Abstract
Reference Expression dataset (RefEx) is a web tool which allows users to search by the gene name, various types of IDs, chromosomal regions in genetic maps, gene family based on InterPro, gene expression patterns, or biological categories based on Gene Ontology. RefEx also provides information about genes with tissue-specific expression, and the relative gene expression values are shown as choropleth maps on 3D human body images from BodyParts3D. Combined with FANTOM dataset, RefEx enables users to draw insights regarding the functional interpretation of unfamiliar genes.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
6.1 Introduction
Reference Expression dataset (RefEx) (Ono et al. 2017) is a web tool which allows users to browse gene expression profiles by genes collected from public databases. It can be searched by various types of IDs including gene names, chromosomal regions, gene family based on InterPro (Mitchell et al. 2015), gene expression patterns, or gene annotations based on Gene Ontology (Ashburner et al. 2000). Information about genes with tissue-specific expression are also provided, and the relative gene expression values are shown as choropleth maps on 3D human body images from BodyParts3D (Mitsuhashi et al. 2009). RefEx provides insight regarding the functional interpretation of unfamiliar genes through its web interface.
The purpose of RefEx is to provide a web tool for visualization of reference gene expression pattern of mammalian tissues and cell lines measured using different methods, which can facilitate the reuse of the precious data archived in several public databases.
RefEx provides suitable datasets as a reference for gene expression data from 40 normal tissues from human, mouse, and rat collected from public gene expression databases. The collected gene expression data are classified based on four different measurement strategies (Expressed Sequence Tags (ESTs), GeneChip, Cap Analysis of Gene Expression (CAGE), and RNA-Seq). These four types of data were linked based on the NCBI gene IDs in the dataset in RefEx. In addition to these datasets, RefEx currently includes quantified gene expression data from Functional Annotation of the Mammalian genome 5 (FANTOM5) dataset for human and mouse (The FANTOM Consortium & the RIKEN PMI and CLST (DGT) 2014).
Target user group of RefEx is biologists who wish to reuse public data, but accessing the data remains difficult due to its sheer magnitude and complicated access.
6.2 Database Overview
6.2.1 Importance of Reference Gene Expression Datasets
Gene expression data are exponentially accumulating after the advent of gene expression measurement methods on a genomic scale. Many datasets are now archived in the public gene expression databases [NCBI Gene Expression Omnibus (GEO) (Barrett et al. 2013) and EBI ArrayExpress (Kolesnikov et al. 2015)]. Because the description about datasets is written by different researchers who produced the gene expression data, they are so different that it is not machine readable currently.
Nevertheless, there is strong demand for a comprehensive set of reference gene expression data from huge gene expression data in public. The availability of such data is of benefit to biologists who wish to reuse it, but accessing the data remains difficult due to its sheer magnitude and complicated access. Recently, a meta-analysis of RNA-Seq expression data across various species, tissues, and studies was reported (Sudmant et al. 2015). However, the interpretation of such data is not easy. Biologists are often at a loss because of the sheer number of datasets in public databases provided by numerous researchers. From such situations, reference expression datasets are needed for the inference of functions of genes, and a proper web interface for visualizing such data is essential.
In addition, concerted patterns of gene expression profiles for different quantification methods can strengthen the evidence of these patterns. Also, tissue-specific expression can be a key feature to examine the function of genes of interest, and lists of genes with tissue-specific expression can help biologists to explore unannotated genes with prominent expression patterns. Thus, the functional annotation of genes from meta-analysis and the interface to access the data with graphical visualization are urgently required.
6.2.2 Current Status of Reference Gene Expression Data
As a reference gene expression data for a genomic scale, expression profiles in normal mammalian tissues by GeneChip were first maintained by researchers at the Genomics Institute of the Novartis Research Foundation (GNF). Users can access the microarray data produced in that project at the GNF Expression Atlas (also known as GNF SymAtlas), now called BioGPS (Wu et al. 2016). EBI also maintains the Expression Atlas which provides gene expression patterns under various biological conditions based on data archived in ArrayExpress (Petryszak et al. 2016). Recently, using RNA sequencing and CAGE (Shiraki et al. 2003), the FANTOM collaboration consortium released terabytes of transcriptome sequencing data from adult and fetal human and mouse tissue primary cell lines that can be used as the reference gene expression data (Lizio et al. 2015).
6.2.3 The Main Feature of RefEx
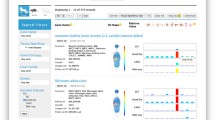
The main feature of RefEx is a simple web interface, which allows users to compare expression profiles by different methods at a glance (Fig. 6.1). It provides access to curated data from several other public databases, with expression levels in 40 tissues measured by four well-established gene expression quantification technologies (ESTs, Affymetrix GeneChip, CAGE, and RNA-Seq). The web interface allows users to browse the expression profiles by the gene name, various types of IDs, chromosomal regions in genetic maps, gene family based on InterPro, gene expression patterns, or biological categories based on Gene Ontology. The web interface also includes the way to browse the expression profile for adult and fetal human and mouse tissues obtained by the FANTOM5 project using CAGE for gene expression quantification. All the data provided through the RefEx web interface is listed with corresponding digital object identifiers (DOI) in Table 6.1.
The top page of RefEx web tool (https://refex.dbcls.jp/)
6.2.4 Future Update and Availability of the Database
RefEx is planned to be updated when the useful dataset for gene expression is publicly available. Data in RefEx is freely available under a Creative Commons Attribution 4.0 International License (CC BY 4.0). Users can download a concatenated version of all the data at the RefEx download page (https://refex.dbcls.jp/download.php?lang=en), including the log-transformed ratios of the gene expression, the functional annotation of the genes, the list of tissue-specific genes, and the sample information, in a tab-delimited text format (Table 6.1). Data in RefEx is also available at figshare (https://doi.org/10.6084/m9.figshare.c.3812815) and the scripts to make RefEx data are available at GitHub (https://github.com/dbcls/RefEx/).
6.3 Content and Architecture of the Database
The type of data stored in RefEx is the processed and quantified gene expression data for human, mouse, and rat. All data in RefEx is originally from the public database. Table 6.2 shows original data sources for RefEx dataset.
The data in RefEx were manually collected by RefEx curators from public databases, including the International Nucleotide Sequence Database (INSD, consisting of GenBank/DDBJ/ENA) (Cochrane et al. 2016), the NCBI Gene Expression Omnibus (GEO), and RNA-Seq data in the Sequence Read Archive (SRA) (Kodama et al. 2012). The raw data from the public databases were re-organized and compared against each other. Four types of data were linked based on the NCBI gene IDs, while the EST data were based on the Unigene IDs, and the GeneChip data were based on the Probe set IDs. Detailed information regarding four data extraction methods are described below. All scripts used to produce the data and additional descriptions are available on the GitHub site at https://github.com/dbcls/RefEx.
6.3.1 EST
The original EST data were retrieved from the EST division of the INSD. The number of ESTs was counted by source organ based on the BodyMap method (Okubo et al. 1992) according to the cDNA annotation of each EST entry. The EST data in RefEx originated from the BodyMap-Xs database, which contains previously compiled gene expression data from the INSD EST division for reuse (Ogasawara et al. 2006). After counting the number of ESTs, gene expression data were obtained for the 40 normal tissues stored in the BodyMap-Xs database (https://doi.org/10.6084/m9.figshare.4028721). For visualization purposes, the data were grouped into ten subsets (i.e., brain, blood, connective, reproductive, muscular, alimentary, liver, lung, urinary, and endo/exocrine; https://doi.org/10.6084/m9.figshare.4028715). This categorization of the organs was also applied to the gene expression data that were obtained by the other methods.
6.3.2 GeneChip
The GeneChip data deposited in the NCBI GEO database were selected for the reference dataset (tissue-specific patterns of mRNA expression) (Table 6.2). Those data were analyzed based on a typical microarray data analysis method (Wu et al. 2016). The expression values of the genes were calculated from the original CEL files after robust multi-array averaging (RMA) normalization (Irizarry et al. 2003) by the affy package (Gautier et al. 2004) in R (ver.3.0.3)/BioConductor (ver.2.12) (Gentleman et al. 2004).
6.3.3 CAGE
CAGE is a technique that produces a snapshot of the 5′ end of the mRNA population in a biological sample, and the CAGE data collected in the RIKEN FANTOM5 project were counted by source organ based on the original data, the FANTOM5 CAGE peak expression, and the annotation tables (Lizio et al. 2015). The CAGE tag counts were mapped onto the reference genome sequences (hg19 for human and mm9 for mouse) and reflect the intensity of the gene expression of the corresponding transcripts. The tag counts are normalized by tag per million (TPM). The processed data in RefEx is converted to log 2 for each TPM value of the original FANTOM 5 CAGE data and then organized for each sample classification and the data to which the same GeneID is assigned are added up and averaged. In addition to the 40 normal tissues, the FANTOM5 project collected hundreds of samples from cell lines, primary cells, and adult and fetal tissues of human (https://doi.org/10.6084/m9.figshare.4028685) and mouse (https://doi.org/10.6084/m9.figshare.4028688).
6.3.4 RNA-Seq
For RNA-Seq data, the normal tissue transcriptome sequence data were selected from the SRA. Utilizing human and mouse reference genome sequences (hg19 for human and mm9 for mouse), these data were processed using a typical RNA-Seq data analysis pipeline with TopHat (ver.2.0.7) (Trapnell et al. 2009) and Cufflinks (ver.2.0.2) (Trapnell et al. 2010), and the transcript abundances were calculated and normalized to fragments per kilobase of transcript per million reads (FPKM).
Four types of data above were linked based on the NCBI gene IDs in the dataset in RefEx. The EST data were clustered by sequence similarity, and the NCBI UniGene IDs were added to those clusters. The GeneChip data were based on Affymetrix probe IDs, which were originally designed based on the UniGene database (Wagner and Agarwala 2013). The remaining two methods were based on direct sequencing and were developed after the completion of the human and mouse genome sequencing projects; the data obtained by these methods can be mapped to the reference genomes by the genomic position. Thus, the NCBI Gene IDs were adopted, which are currently widely used to integrate other gene IDs, as a standard. Mapping the various gene IDs (UniGene ID, Affymetrix probe ID, and NCBI Gene ID) onto the various genomes was performed using the Biomart REST API (http://www.biomart.org/martservice.html).
The quality of expression data above is guaranteed by the biological replicates. For example, the RNA-Seq data used were generated from multiple reads (single-read and paired-end read) and those of mouse from triplicate sequence reads were averaged to represent the gene expression value.
6.4 Database Access and Mining Methods
6.4.1 Gene Expression Visualization Tool in RefEx
The relative gene expression values are shown in RefEx as choropleth maps on 3D human body images from BodyParts3D (Mitsuhashi et al. 2009). BodyParts3D has been developed by the Database Center for Life Science (DBCLS) as a dictionary-type anatomy database in which anatomical concepts are represented by 3D structural data that specify the corresponding segments on a 3D whole-body model of an adult human male. Foundational Model of Anatomy (FMA) ontology (https://bioportal.bioontology.org/ontologies/FMA) was used to map the gene expression data onto the corresponding tissues. Because drawing the choropleth maps dynamically on a 3D human body is quite labor-intensive, still images were prepared for only the GeneChip data for the whole entries. Figure 6.2 clearly illustrates that the selected transcript is highly expressed in the liver tissue. On the right (Fig. 6.2), the relative expression levels in 40 types of normal tissues that were more precisely classified are displayed. The visualization can help users to understand the differences in the gene expression patterns among tissues more intuitively.
The search results for liver-specific genes. This view can easily be viewed by clicking the liver icon at the top of the RefEx page (Fig. 6.1)
6.4.2 How to Query RefEx
Users can easily query RefEx with an effective filter to extract genes with concerted gene expression profiles. For example, genes with liver-specific gene expression can be retrieved only a single click. Details are available in a video tutorial (https://doi.org/10.7875/togotv.2016.068).
After the publication of the FANTOM5 project, pre-calculated gene expression data from the CAGE data in the FANTOM5 project were incorporated into RefEx. The most important benefit of the FANTOM5 CAGE data is that the search targets are much more abundant. The original version of RefEx only had forty tissue search targets (Fig. 6.3a). However, it is now possible to search more than 500 human samples, encompassing cell lines, primary cells, and adult and fetal tissues (Fig. 6.3b). RefEx also enables users to browse high-resolution gene expression data from approximately 800 samples (human plus mouse).
By clicking the tab on the right-hand side, users can switch to a FANTOM5 CAGE data viewer (Fig. 6.3b). This viewer shows the expression patterns of all samples in the lower portion of the screen and displays an enlarged view of a specific area in the upper portion of the screen. Because this is a representation of the expression profile in humans, 556 samples are shown in a bar chart in the lower portion of the screen. Therefore, a user can observe an overview of expression patterns in all the samples. The area displayed in the enlarged box can be moved freely by dragging. When a user enters a keyword into the search window of the viewer, the sample name containing that keyword is highlighted. The FANTOM5 CAGE data correspond to the tissue classification in the original RefEx and are linked to the original FANTOM5 data. The expression values of the samples obtained in the FANTOM5 project are averaged and listed in RefEx.
6.4.3 How to Download Data from RefEx
While the data shown in RefEx is originally from the public database and the sources for all data records are summarized in Table 6.2, the data used in RefEx including processed gene expression data can be downloaded from RefEx download page (https://refex.dbcls.jp/download.php?lang=en). These data are deposited in figshare (Fig. 6.4), which is a repository where users can make all of their research outputs available in a citable, shareable, and discoverable manner. Forty-one datasets uploaded to figshare can be accessible from figshare collection at https://doi.org/10.6084/m9.figshare.c.3812815.
Data in RefEx can be downloaded from figshare. https://doi.org/10.6084/m9.figshare.4028622.v5
6.4.4 Programmatic Technique to Access RefEx
As a member of the integrated database project in Japan, the Resource Description Framework (RDF) version of RefEx resides at the National Bioscience Database Center (NBDC) RDF portal, and the RefEx dataset is ready for use at the NBDC RDF Portal (https://integbio.jp/rdf/).
6.5 Use-Cases and Demo to Utilize the Database
As a simple use-case of RefEx, gene expression profiles for specific genes of interest in normal tissues were often used in medical research. The mRNA expression levels of isocitrate dehydrogenase 3 (NAD(+)) alpha (IDH3α) and vascular endothelial growth factor A (VEGF-A) were used and visualized as main indicators in ten major groups of normal tissues (Fig. 6.5) (Zeng et al. 2015) in conjunction with the calculated prognostic values by the PrognoScan database (Mizuno et al. 2009).
RefEx-based quantification of the mRNA expression levels of VEGF (a, c) gene and IDH3a (b, d) gene in the indicated ten major groups of normal tissues (Zeng et al. 2015)
Another application is used as a reference of tissue-specific information. In a study of murine colon proteomes, colon-specific genes in the mouse version of RefEx were compared to a list of genes from murine colon proteomes that was generated by the researchers’ own results (Magdeldin et al. 2012). A similar example was a study of liver-specific genes to investigate biomarkers indicating liver injury in humans. The gene expression profiles of albumin (ALB), apolipoproootein H (APOH), group-specific component (GC), and α-1 microglobulin/bikunin precursor (AMBP) were used to confirm the liver-specific expression (Okubo et al. 2016).
The data retrieved from RefEx strengthened the authors’ hypotheses without the further confirmation in wet-lab. The gene expression profile of noncardiac myosin, light chain 12A (MRLC) in RefEx was used to confirm the conclusions because noncardiac MRLC was expressed in the heart at the same level as that in the skeletal muscle while it was annotated as “noncardiac” (Mizutani et al. 2016). As an another example, RefEx was used to list the gene expression profiles of all genes previously reported to cause deafness in a review article (Nishio et al. 2015).
As an intermediate use-case of RefEx using the RefEx web interface, users can add up to three genes to their list and compare these genes simultaneously. Users can compare all the detailed information about the genes in that list, including the expression data. This parallel comparison enables users to easily identify the differences among the genes. Overlapped terms, such as the Gene Ontology and the InterPro gene family terms, are arranged in the same row (Fig. 6.6). Therefore, RefEx is also useful as a tool for investigating the relationships of unknown genes found in gene expression analyses.
For the advanced use-case, RefEx can be used in comparative transcriptomic studies as a reliable reference expression dataset of human normal tissues and cell lines. To compare the characteristics between human small intestine and Bombyx mori larval midgut, gene expression profiles from a human colon carcinoma cell line (Caco-2) cells, which is used to estimate human intestinal absorption, human small intestine, and B. mori larval midgut were compared to identify common drug transporters in the human intestine and B. mori larval midgut (Ichino et al. 2018). While the B. mori larval midgut RNA-seq data were originally analyzed, the gene expression data for Caco-2 cells and human small intestine were reused from RefEx [“Processed expression data of all samples for CAGE human PRJDB3010 (FANTOM5)” downloaded from https://doi.org/10.6084/m9.figshare.4028613.v4]. As a result, 26 drug transporter homologs were found and those were common in the B. mori larval midgut and human intestine (Fig. 6.7).
6.6 Summary and Future Development of the Database
RefEx is a tool for an interactive analysis of gene expression patterns on the web via the latest version of web browsers such as Firefox, Safari, and Chrome. RefEx has three main applications. First, users can examine the expression profiles of unfamiliar genes in normal body tissues, cells, and cell lines based on actual measurement data rather than only from a description in a journal article. Second, a search for tissue-specific genes can be performed simply by clicking on the appropriate tissue icon at the top of the RefEx page. Third, users can compare differences in gene expression levels related to the use of different experimental methods.
Currently, transcripts, which are based on RefSeq mRNA records, are used to integrate different types of measurement methods for gene expression. However, according to a high-throughput sequencing data analysis, over 90% of human genes undergo alternative splicing (Pan et al. 2008), and many of these are not yet included in RefSeq. To address this limitation, the definitions of the transcripts need to be redefined to include noncoding RNA in tight collaboration with the FANTOM project. In the upcoming version of RefEx, it is planned to use personalized gene expression data from the Genotype-Tissue Expression database (GTEx) (GTEx Consortium et al. 2015).
References
Ashburner M et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29
Barrett T et al (2013) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41:D991–D995
Cochrane G, Karsch-Mizrachi I, Takagi T (2016) The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res 44:D48–D50
Gautier L et al (2004) Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315
Gentleman RC et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80
GTEx Consortium et al (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–660
Ichino F et al (2018) Construction of a simple evaluation system for the intestinal absorption of an orally administered medicine using Bombyx mori larvae. Drug Discov Ther 12(1):7–15
Irizarry RA et al (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264
Kadota K et al (2006) ROKU: a novel method for identification of tissue-specific genes. BMC Bioinformatics 7:294
Kodama Y, Shumway M, Leinonen R (2012) International Nucleotide Sequence Database Collaboration. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res 40:D54–D56
Kolesnikov N et al (2015) ArrayExpress update—simplifying data submissions. Nucleic Acids Res 43:D1113–D1116
Lizio M et al (2015) Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 16:22
Magdeldin S et al (2012) Murine colon proteome and characterization of the protein pathways. BioData Mining 5:11
Mitchell A et al (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43:D213–D221
Mitsuhashi N et al (2009) BodyParts3D: 3D structure database for anatomical concepts. Nucleic Acids Res 37:D782–D785
Mizuno H, Kitada K, Nakai K, Sarai A (2009) PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genet 2:18
Mizutani T et al (2016) Heterogeneous filament network formation by myosin light chain isoforms effects on contractile energy output of single cardiomyocytes derived from human induced pluripotent stem cells. Regen Ther 3:90–96
Nishio S et al (2015) Gene expression profiles of the cochlea and vestibular endorgans: localization and function of genes causing deafness. Ann Otol Rhinol Laryngol 124:6S–48S
Ogasawara O et al (2006) BodyMap-Xs: anatomical breakdown of 17 million animal ESTs for cross-species comparison of gene expression. Nucleic Acids Res 34:D628–D631
Okubo K et al (1992) Large scale cDNA sequencing for analysis of quantitative and qualitative aspects of gene expression. Nat Genet 2:173–179
Okubo S et al (2016) Albumin and apolipoprotein H mRNAs in human plasma as potential clinical biomarkers of liver injury: analyses of plasma liver-specific mRNAs in patients with liver injury. Biomarkers 21:353–362
Ono H, Ogasawara O, Okubo K, Bono H (2017) RefEx, a reference gene expression dataset as a web tool for the functional analysis of genes. Sci Data 4:170105
Pan Q et al (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415
Petryszak R et al (2016) Expression Atlas update—an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res 44:D746–D752
Shiraki T et al (2003) Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A 100:15776–15781
Sudmant PH, Alexis MS, Burge CB (2015) Meta-analysis of RNA-seq expression data across species, tissues and studies. Genome Biol 16:287
The FANTOM Consortium & the RIKEN PMI and CLST (DGT) (2014) A promoter-level mammalian expression atlas. Nature 507:462–470
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111
Trapnell C et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Wagner L, Agarwala R (2013) The NCBI handbook, 2nd edn. National Center for Biotechnology Information
Wu C et al (2016) BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res 44:D313–D316
Zeng L et al (2015) Aberrant IDH3α expression promotes malignant tumor growth by inducing HIF-1-mediated metabolic reprogramming and angiogenesis. Oncogene 34:4758–4766
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
This work was supported by the Integrated Database Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the National Bioscience Database Center (NBDC) of the Japan Science and Technology Agency (JST). Computations were partially performed on the NIG supercomputer at the ROIS National Institute of Genetics.
Rights and permissions
Copyright information
© 2021 The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Ono, H., Bono, H. (2021). RefEX: Reference Expression Dataset. In: Abugessaisa, I., Kasukawa, T. (eds) Practical Guide to Life Science Databases. Springer, Singapore. https://doi.org/10.1007/978-981-16-5812-9_6
Download citation
DOI: https://doi.org/10.1007/978-981-16-5812-9_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-5811-2
Online ISBN: 978-981-16-5812-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)