Abstract
The whole world is facing a pandemic situation ever since the outbreak of SARS-CoV-2 in Wuhan, China in December 2019. The coronavirus disease (COVID-19) presents a wide spectrum of clinical manifestations ranging from asymptomatic to severe pneumonia-like situations followed by multisystem failure leading to the death of the individual. Studies from the past coronavirus outbreaks, the SARS, and MERS-CoV, have helped us understand the current SARS-CoV2 to a large extent. Once the host encounters the virus, an innate immune response is generated which subsequently leads to activation of the adaptive immune response to eliminate the virus. However, this immune response is misbalanced in some individuals and is the main factor causing the pathological manifestation of COVID-19. In this chapter, we have addressed the humoral and cellular immune changes induced by the virus along with the role of cytokine storm in disease progression.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
14.1 Introduction
December 2019 saw an outbreak of severe acute respiratory syndrome coronavirus-2 (SARS-CoV2) in Wuhan city of China, which was declared as a pandemic by the World Health Organization in late March 2020, due to its rapid transmission to almost all parts of the world. It is causing concerns globally due to the complications associated with it, such as acute respiratory distress syndrome (ARDS), pneumonitis, shock, respiratory failure, and death. In 1996, the first pathogenic coronavirus (CoV) was discovered that could cause interspecies infection. The CoV strains, E229 and OC43 came from rodents, and NL63 and HKU1 came from bats and infected the humans (Cao et al. 2020). These viruses remained limited in their spread causing mild seasonal common cold-like symptoms. In November 2002, an outbreak of severe acute respiratory syndrome (SARS) caused by a CoV from bats, also originated in Foshan city of China and was named SARS (now termed as SARS-CoV or SARS-CoV-1 to distinguish it from currently pandemic SARS-CoV-2). The SARS-CoV-1 epidemic lasted for over a year and ended in July 2003 (Wang et al. 2020b). The second CoV outbreak took place in June 2012, first detected in Jeddah, Saudi Arabia and so termed as Middle East Respiratory Syndrome (MERS), mimicked several clinical manifestations of SARS (Grein et al. 2020). The current spillover is the third one from animal CoVs to humans, first detected in Wuhan city of China in October 2019 and quickly spread over six continents of the world including 66 countries by March 1, 2020, when the World Health Organization (WHO) declared it as a pandemic. The novel coronavirus (initially termed as 2019-nCov and subsequently as SARS-CoV-2) causes severe coronavirus disease, now termed as COVID-19. All these coronaviruses (SARS-CoV-1, MERS, and SARS-CoV-2) have been found to have jumped to humans from bats, but some researchers believe that there is an intermediary host. In the case of SARS-CoV1, the intermediary animal is thought to be civet cats which are sold in abundance in the live-animal markets of China (Chen et al. 2020a). However, the origin of SARS-CoV2 is not clear but it shares around 96% of its genetic material with CoV found in bats (Lynch et al. 2016). These three coronaviruses share genetic and structural similarities but differ significantly at the epidemiological level. SARS-CoV and MERS-CoV have high lethality and low transmissibility while SARS-CoV2 has high transmissibility and the level of lethality has not yet been established globally. Thus, its tremendous spread has brought extreme pressure and disastrous consequences on the public health and medical setup worldwide.
14.2 General Characteristics of the Virus
Novel SARS-CoV2 has been placed under the family Coronaviridae. The virus contains single-stranded positive RNA of nearly 30 kbp as its genetic material. The genetic material is protected by an outer fatty layer of envelope containing spike –S, membrane –M and envelope –E- proteins. The subfamily Coronaviridaeis further subdivided into four genera- the alpha, beta, gamma, and delta CoV. Viruses having the ability to infect humans are categorized under α-CoV and β-CoV (SARS-CoV and MERS-CoV) and viruses infecting avians and pigs belong to ϒ-CoV and δ-CoV genera. The novel SARS-CoV2 has been placed under the genus β-CoV.
The entry of enveloped viruses into the host cells is usually mediated through the attachment of proteins expressed on the surface of host cells. To gain entry into the host cell, the S-glycoprotein present on the surface of the virus engages with the receptors on the host cells. Based on sequence similarities between the receptor-binding domains (RBD) in the S-protein of SARS-CoV-1 and SARS-Cov-2, it is now established that the angiotensin-converting enzyme 2 (ACE2), serves as the receptor for both the viruses. The ACE-2 is a type-1 transmembrane metallocarboxypeptidase molecule, which is highly expressed on vascular endothelial cells, the renal tubular epithelium, and Leydig cells in the testes. The ACE-2 is homologous to ACE, which is an enzyme and a key player in the Renin-Angiotensin system (RAS). For entry of the virus into the host cells, the spike protein on the surface of the virus, which binds to its receptor on the host cell, needs to be cleaved first.
The S protein is cleaved into subunits S1 and S2 during the infection stage, where the receptor-binding domain (RBD) present in the S1 subunit binds directly to the peptidase domain of the ACE2 molecule and the S2 subunit facilitates membrane fusion between host cell and virion (Kucharski et al. 2020). RBD-ACE2 binding induces conformational change on S-protein which exposes a cleavage site on the S2 subunit, which is cleaved by host serine proteases TMPRSS2. This step is a critical process mediating the fusion of the virus envelope with the cell membrane and thus allowing the viral RNA to enter into the target cell’s cytoplasm (Hoffmann et al. 2020). Subsequently, viral RNA serves as a template for the translation of the polyproteins pp1a and pp1b that are cleaved into smaller proteins which join to form a replicase-transcriptase complex (RTC). In this complex, several copies of negative-strand RNA are made and used as the template to form complete positive-strand RNA. The structural proteins (S, E, M & N) are translated and are transported to the lumen of the intermediate compartment of the endoplasmic reticulum golgi intermediate complex (ERGIC). Along with the genomic RNA, virion formation occurs and mature virions are released from the cell by exocytosis, which then infect the neighboring healthy cells as well as released into the surrounding environment via respiratory droplets. The droplets carrying the infectious virus are highly contagious and cause the potential spread of the disease in healthy individuals (Fung and Liu 2019). Thus, ACE2 bearing cells are most vulnerable against SARS-CoV2. The majority of the ACE-2 bearing cells are alveolar epithelial type II cells, thus making the lung as primary target tissue and the most common entry route. Other extrapulmonary tissues expressing ACE-2 are kidneys, heart, endothelium, intestine, and tongue.
14.3 Clinical Manifestation of COVID-19
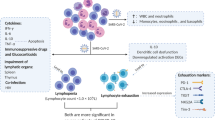
Depending on the individual immune response, the clinical manifestation ranges from asymptomatic (positive for SARS-CoV-2 virus but no symptoms), mild symptomatic (positive for virus with mild clinical manifestation), and severe (positive for the virus and a high degree of symptoms). In SARS-CoV-2, the mild symptomatic individuals present with dry cough, fever, and fatigue, difficulty in breathing, some have diarrhea, sore throat, congestion, and runny nose. The severe symptomatic patients have difficulty in breathing (greater than 30 times/minute), oxygen saturation less than 93%, the ratio between partial pressure of arterial oxygen and oxygen concentration in arterial blood is less than 300 mmHg) (Kimball et al. 2020). Critical individuals have a respiratory failure with the need for mechanical ventilation, organ failure, or need for treatment in the Intensive care unit (ICU) of a hospital. The wide range of clinical manifestations found in COVID-19 are associated with risk factors such as age and gender. Diabetes, cardiovascular diseases, high BP, lung diseases, treatments affecting the immune system are considered comorbidities that may result in the highest risk of severe disease and death in COVID-19 (Chen et al. 2020b) (Fig. 14.1).
14.4 Immune Responses in COVID-19
14.4.1 Innate Immune Response
The innate immune system is made up of barriers that can prevent or limit the entry and spread of various foreign particles. Innate immune cells include DCs, neutrophils, macrophages, parenchymal cells like epithelial and fibroblast cells. Antigens related to viruses are recognized by receptors of innate immune cells known as pattern recognition receptors (PRR). The toll-like receptors (TLRs) recognizing pathogen-associated molecular patterns (PAMPs), RIG-I-like receptors (RLRs) recognizing nucleic acids, C type lectin-like receptors (CLRs), and NOD-like receptors (NLRs) are few other pattern recognition receptors that are responsible for identifying the viral antigens (Li et al. 2020). The RLRs and NLRs are usually expressed by epithelial cells and some local cells of the innate immune response like alveolar macrophages. Once the recognition of the pathogen is executed, the PRR’s recruit adaptor proteins which lead to downstream activation of transcription factors like IFN, AP-1, NF-κB, which lead to the secretion of critically important type I and type III antiviral interferon (IFN). After activation of IFN signaling, an entire cascade of events occur that leads to the production of pro-inflammatory cytokines which further activate the endothelial cells, and produce chemokines. These chemokines attract other immune cells like monocytes, NK cells, dendritic cells, and polymorphonuclear leukocytes (PMN) including Neutrophils. These cells further release reactive oxygen species and directly kill the infected cells and produce more chemokines like monokine induced gamma interferon (MIG), Interferon-gamma inducible protein-10 (IP-10), monocyte chemotactic protein-1 (MCP-1), which recruit lymphocytes that can recognize the viral antigen presented by the DCs, thereby activating a pathogen-specific adaptive immune response (Dandekar and Perlman 2005). Apart from IFNs other cytokines like TNF-α, IL-1, IL-6, and IL-18 are also released. Thus, together these cytokines induce an antiviral defense mechanism and also activate the adaptive immune response. CoV infection can be limited in early stage if the IFNs are properly localized. The IFN-stimulated genes (ISG) like lymphocyte antigen-6 complex locus E (LY6E) interfere with S-protein mediated membrane fusion (Huang et al. 2011). However, in the later phase of infection, IFN can cause immune-pathogenesis by induction of other cytokines.
14.4.2 Immunopathogenesis of COVID-19 and Immune Escape Mechanisms
In-vitro infection of human lung explant by SARS-CoV2 leads to viral infection and replication in type I and type II pneumocytes and alveolar macrophages, although it failed to induce expression of IFN-I, IFN-II, and IFN-III (Chu et al. 2020). CoV has also evolved a mechanism to inhibit IFN-I induction and signaling. It has been observed that patients with severe COVID-19 have impaired IFN-I production as compared to patients with mild symptoms (Blanco-Melo et al. 2020). Thus, it is inferred that CoV uses various evasion mechanisms like avoiding PRR sensing either by avoiding recognition or antagonizing PRR activation. For avoiding recognition by PRR, ssRNA CoV forms dsRNA-intermediate during replication, which is shielded by membrane-bound compartments and nonstructural proteins help in methylation of the viral RNA at 5′ end (Bouvet et al. 2010). In addition, in SARS-CoV2 it has been demonstrated that the viral proteins ORF9b, NSP13 & NSP15 interact with the downstream signaling molecules like Mitochondrial antiviral signaling proteins(MAVs), signaling intermediate like Tank-binding kinase-2 (TBK2) and Rosetta Net Implementation Framework (RNIF41), respectively, and they interfere with the TBK1 signaling complex (Gordon et al. 2020). SARS-CoV1 and MERS-CoV are also known to inhibit the TBK1 signaling complex (Gordon et al. 2020). MERS-CoV encodes matrix and accessory proteins from the open reading frame, ORF4a, 4b, and 5 which directly inhibit IFN promoter and nuclear localization of interferon regulatory factor-3 (IRF3) (Yang et al. 2013). Thus, pathogenic CoVs can block or inhibit IFN pathways but can also activate other inflammatory pathways that contribute to disease pathogenesis. For instance, SARS-CoV2 non-structural proteins, NSP-9 and NSP-10 might induce the IL-6 and IL-8 production and drive pathological inflammation. In fact, IL-6 has emerged as the dominant cytokine during immunopathogenesis. Thus, a decrease in innate antiviral response and hyper inflammation causing a “cytokine storm” could be a leading cause of COVID-19 severity.
14.4.3 Role of Innate Immune Cells in COVID-19 Pathogenesis
Neutrophils, monocytes, NK cells are the innate immune cells that play an important role in viral clearance as well as the pathogenesis of COVID-19. The cytotoxicity mediated by the NK cells is regulated by the expression of inhibitory and activating receptors expressed on them. The frequency of NK cells has been reported to be lower in severe COVID-19 cases as compared to mild cases. The NK cells from the peripheral blood of severe COVID-19 patients showed a reduced expression of CD107a (a marker for degranulation), Granzyme B (a marker of killer activity), and Ksp37 (a marker co-expressed with perforin, also a marker of killing activity), thereby indicating impaired cytotoxicity. In addition, there is impaired production of chemokines, IFN-ϒ and TNF-α (Zheng and Song 2020). The lower frequency of NK cells can be correlated with the increased concentration of IL-6 in the plasma. An in-vitro data suggests that stimulation by IL-6 and soluble IL-6 receptors can impair perforin and Granzyme B production in healthy donor NK cells, which can be restored by blocking IL-6R with tocilizumab (Wenjun et al. 2020). Further, this heterodimeric inhibitory receptor prevents NK cells from releasing IFN-ϒ. Thus, targeting this receptor by monalizumab can boost antiviral immunity. Reduced production of IFN-ϒ can also lead to the infiltration of the neutrophils in the alveoli and the high neutrophil to lymphocyte (N:L) ratio is an indicator of the severe stage of COVID-19. These accumulating neutrophils cause sustained neutrophil extracellular traps (NET) formation which leads to stimulation of cascade of damaging inflammatory reactions. This has been observed in COVID-19 patients, with increased levels of NET-specific markers like myeloperoxidase DNA and citrullinated histone H3 (Kawasaki et al. 2018). In addition, the immune checkpoint molecule, NKG2A was also found to be increased in severe COVID-19 patients, indicating viral escape and NK cell exhaustion. Monocytes-derived DCs (Mo-DC), plasmacytoid DCs (PDC) also get dysregulated and potentially drive cytokine release syndrome (CRS), acute respiratory distress syndrome (ARDS), and lymphopenia. Single-cell transcriptome analysis done on the pulmonary tissue samples and peripheral blood samples of severe COVID-19 disease have revealed expansion of CD14+HLA-DRlow inflammatory monocytes, which are known to be an immunosuppressive phenotype of these monocytes and now clubbed as a broad category of Monocyte-derived suppressor cells or MDSCs. Thus, the innate immune system is unable to strike a balance to control the infection in a timely manner.
14.4.4 Complement Activation in SARS-CoV2 Infection
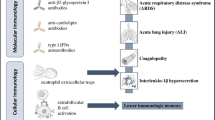
The complement system is an important part of innate immunity and acts as a bridge between innate and adaptive immunity. Complement recognizes a foreign pathogen and leads to the initiation of complement activation pathway using one of the three mechanisms. The classical pathway- characterized by C1q mediated antigen-antibody complex, the lectin pathway is mediated by mannose present on the microbial surface (also known as “Mannose-binding pathway”) and third is the alternative pathway, which is mediated by spontaneous cleavage of C3 binding with pathogens’ cell surface components. An imbalance in the complement system function can result in enhanced inflammatory and degenerative responses in various pathological conditions. Usually, C3a and C5a are the molecules that are increased in case of over-activation of complement pathways. The C5a being a chemoattractant, recruits inflammatory cells and leads to the production of granular enzymes and free radicals. It has been observed in past, that SARS-CoV leads to overactivation and production of C3a, which causes excessive alveolar air sac infiltration by neutrophils and monocytes and subsequently leads to SARS-CoV associated ARDS (Gralinski et al. 2018). In an animal model, these results were validated, wherein the C3 knockout mice were infected with SARS-CoV and a lower infiltration of neutrophils and monocytes was observed with a low level of cytokine and chemokine production in lungs as compared to wild-type mouse. However, the level of viral load was not changed in the lung alveoli (Bosmann and Ward 2014). In SARS-CoV2, a C5a mediated cell activation and cytokine release has been observed, which results in increased vascular permeability and epithelial cell degradation leading to an increase in the number of infiltrating cells in the lungs causing respiratory distress (Wang et al. 2015).
14.4.5 Adaptive Immune Response
The innate immune response invariably leads to an adaptive immune response against a pathogen, which is specific and longer lasting. Many immune cells like T cells, B cells, NK cells, all play a central role in the functioning of the immune system. The antigen-specific cell-mediated response, mainly governed by the helper T-cells and cytotoxic T cells is directed towards the elimination of the viral-infected cells, while the B cells help in antibody synthesis and long-lived memory cells are some of the components of the adaptive immune system. The pathogen taken up by the phagocytic cells (the DCs, macrophages, and neutrophils) of the innate immune system is processed and expressed on the surface of these cells in conjunction with molecules of major histocompatibiliy complex (the MHC-I and MHC-II). The antigens in this form are presented by these so-called antigen-presenting cells or APCs to the naive T cells in the lymphoid organs. So, the naive T-cells carrying specific T-cell receptors recognize the antigen along with the MHC molecule and get activated, which further differentiate into effector/helper CD4+ T cells (Th1, Th2, or Th17 cells). Subsequently, the activated helper T-cells further pass activation signals to antigen-specific B cells and CD8+ cytotoxic T lymphocytes (CTLs) through cell-to-cell interaction and release of cytokines. Activated B cells differentiate further to antibody-forming plasma cells, which can then produce antiviral antibodies and act through various mechanisms (opsonization, neutralization, complement activation) for clearance of the virus. In addition, activated CD8+ CTLs cause the direct lysis of the target infected cells. However, the dysregulated T-cell response can lead to the immunopathogenesis of the disease.
14.4.6 T-Cell Responses
In COVID-19 patients, particularly with moderate and severe disease, reduced frequency of CD4+ and CD8+ cells (lymphopenia) has been observed which correlated with disease severity and mortality (Wang et al. 2020a). Though this phenomenon is observed in other viral infections also but the exact mechanism is unclear since the direct infection of T cells by SARS-CoV2 has not yet been reported (Xiong et al. 2020). Few underlying mechanisms have been proposed for lymphopenia in COVID-19 patients, one such is the contribution of inflammatory cytokines. Increased serum levels of IL-6 have been correlated to lymphopenia in COVID-19, while in recovered patients the level of lymphopenia recovers to normal limits as the level of IL-6 in serum decreases. A similar trend was seen with other cytokines like TNF-α and IL-10 (Xiong et al. 2020). Multiple studies suggest that IL-6 leads to the down-regulation of HLA Class II molecules on CD14+ monocytes and B-cells (Wilk et al. 2020). While no such effect was seen on HLA Class I molecules. Thus, with low HLA class II molecules, the severe COVID-19 patients will not be able to mount a sufficient level of T cell response due to the decreased capacity of the antigen-presenting cells to present the antigen to the TCR, thereby such T cells are removed by apoptosis. These observations were reported in the autopsy samples of spleen and hilar lymph nodes, where massive death of lymphocytes is linked to high IL-6 level and fas-induced apoptosis (Feng et al. 2020). Since CD8+ T cells require HLA-Class I for their activation via TCR and IL-6 doesn’t affect the expression level of HLA-Class I molecules, so similar mechanism of apoptosis doesn’t seem to operate in this condition. However, T cell exhaustion and activation can be the underlying mechanism for the elimination of CD8+ T cells (Diao et al. 2020). Higher levels of co-stimulatory and inhibitory molecules like CTLA-4, CD137, TIM-3, PD-1, NKG2a, etc. have been reported in several studies. In addition, the levels of these markers were found high in severe disease as compared to mild disease. Besides the frequency, the functionality of CD4 and CD8 T cells has also been shown to be altered, as these cells produced low quantities of IFN-ϒ and TNF-α on PMA stimulation and the CD8 cells expressed lower levels of cytotoxic molecules Granzyme B, Granzyme A, perforin, and CD107a (Yang et al. 2020). While the frequency of Th-17 cells is increased in the severe COVID-19 patients (He et al. 2020), the naive as well as induced T-regulatory cells, are reported to be decreased (Qin et al. 2020). Apart from effector T-cell subsets, the memory T-cells are also generated that fight against reinfection. Usually, the memory CD4+ T cells upon re-stimulation trigger B cells and other cells via cytokine production, and the CD8+ cytotoxic memory T cell destroy the virus-infected cells on re-exposure. In recent reports, the COVID-19 patients show a reduced level of memory T cells, which may aggravate the inflammatory response leading to cytokine storm and enhanced tissue damage and organ failure (Qin et al. 2020). In an animal model, when CD4+ memory T cells were administered through the intranasal route, it leads to a protective affect against human coronavirus. In addition, upon re-stimulation, these memory T cells were able to produce IFN-ϒ and also recruit CD8+ cells and cause rapid clearance of SARS-S366 peptides (Zhao et al. 2016). Thus, various studies reveal the dynamics of T-cell number and functional status during SARS-CoV2 infection and also indicate the important relationship between these parameters and the disease profile. Large-scale multi-centric studies are however warranted from different geographical locations in the world to precisely understand the role of T-cells in the pathogenesis of COVID-19.
14.4.7 B-Cell Responses
Apart from T-cell responses, humoral responses also play an important role in the clearance of viruses and it has been observed that B-cell responses start to appear within the first week following the onset of the COVID-19 symptoms. Similar to SARS-CoV1 infection, seroconversion occurs in COVID-19 patients between 7 and 14 days after the infection. The initial response occurs against the nucleocapsid (N) protein and later the antibody production continues to occur against the S-proteins (Dienz et al. 2009). Many studies support the fact that antibodies against the S-proteins can block the virus attachment to the ACE2+ cells (Tai et al. 2020). However, there are many questions, which are still unanswered with regard to the cross-reactivity of antibodies against alpha and beta coronaviruses, although it seems that most likely the cross-reactivity seems to appear between SARS-CoV and SARS-CoV2, which share 90% of the amino acid sequence in S1 (Walls et al. 2020). IgM and IgA are usually detected within the first week of symptoms onset while IgG antibodies are detected around 14 days after the initial symptoms (Guo et al. 2020). Comparing the mild and severe cases, there was no significant difference in the production of IgG and IgA levels, while there was a slight reduction in the level of IgM in severe cases (Zhang et al. 2006). The B cell responses provide both immediate protection from the initial challenge besides exerting extended immunity against reinfection. Memory B cells can quickly respond to the reinfection by generating new high-affinity plasma cells while long-term protection is achieved through long-lived plasma cells and memory B cells. Thus, understanding how long the lifespan of B cells memory remains against SARS-CoV2 will be interesting. Due to the short time elapsed it has not been possible yet to understand but from the past human CoVs we have understood that in the case of SARS-survivors who were followed up for 6 years, the majority of patients had undetectable levels of anti-SARS CoV antibody and none of them showed the level of memory B cells. In contrast, the T-cell specific memory was present in about half of the SARS survivors (Abbasi 2020). In the case of MERS-CoV, the antibodies were detected in most of survivors after even 3 years of infection (Payne et al. 2016). Thus, studies from SARS-CoV1 and MERS-CoV indicate that virus-specific antibodies wane out with time and results in partial protection in case of reinfection. In SARS-CoV-2 infection, however, it has recently been shown that the virus causes an early disruption of germinal centers due to loss of Bcl6 expression on T-follicular helper cells (Tfh cells), which are absolutely required for the maintenance of long-living antibody-secreting plasma cells in the germinal center (Kaneko et al. 2020). This explains the dysregulated humoral immune induction in COVID-19 disease besides providing a mechanistic explanation for the limited durability of antibody responses in coronavirus infections. So, further long-term studies are needed to check on the degree and extent of the long-term protection.
A few research groups are beginning to question whether anti-SARS-CoV2 antibodies might be detrimental to the patients. It is postulated that antibodies can bind to viral fragments and can cause complement activation, which can exacerbate the disease. Secondly, antibody response to CoV might contribute to antibody-dependent enhancement (ADE). This phenomenon is observed when non-neutralizing virus-specific IgG antibodies facilitate the entry of the virus particles into the Fc receptor-expressing cells (macrophages and monocytes) and thereby leads to the activation of these cells. However, direct evidence for ADE in COVID-19 patients has not been reported so far. Thus, during the development of therapeutic targets for SARS-CoV2, the possibility of ADE should be considered (Fig. 14.2).
14.5 Viral Response During the Infection by SARS-CoV-2
14.5.1 Cytokine Storm in COVID-19
The term “Cytokine Storm” dates back to 1993 when it was initially reported in case of graft vs host disease (GVHD) and was used to describe an overactive immune response, which can be triggered by a variety of factors like virus infection, autoimmune disease, and immunotherapies. Under normal conditions, a balance is maintained between pro-inflammatory and anti-inflammatory cytokines, which can be broken under viral infection by abnormal activation of immune cells like dendritic cells, macrophages, and lymphocytes. These abnormally activated immune cells could lead to the overproduction of cytokines, among which the pro-inflammatory cytokines in a positive feedback loop mechanism could further activate more immune cells. Thus, the formation of a “cytokine storm” leads to a sort of suicidal attack that not only contributes to the elimination of pathogenic microorganisms but also causes tissue injury affecting many organs. The cytokine storm leads to a systemic inflammation syndrome known as cytokine release syndrome (CRS), which has previously been reported in SARS-CoV-1 and MERS-CoV patients, as well as in leukemia patients receiving engineered T cell therapy (Chen H et al. 2019). In SARS-CoV-1, immune cells triggered the production of pro-inflammatory cytokines like IL-6, IFN-α/γ, TNF-α and in MERS-CoV though delayed but increased production of IL-6, IL-8, IL-1β, IFN-α has been observed. In addition, one of the main causes reported for the deaths of patients with SARS-CoV and MERS-CoV was the “cytokine storm.” A large number of factors are responsible for the dysregulated release of cytokines in case of these infections. It is assumed that once SARS-CoV2 binds to the ACE2+ epithelial cells in the lungs, rapid viral replication in the first stages of the infection results in high pro-inflammatory responses. In addition, the virus induces the release of large amounts of proteins that are known to delay IFN responses, which further provoke an accumulation of pathogenic inflammatory monocyte-macrophages. This, in turn, results in even higher production of cytokines in the lungs and the recruitment of more immune cells. The consequences of this hyper-inflammatory cascade are diverse, ranging from the dampening of T-cell responses, which leads to an even less controlled inflammatory response, to the apoptosis of epithelial cells, vascular damage, and ARDS (Fig. 14.3) (Channappanavar and Perlman 2017).
In the case of COVID-19 the patients in the critical stage, needing intensive care hospitalization and oxygen therapy, had higher levels of plasma inflammatory cytokines IL-2, IL-7, IL-10, G-CSF(granulocyte colony-stimulating factor), IFN-γ and MCP, and TNF-α circulating in the blood as compared to patients with mild or no symptoms, thus indicating a positive correlation between cytokine storm and disease severity. These listed cytokines suggest that both Th1 and Th2 responses are involved in COVID-19 pathogenesis, unlike SARS-CoV infection. It is becoming evident from many published reports that the IL-6 and GM-CSF cytokines released by T lymphocytes and mononuclear cells are the key link of cytokine storm in COVID-19 in adult patients (Siddiqi and Mehra 2020). Although not much data is yet available from pediatric patients, yet the available literature indicates similar involvement of IL-6, IL-10, and IFN-γ in the severity of disease in children below 15 years of age. Thus, early recognition and prompt treatment can lead to better outcomes. Several biological agents targeting cytokines have been proposed for treating Cytokine storms. Anakinra, an IL-1 receptor (IL-1R) antagonist, which is used in the treatment of rheumatoid arthritis, has also proven to be helpful in cytophagic histiocytic panniculitis with secondary hemophagocytic lymphohistiocytosis, a disease associated with severe cytokine storm, is been proposed and under trial (NCT04324021). Tocilizumab, a recombinant humanized IL-6 receptor antagonist that interferes with IL-6 binding to its receptor and blocks signaling, being the other one. Tocilizumab is already being used in the treatment of rheumatoid arthritis, juvenile idiopathic arthritis, and has proven valuable in the treatment of Cytokine storm triggered by CAR-T cell therapy for hematological malignancies. Thus, tocilizumab can be used as a candidate drug in managing the cytokine storm accompanying COVID-19 (Xu et al. 2020). Sarilumab is another IL-6R blocker that is being investigated in SARS-CoV-2 infection, which may reflect a possible clinical benefit regarding early intervention with IL-6-modulatory therapies for COVID-19. Thus, targeting cytokines during the management of COVID-19 patients could improve survival rates and reduce mortality.
14.5.2 Emergence of Antigenic Drift in SARS-CoV-2
Several approaches and vaccines are being developed worldwide to combat the COVID-19 pandemic. The major area of focus has been toward antibody-based immunity to coronaviruses. In this context, based on some recent findings regarding the detection of some mutations in the viral genome that may lead to the emergence of viral species which are more infectious and highly transmissible in nature, the attention has diverted to the fact whether these viruses evolve to escape recognition by the immune system and if they do so what is the rate at which they change. Such kind of a phenomenon is known as antigenic evolution. For example, infection with the influenza virus produces antibodies that generally protect humans against that same viral strain for at least several decades. Unfortunately, the influenza virus undergoes rapid antigenic evolution to escape these antibodies, meaning that although immunity to the original viral strain lasts for decades, humans are susceptible to infection by its descendants within about 5 years (Couch and Kasel 1983). This continual antigenic evolution is the reason that the influenza vaccine is periodically updated. Similarly, in the case of SARS-CoV2, recently a new strain has been detected in a sizable number of infected individuals in the United Kingdom and has been given the code name B.1.1.7. Though only limited data is available on coronavirus antigenic evolution but a few studies conducted in 1980s on 229E strain showed that individuals infected with one strain of 229E were resistant to reinfection with that same strain but partially susceptible to a different strain, which evolved from the original strain of virus (Reed 1984). Additional experimental studies suggest that sera or antibodies can differentially recognize spike proteins from different 229E strains (Shirato et al. 2012). Thus, consideration of antigenic drift in the different sub-strains of the virus is imperative in the design of a “one size fits all” universal vaccine to offer protection against the deadliest outbreak of this century. Since many leading SARS-CoV-2 vaccines use new technologies such as mRNA-based delivery that should make it easy to update the vaccine if there is antigenic evolution in spike. So for these reasons, the antigenic mutation in SARS-CoV-2 should be monitored as it might help in periodically updating the vaccine.
14.5.3 Vaccine Strategies
The widespread transmission of SARS-CoV2 infection across the globe has accelerated the development of vaccines using S-protein as the target antigen. Various vaccine platforms are being used, from traditional whole pathogen vaccine (live-attenuated vaccine, inactivated vaccine) to new generation vaccines (recombinant protein vaccine, vector-based vaccine) (Table 14.1). Some of these vaccines have already completed clinical trials showing a high level of immunogenicity and safety. Looking at the satisfactory levels of efficacy of these vaccines against the severe forms of the disease the manufacturers of these vaccines have submitted the interim analysis data to the regulatory bodies of different countries including WHO and got “Emergency use approval” in many countries world over. The vaccination drives have already started in these countries with the United States and India top-listing.
14.6 Summary
In December 2019, an outbreak of COVID-19 emerged in Wuhan city of China and soon it spread to different parts of the world. The rapid spread and increasing mortalities from this disease have alarmed the whole world and demanded urgency in both basic science and clinical research and each nation has met the current need with remarkable productivity. Within a short span of time, there has been a generation of enormous data which helped us to understand the basic pathophysiology of SARS-CoV2, but still, there remain many grey areas. Studies from the past on SARS-CoV and MERS-CoV have helped us better understand the current new virus but besides many similarities, there is considerable difference in the host immune response and disease profile as far as the SARS-CoV2 infection is concerned. The most devastating response to the viral infection studied so far is the excessive inflammation and cytokine release syndrome in COVID-19. Therefore, it is important that these immune mechanisms are further studied in larger group of patients so that better therapeutic strategies for COVID-19 can be developed. Currently, few vaccine candidates have been approved for administering in healthy volunteers but mass production and making the vaccine available world-over and administrating it globally is another major challenge, which is going to be faced in coming times. The success of the vaccine or any other therapy would further depend on the emergence of escape mutants of the virus. Hence, further investigations are required in order to reach precise and most efficient ways to combat this global health issue.
References
Abbasi J (2020) The promise and peril of antibody testing for COVID-19. JAMA 323(19):1881–1883. https://doi.org/10.1001/jama.2020.6170
Blanco-Melo D et al (2020) Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181(5):1036–1045.e9. https://doi.org/10.1016/j.cell.2020.04.026
Bosmann M, Ward PA (2014) Protein-based therapies for acute lung injury: targeting neutrophil extracellular traps. Expert Opin Ther Targets 18(6):703–714. https://doi.org/10.1517/14728222.2014.902938
Bouvet M et al (2010) In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLOS Pathog 6(4):e1000863. https://doi.org/10.1371/journal.ppat.1000863
Cao B et al (2020) A trial of Lopinavir–ritonavir in adults hospitalized with severe Covid-19. N Engl J Med 382(19):1787–1799. https://doi.org/10.1056/NEJMoa2001282
Channappanavar R, Perlman S (2017) Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol 39(5):529–539. https://doi.org/10.1007/s00281-017-0629-x
Chen H et al (2019) Management of cytokine release syndrome related to CAR-T cell therapy. Front Med 13(5):610–617. https://doi.org/10.1007/s11684-019-0714-8
Chen L et al (2020a) Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis 20(4):398–400. https://doi.org/10.1016/S1473-3099(20)30141-9
Chen T et al (2020b) Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ 368:m1295. https://doi.org/10.1136/bmj.m1091
Chu H et al (2020) Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis 71(6):1400–1409. https://doi.org/10.1093/cid/ciaa410
Couch RB, Kasel JA (1983) Immunity to influenza in man. Annu Rev Microbiol 37(1):529–549. https://doi.org/10.1146/annurev.mi.37.100183.002525
Cui F, Zhou HS (2020) Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens Bioelectron 165:112349. https://doi.org/10.1016/j.bios.2020.112349
Dandekar AA, Perlman S (2005) ‘Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol 5(12):917–927. https://doi.org/10.1038/nri1732
Diao B et al (2020) Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol 11:827. https://doi.org/10.3389/fimmu.2020.00827
Dienz O et al (2009) The induction of antibody production by IL-6 is indirectly mediated by IL-21 produced by CD4+ T cells. J Exp Med 206(1):69–78. https://doi.org/10.1084/jem.20081571
Feng Z et al. (2020) The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv, p. 2020.03.27.20045427. https://doi.org/10.1101/2020.03.27.20045427
Fung TS, Liu DX (2019) Human coronavirus: host-pathogen interaction. Annu Rev Microbiol 73(1):529–557. https://doi.org/10.1146/annurev-micro-020518-115759
Gordon DE et al (2020) A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583(7816):459–468. https://doi.org/10.1038/s41586-020-2286-9
Gralinski LE et al (2018) ‘Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio 9(5):e01753–e01718. https://doi.org/10.1128/mBio.01753-18
Grein J et al (2020) Compassionate use of Remdesivir for patients with severe Covid-19. N Engl J Med 382(24):2327–2336. https://doi.org/10.1056/NEJMoa2007016
Guo L et al (2020) Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis 71(15):778–785. https://doi.org/10.1093/cid/ciaa310
He R et al (2020) The clinical course and its correlated immune status in COVID-19 pneumonia. J Clin Virol 127:104361. https://doi.org/10.1016/j.jcv.2020.104361
Hoffmann M et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271–280.e8. https://doi.org/10.1016/j.cell.2020.02.052
Huang I-C et al (2011) Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog 7(1):e1001258. https://doi.org/10.1371/journal.ppat.1001258
Kaneko N et al (2020) Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell 183(1):143–157.e13. https://doi.org/10.1016/j.cell.2020.08.025
Kawasaki T et al (2018) DPP4 inhibition by sitagliptin attenuates LPS-induced lung injury in mice. Am J Physiol Lung Cell Mol Physiol 315(5):L834–L845. https://doi.org/10.1152/ajplung.00031.2018
Kimball A et al (2020) Asymptomatic and Presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility—King County, Washington, March 2020. Morbidity Mortality Weekly Report 69(13):377–381. https://doi.org/10.15585/mmwr.mm6913e1
Kucharski AJ et al (2020) Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis 20(5):553–558. https://doi.org/10.1016/S1473-3099(20)30144-4
Li G et al (2020) Coronavirus infections and immune responses. J Med Virol 92(4):424–432. https://doi.org/10.1002/jmv.25685
Lynch M et al (2016) Genetic drift, selection and the evolution of the mutation rate. Nat Rev Genet 17(11):704–714. https://doi.org/10.1038/nrg.2016.104
Payne DC et al (2016) Persistence of antibodies against Middle East respiratory syndrome coronavirus. Emerg Infect Dis 22(10):1824–1826. https://doi.org/10.3201/eid2210.160706
Qin C et al (2020) Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis 71(15):762–768. https://doi.org/10.1093/cid/ciaa248
Reed SE (1984) The behaviour of recent isolates of human respiratory coronavirus in vitro and in volunteers: evidence of heterogeneity among 229E-related strains. J Med Virol 13(2):179–192. https://doi.org/10.1002/jmv.1890130208
Shirato K et al (2012) Differences in neutralizing antigenicity between laboratory and clinical isolates of HCoV-229E isolated in Japan in 2004-2008 depend on the S1 region sequence of the spike protein. J Gen Virol 93(Pt 9):1908–1917. https://doi.org/10.1099/vir.0.043117-0
Siddiqi HK, Mehra MR (2020) COVID-19 illness in native and immunosuppressed states: a clinical–therapeutic staging proposal. J Heart Lung Transplant 39(5):405–407. https://doi.org/10.1016/j.healun.2020.03.012
Tai W et al (2020) Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol, pp 1–8. https://doi.org/10.1038/s41423-020-0400-4
Walls AC et al (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181(2):281–292.e6. https://doi.org/10.1016/j.cell.2020.02.058
Wang R et al (2015) The role of C5a in acute lung injury induced by highly pathogenic viral infections. Emerg Microbes Infect 4(5):e28. https://doi.org/10.1038/emi.2015.28
Wang F et al (2020a) Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis 221(11):1762–1769. https://doi.org/10.1093/infdis/jiaa150
Wang M et al (2020b) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30(3):269–271. https://doi.org/10.1038/s41422-020-0282-0
Wenjun W et al (2020) ‘The definition and risks of cytokine release syndrome-like in 11 COVID-19-infected pneumonia critically ill patients: disease characteristics and retrospective analysis. medRxiv, p. 2020.02.26.20026989. https://doi.org/10.1101/2020.02.26.20026989
Wilk AJ et al (2020) A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med 26(7):1070–1076. https://doi.org/10.1038/s41591-020-0944-y
Xiong Y et al (2020) Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect 9(1):761–770. https://doi.org/10.1080/22221751.2020.1747363
Xu X et al (2020) Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci U S A 117(20):10970–10975. https://doi.org/10.1073/pnas.2005615117
Yang Y et al (2013) The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 4(12):951–961. https://doi.org/10.1007/s13238-013-3096-8
Yang X et al (2020) Analysis of adaptive immune cell populations and phenotypes in the patients infected by SARS-CoV-2. medRxiv, p. 2020.03.23.20040675. https://doi.org/10.1101/2020.03.23.20040675
Zhang L et al (2006) Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J Med Virol 78(1):1–8. https://doi.org/10.1002/jmv.20499
Zhao J et al (2016) Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity 44(6):1379–1391. https://doi.org/10.1016/j.immuni.2016.05.006
Zheng M, Song L (2020) Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell Mol Immunol 17(5):536–538. https://doi.org/10.1038/s41423-020-0385-z
Acknowledgments
The authors would like to acknowledge the COVID-19 management team of healthcare workers in Post Graduate Institute of Medical Education and Research (PGIMER) Chandigarh, Director Health Services, Chandigarh Administration, and Indian Council of Medical Research, Ministry of Health & Family Welfare, Govt. of India for their unending efforts in the management of this disease. The authors learnt a lot from the online training programs being conducted at various levels, which has helped in many ways, and Biorender.com for helping us create the figures for the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive licence to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Vohra, M., Arora, S.K. (2021). Immunology and Pathogenesis of COVID-19. In: Sobti, R.C., Dhalla, N.S., Watanabe, M., Sobti, A. (eds) Delineating Health and Health System: Mechanistic Insights into Covid 19 Complications. Springer, Singapore. https://doi.org/10.1007/978-981-16-5105-2_14
Download citation
DOI: https://doi.org/10.1007/978-981-16-5105-2_14
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-5104-5
Online ISBN: 978-981-16-5105-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)