Abstract
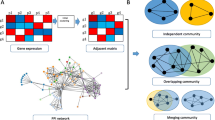
Liver hepatocellular carcinoma (LIHC) is the most common primary malignancy of the liver and is one of the primary contributors to cancer-related death worldwide. The present work proposed a computational framework to discover functional gene communities in LIHC by integrating RNASeq gene expression profiles with protein-protein interaction data to elucidate the inherent complexities of biomolecular mechanisms in LIHC. Here, we have proposed an offsetNMF-based module integration technique that incorporates characteristics of both gene co-expression modules discovered through a refined WGCNA-based algorithm and protein complexes predicted through a parameter-free greedy approximation algorithm PC2P. Biological significance analysis of the integrated gene communities discovers several highly LIHC-associated pathways.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Nat Precedings 1–1
Arista-Nasr J, Fernández-Amador JA, Martínez-Benítez B, de Anda-González J, Bornstein-Quevedo L (2010) Neuroendocrine metastatic tumors of the liver resembling hepatocellular carcinoma. Annals Hepatol 9(2):186–191
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25–29
Badea L (2008) Extracting gene expression profiles common to colon and pancreatic adenocarcinoma using simultaneous nonnegative matrix factorization. In: Biocomputing. World Scientific, pp 267–278
Bai KH, He SY, Shu LL, Wang WD, Lin SY, Zhang QY, Li L, Cheng L, Dai YJ (2020) Identification of cancer stem cell characteristics in liver hepatocellular carcinoma by WGCNA analysis of transcriptome stemness index. Cancer Med 9(12):4290–4298. https://doi.org/10.1002/cam4.3047
Botía JA, Vandrovcova J, Forabosco P, Guelfi S, Sa D, Hardy K, Lewis J, Ryten CM, Weale M (2017) An additional k-means clustering step improves the biological features of WGCNA gene co-expression networks. BMC Syst Biol 11(1):1–16
Charrad M, Ghazzali N, Boiteau V, Niknafs A (2014) NbClust: an R package for determining the relevant number of clusters in a data set. J Stat Softw 61(6):1–36
Ding C, He X, Simon HD (2005) On the equivalence of nonnegative matrix factorization and spectral clustering. In: Proceedings of the 2005 SIAM international conference on data mining. SIAM, pp 606–610
Edwards AW, Cavalli-Sforza LL (1965) A method for cluster analysis. Biometrics 362–375
Friedman HP, Rubin J (1967) On some invariant criteria for grouping data. J Am Stat Assoc 62(320):1159–1178
Gu Y, Li J, Guo D, Chen B, Liu P, Xiao Y, Yang K, Liu Z, Liu Q (2020) Identification of 13 key genes correlated with progression and prognosis in hepatocellular carcinoma by weighted gene co-expression network analysis. Front Genet 11:153. https://doi.org/10.3389/fgene.2020.00153
Hossain SMM, Halsana AA, Khatun L, Ray S, Mukhopadhyay A (2021) Discovering key transcriptomic regulators in pancreatic ductal adenocarcinoma using Dirichlet process Gaussian mixture model. Sci Rep 11(1):7853. https://doi.org/10.1038/s41598-021-87234-7
Hossain SMM, Khatun L, Ray S, Mukhopadhyay A (2021) Identification of key immune regulatory genes in hiv-1 progression. Gene 792:145735. https://doi.org/10.1016/j.gene.2021.145735
Hossain SMM, Mahboob Z, Chowdhury R, Sohel A, Ray S (2016) Protein complex detection in PPI network by identifying mutually exclusive protein-protein interactions. Procedia Comput Sci 93:1054–1060. https://doi.org/10.1016/j.procs.2016.07.309
Hossain SMM, Ray S, Mukhopadhyay A (2019) Identification of hub genes and key modules in stomach adenocarcinoma using nsnmf-based data integration technique. In: IEEE 2019 international conference on information technology (ICIT), pp 331–336
Hossain SMM, Ray S, Mukhopadhyay A (2017) Preservation affinity in consensus modules among stages of HIV-1 progression. BMC Bioinform 18(1):181
Hossain SMM, Ray S, Mukhopadhyay A (2020) Detecting overlapping gene communities during stomach adenocarcinoma: a discrete nmf-based integrative approach. In: 2020 IEEE international conference on advent trends in multidisciplinary research and innovation (ICATMRI), pp 1–6. https://doi.org/10.1109/ICATMRI51801.2020.9398458
Hossain SMM, Ray S, Tannee TS, Mukhopadhyay A (2017) Analyzing prognosis characteristics of Hepatitis C using a biclustering based approach. Procedia Comput Sci 115(Supplement C):282 – 289
Krstic J, Galhuber M, Schulz TJ, Schupp M, Prokesch A (2018) p53 as a dichotomous regulator of liver disease: the dose makes the medicine. Int J Mol Sci 19(3):921
Langfelder P, Horvath S (2007) Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 1(1):1–17
Langfelder P, Horvath S (2008) Wgcna: an r package for weighted correlation network analysis. BMC Bioinform 9(1):1–13
Li X, Wang X, Gao P (2017) Diabetes mellitus and risk of hepatocellular carcinoma. BioMed Res Int
Masserot-Lureau C, Adoui N, Degos F, de Bazelaire C, Soulier J, Chevret S, Socié G, Leblanc T (2012) Incidence of liver abnormalities in Fanconi anemia patients. Am J Hematol 87(5):547–549
Milligan GW, Cooper MC (1985) An examination of procedures for determining the number of clusters in a data set. Psychometrika 50(2):159–179
Omranian S, Angeleska A, Nikoloski Z (2021) Pc2p: parameter-free network-based prediction of protein complexes. Bioinformatics
Ray S, Hossain SMM, Khatun L (2016) Discovering preservation pattern from co-expression modules in progression of HIV-1 disease: an eigengene based approach. In: 2016 IEEE international conference on advances in computing communications and informatics, ICACCI 2016, Jaipur, September 21–24, 2016. IEEE, pp 814–820
Ray S, Hossain SMM, Khatun L, Mukhopadhyay A (2017) A comprehensive analysis on preservation patterns of gene co-expression networks during Alzheimer’s disease progression. BMC Bioinform 18(1):579
Robinson MD, McCarthy DJ, Smyth GK (2010) Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140
Saitta C, Pollicino T, Raimondo G (2019) Obesity and liver cancer. Annals Hepatol 18(6):810–815
Song E, Song W, Ren M, Xing L, Ni W, Li Y, Gong M, Zhao M, Ma X, Zhang X, An R (2018) Identification of potential crucial genes associated with carcinogenesis of clear cell renal cell carcinoma. J Cell Biochem 119(7):5163–5174. https://doi.org/10.1002/jcb.26543
Sun M, Song H, Wang S, Zhang C, Zheng L, Chen F, Shi D, Chen Y, Yang C, Xiang Z, Liu Q, Wei C, Xiong B (2017) Integrated analysis identifies microrna-195 as a suppressor of hippo-yap pathway in colorectal cancer. J Hematol Oncol 10(1):79. https://doi.org/10.1186/s13045-017-0445-8
Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Gene Mol Biol 4(1). https://doi.org/10.2202/1544-6115.1128
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Hossain, S.M.M., Halsana, A.A. (2022). Identification of Gene Communities in Liver Hepatocellular Carcinoma: An OffsetNMF-Based Integrative Technique. In: Hemanth, D.J., Pelusi, D., Vuppalapati, C. (eds) Intelligent Data Communication Technologies and Internet of Things. Lecture Notes on Data Engineering and Communications Technologies, vol 101. Springer, Singapore. https://doi.org/10.1007/978-981-16-7610-9_30
Download citation
DOI: https://doi.org/10.1007/978-981-16-7610-9_30
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-7609-3
Online ISBN: 978-981-16-7610-9
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)