techniques such as convolutional neural networks (CNN) have been seeing increased attention in medical imaging [2,3,4,5]. Most of these applications have been focusing on using CNN for classification tasks such as pathology identification [6] and tumor segmentation [7]. However, the application of CNNs in low-level medical imaging tasks is an understudied area. When using CNN for CT, there are two main strategies used for model training due to the fact that CT images have two domains. These two strategies differ because one uses sinogram data3 for training and the other uses reconstruction data. A sinogram is the direct output of the CT detector, which consists of all the X-ray projections taken from different angles (See Fig. 1). The reconstruction is the result after a reconstruction algorithm [8] has been applied on the sinogram, and this three-dimensional image is what allows professionals to analyze the internal structure of objects or patients.
A benchmark of the performance of these two strategies is missing, and the purpose of this study is to compare these strategies through three different CT imaging tasks. The tasks consist of sinogram completion (See second column of Fig. 9), Poisson noise removal (See second column of Fig. 11), and focal spot deblurring (See second column of Fig. 13). These are all common CT artifacts, which lower the image quality and make analysis more difficult. The cause of these artifacts is due to the nature of CT scans requiring the use of X-ray beams. Since X-ray is a form of radiation, exposure to it also increases the likelihood of developing cancer. Therefore, doctors must find a careful balance when selecting the radiation dose. While a high dose produces a high-quality image, it also increases the patient’s chance of developing cancer. On the other hand, having a lower dose reduces the risk for cancer, but it also introduces the aforementioned image artifacts, lowering the image quality and making diagnosis harder. Sinogram completion and Poisson noise artifacts are introduced from lower dose images, while focal spot deblurring comes from the X-ray source itself.
The first task, sinogram completion, stems from a scanning method named sparse-sampling, where the CT scanner produces X-ray beams at fewer angles than a full sample. For example, a sparse sample’s sinogram could include 180 angles around the patient’s body while a full sample includes 360. A full sample sinogram produces a higher quality image than the sparse sampled one, but a sparse sample scan reduces the patient’s radiation exposure by a large margin. Another way to reduce radiation dose is to lower the scanner’s tube current (mA), causing it to emit a lower amount of X-ray photons in each projection, which produces Poisson noise throughout the image. The final task isn’t controlled by the radiation dose; it is due to the size of the X-ray source. An image with no blur requires the source to be infinitely small, which isn’t possible in the real world. Even then, extremely small sources will not be able to handle large amounts of X-ray output due to overheating. Therefore, normal X-ray sources will produce images with a reasonable amount of blur, and thus, the need for deblurring arises. Success of this work will help others understand the benefits and downsides of the two strategies (Fig. 1).
2 Materials and Methods
2.1 Dataset
Experimentation involved the use of sinogram data obtained from the FIPS Open X-Ray Tomographic Datasets. Four sinograms were used, and they were created from four different objects: a walnut, carved cheese, a lotus, and a cross phantom. The walnut data was a 1200 × 2296 sinogram, signifying that it consisted of projections taken from 1200 angles. The rest of the data had a significantly lower amount of projections, with the lotus sinogram having dimensions of 366 × 2240, and the carved cheese and cross phantom being 360 × 2240 (Fig. 2).
2.2 Network Architecture
The model used to conduct this study is a modified U-Net [9], which was initially used for segmentation in biomedical images [10, 11]. The original structure used 2 × 2 max pooling to down sample the input data. One of the modifications involved replacing the max pooling layer with a convolutional layer with a stride of 2. This allows the down sampling process to be learn-able, which improves the overall ability of the network. The second modification was an addition of a residual block in the last layer. This allows the network to learn how to approximate the difference between the input and target and also accounts for the vanishing gradient issue common in networks with many layers. Both of these modifications improve the approximation ability of the model, making it a better fit for regression tasks as opposed to its original segmentation purpose. The architecture of the modified model is shown in Fig. 6. In order to generate input images to train the models for the sinogram domain, image artifacts were simulated over the original sinogram, and the ground truth sinogram was used as the desired output. For the reconstruction domain, the Poisson noise and focal spot deblurring tasks used the reconstruction of the estimated sinogram from the sinogram domain model as input and the ground truth sinogram’s reconstruction as output. The sinogram completion model used the sparse sinogram’s reconstruction as input and the ground truth sinogram’s reconstruction as output, and an explanation of this difference is in the Focal Spot Deconvolution section. Training and testing data was split by using the walnut, carved cheese, and lotus as the training set, while the cross phantom was used as the test set. Since using the entire image data for training would be too computationally expensive and only net four sets of data for training, the model utilizes patch-based training, each patch being a 64 × 64 portion of the entire image. This results in train and test sets consisting of 66,848 and 12,672 patches, respectively. The final predicted result would then be all the patches pieced back together after going through the model. During training, an Adam optimizer with learning rate 0.001 was used, along with a callback that reduced the learning rate by half if no improvement was seen over three epochs. A mean squared loss function calculated the average of how far off each predicted pixel was from the ground truth, its equation given by (Fig. 3): scikit-image
Afterwards, the model was trained for 100 epochs, and the model was able to learn very quickly, as shown by the learning curve (taken from the sinogram completion model that was trained in the sinogram domain) in Fig. 7.
2.3 Image Artifact Simulation
Sinogram Completion.
To generate the input images for this task, a python for loop with a step of 10 was used over the original sinogram, and the input image only contained values at these slices, with the rest of the image having a zero-pixel value. This method successfully reduced the sinogram to only 10% of the original amount of slices (Fig. 4).
Poisson Noise Removal.
In order to add noise to the original images, a python function modeled after [12] was used. The function takes 3 parameters that determine the amount of noise, along with a fourth parameter which is the original low-noise sinogram. The function then returns an array that represents a high-noise version of the original sinogram (Fig. 5).
Focal Spot Deconvolution.
The method introduced in [13] was used for the focal spot deconvolution. The simulation of focal spot deconvolution is very straightforward with the help of the gaussian function from the scikit-image library. The [14] function takes a parameter named σ, which is the standard deviation of the gaussian kernel, to determine the amount of blur that will be applied to the image. The higher the σ the blurrier the image. We used σ = 2 for the input image simulation. Finally, the function returns the desired blurry image that simulates the focal spot blur artifact (Fig. 6).
3 Results
3.1 Sinogram Completion
Looking at the walnut data (Fig. 8), both predicted reconstructions perform well in removing the streak artifacts prevalent throughout the sparse reconstruction. However, looking at the zoomed in portions, the quality at the center of the walnut for the sinogram domain reconstruction has decreased substantially when compared to the ground truth. The reconstruction domain result, on the other hand, is very close to the ground truth, quality wise, and it outperforms the sinogram domain for the walnut data. Without looking closely, it is hard to find noticeable differences between the reconstruction domain image and the ground truth, but when magnifying the center of the walnut, it is clear that the model smooths over intricate details, which could prove troublesome when applied on complex structures like the heart. The remaining data (Fig. 9) followed a similar trend, the predicted reconstructions successfully removed the streak artifacts (with one exception), but the centers of the sinogram domain images are heavily warped, making details and edges of the image difficult to see, which render the images useless for diagnosis in a clinical setting. After inspection, the reconstruction domain achieves better performance than the sinogram domain, maintaining higher quality images that are close to the ground truth. However, it still isn’t the same as a full sinogram due to the bright spots introduced in the cheese and lotus data, and the small streak artifacts still present in the cross phantom data. These issues are small compared to the warping in the sinogram domain and the streak artifacts covering the image in the sparse reconstructions. We can conclude that training in the reconstruction domain works best for the sinogram completion task.
3.2 Poisson Noise Removal
At first, it is difficult to tell the difference between the four cross phantom images (See Fig. 10), but magnifying a portion of the image helps us easily differentiate and compare the results. The noisy reconstruction is covered with tiny black spots that make up Poisson noise, and the noise gives the image a static texture. Although the noise level isn’t extremely high in this experiment, the results can still tell us which strategy works better for denoising. Both models perform very similarly in removing the actual noise artifacts from the image, but the sinogram domain’s result changes the quality of the image, making it blurrier than the reconstruction domain and noisy reconstructions. Because of this, the reconstruction domain strategy outperforms the sinogram domain strategy for the cross phantom data, due to a closer image quality to the ground truth.
Some of the remaining results (Fig. 11) differ from that of the cross phantom. For the walnut data, the two predicted reconstructions appear to be virtually the same, meaning in some instances, the two strategies achieve the same performance. This could be due to the fact that the walnut data makes up a large amount of the training data set, and this shows that the reconstruction domain strategy for denoising works better in cases with less data provided. A similar trend to the cross phantom data is seen in the rest of the images, with the sinogram domain result tending to be a blurrier and lower quality image than the reconstruction domain, but overall, both training strategies successfully remove most of the noise from the image. Again, the reconstruction domain has a better performance than the sinogram domain overall, though in some instances, the two strategies have similar results.
3.3 Focal Spot Deconvolution
The focal spot deconvolution artifact proved to be harder for the models to remove than the other two tasks. In the previous two tasks, the models removed virtually all of the streak artifacts or noise, however, in the deblurring task, blur was still present, even in the best result. At first glance, the blurred reconstruction, the sinogram domain result, and the reconstruction domain result all have a clear difference in quality for the carved cheese (See Fig. 12). The image with nothing done to it is the lowest in quality, while the sinogram domain image is in the middle, and the reconstruction domain image is the highest in quality. Upon magnification of the hole to the left of the “C”, the difference is even more pronounced. The circle’s edges are very hard to define in the blurred reconstruction, and its shape has been slightly altered. The texture of the cheese itself has changed as well, looking smooth instead of textured like the original. The circle shape is easier to see in the predicted results, and the background isn’t as devoid of texture, but the edges are still hard to define. The difference in quality between the blurry and predicted results is clear even without the magnification, but the magnification helps clarify that the predicted results do a lot better at approximating texture, and it makes the difference in quality between the two predicted results clearer.
As for the rest of the results (Fig. 13), the two strategies have little difference in performance for the walnut and lotus data. Both strategies achieve better image clarity than the blurred image, meaning that for these two cases, it doesn’t matter which type of training is performed. The cross-phantom’s results are the same case as the carved cheese, where the sinogram domain was blurrier than the reconstruction domain.
The PSNR values of predicted results, when compared to the original, for all three tasks are shown in Table 1 for the walnut data. They demonstrate the edge that the reconstruction domain has over the other methods, and it verifies the difficulty for the models to remove the blur artifact. The lotus, cheese, and cross phantom values followed similar trends, and they were omitted to avoid redundancy.
4 Conclusions
In this work, the performance of different CNN training strategies in medical CT was compared through three common low level image-processing tasks, and the target of the study to benchmark the different strategies in terms of image quality and clinical viability was successfully met. The sinogram domain results under-performed the majority of the time, and its predicted reconstructions for the sinogram completion task were not even used as training input for the reconstruction domain due to the fact that the results achieved by using the sparse reconstruction as training input were better. On the contrary, the reconstruction domain never performed worse than the sinogram domain, making it the better strategy to pick when training a model for CT. However, a surprising outcome was that the two strategies had the same performance in some cases. Because the reconstruction domain’s input has already been through a model, it should already have a slight advantage over the sinogram domain, but in most of the walnut results and the lotus results for focal spot deconvolution, the two strategies had virtually no difference in performance. This interesting result shows that in a few cases, the extra step included in the reconstruction domain wasn’t necessary, and the less computationally expensive strategy, he sinogram domain, achieved the same results. In conclusion, this paper successfully bench-marked and compared the two main training strategies, and as a result discovered that the reconstruction domain strategy consistently achieves higher performances than the sinogram domain, with few exceptions.
References
Bushberg, J.T., Boone, J.M.: The essential physics of medical imaging. Lippincott Williams and Wilkins (2011)
Maier, J., et al.: Focal spot deconvolution using convolutional neural networks, vol. 03, p. 25 (2019)
Lee, H., Lee, J., Kim, H., Cho, B., Cho, S.: Deep-neural-network-based sinogram synthesis for sparse-view ct image reconstruction. IEEE Trans. Radiation Plasma Med. Sci. 3(2), 109–119 (2019)
Lin, W.A., et al: Dudonet: Dual domain network for ct metal artifact reduction (2019)
Zhao, Z., Sun, Y., Cong, P.: Sparse-view ct reconstruction via generative adversarial networks. In: 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC), pp. 1–5 (2018)
Bar, Y., Diamant, I., Wolf, L., Greenspan, H.: Deep learning with non-medical training used for chest pathology identification. In: Medical Imaging 2015: Computer-Aided Diagnosis. International Society for Optics and Photonics, vol. 9414, p. 94140V (2015)
Iqbal, S., Ghani, M.U., Saba, T., Rehman, A.: Brain tumor segmentation inmulti-spectral mri using convolutional neural networks (cnn). Microscopy Res. Tech. 81(4), 419–427 (2018)
Lucas, L., et al.: State of the art: iterative ct reconstruction techniques. Radiology, 276(2), 339–357 (2015) PMID: 26203706
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation (2015)
Hartmann, A., et al.: Bayesian u-net for segmenting glaciers in sar imagery, (2021)
Bandyopadhyay, H., Dasgupta, T., Das, N., Nasipuri, M.: A gated and bifurcated stacked u-net module for document image dewarping (2020)
Zeng, D., et al.: A simple low-dose x-ray ct simulation from high-dose scan. IEEE Trans. Nucl. Sci. 62(5), 2226–2233 (2015)
Sawall, S., Backs, J., Kachelrieß, M., Kuntz, J.: Focal spot deconvolution using convolutional neural networks. In: Medical Imaging 2019: Physics of Medical Imaging International Society for Optics and Photonics, vol. 10948, p. 109480Q (2019)
van der Walt, S.J., et al.: Scikit-image: image processing in python. PeerJ, 2, e453 (2014)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Huang, A. (2021). Comparing Different CNN Training Strategies in Low-Level CT Image-Processing Tasks. In: Cao, W., Ozcan, A., Xie, H., Guan, B. (eds) Computing and Data Science. CONF-CDS 2021. Communications in Computer and Information Science, vol 1513. Springer, Singapore. https://doi.org/10.1007/978-981-16-8885-0_7
Download citation
DOI: https://doi.org/10.1007/978-981-16-8885-0_7
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-8884-3
Online ISBN: 978-981-16-8885-0
eBook Packages: Computer ScienceComputer Science (R0)