Abstract
With the rapid development of DNA nanotechnology and the continuous improvement of DNA editing technology, various DNA nanostructures have been constructed. DNA is the carrier of biological genetic information, and almost all organisms contain DNA, so DNA nanostructure has good biocompatibility. Benefiting from its excellent biocompatibility and editable properties, this emerging material is showing promising applications in many fields. Tetrahedral framework nucleic acids (tFNAs) are one of the most widely used typical structures of DNA nanostructures. In recent years, DNA tetrahedral frame nucleic acids have become a focus of biomedical research because of its stable structure, nanometer size, excellent mechanical properties, convenient synthesis, and high yield. Besides, they have good biocompatibility and biodegradability and are rich in modification sites. Moreover, because they can cross cell membranes without any help, they have promising applications in building intelligent drug transport systems. In this paper, the development of DNA nanostructures, the application of DNA tetrahedral frame nucleic acid in drug delivery, and the current problems are reviewed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
5.1 Introduction
Of late years, DNA nanotechnology is advancing by leaps and bounds [1, 2]. In addition to the structural characteristics brought about by the nanoscale, such as surface effect, tunnel effect, small size effect, and so on, DNA nanostructures also possess the characteristics of good biocompatibility, editable property, and strong stability brought about by the nature of DNA structure. DNA nanotechnology has made remarkable progress since professor Seeman first prepared DNA nanostructures in 1982 [3]. DNA nanotechnology has come a long way since it was developed in the 1990s, from simple modular assembly to multiple origami structures. At present, various DNA nanostructures are applied in various fields such as molecular detection, tumor diagnosis, biomedicine, drug delivery, biomolecular assembly, biosensors, nanomolecular machines, targeted therapy, and so on [4, 5]. Tetrahedral framework nucleic acids (tFNAs) are a typical representative of DNA nanostructure, which is made up of four single strands of DNA that self-assemble. Tetrahedral framework nucleic acids have been widely studied because of its simple preparation method and high yield [6, 7]. Tetrahedral framework nucleic acid, due to its ability to enter cells, makes the biological imaging and intelligent drug transportation in cells and animals based on DNA nanotechnology become new development opportunities [8]. This paper reviews the latest research on DNA nanomaterials and introduces different functional modification methods of DNA tetrahedral nanomaterials, and the application of DNA tetrahedral frame nucleic acid in drug delivery and the problems faced are reviewed.
5.2 DNA Nanostructures
5.2.1 The Concept of DNA Nanostructures
DNA nanostructures are two-dimensional or three-dimensional nanomaterials composed of single strands of DNA following the principles of Watson-Crick base pairing. DNA molecule has remarkable molecular recognition performance and remarkable structural characteristics, which makes it have unique advantages in the nanoscale regulation of materials, and also shows a broad application prospect in many fields. DNA is stored in the nucleus as a vehicle for carrying genetic information, which is made up of four different deoxynucleotide molecules. It was Watson and Crick who first proposed in 1953 that DNA is a large molecule with a double helix structure [9]. Soon after, researchers have focused on DNA that can make accurate base complementary pairs and gradually applied it to fields such as medicine, genetics, and ecology. DNA is not only a vehicle for carrying genetic information of living organisms but also the ideal component of biological functional materials [10]. DNA nanostructures are composed mainly by the self-assembly of DNA molecules. Self-assembly is one of nature’s main methods for assembling highly complex materials [11, 12]. DNA strand is assembled into a double helix structure with its complementary strand under the principle of exact base pairing by the synergistic action of hydrogen bond, stack, electrostatic, and hydrophobic. In the preparation of DNA nanostructures, the first step is to have a positive sequence design, and then the DNA molecules are spontaneously assembled into DNA nanostructures by intermolecular or intramolecular hybridization under appropriate solution conditions [13,14,15]. DNA self-assembles into DNA nanostructures following the principle of base complementary pairing. As a kind of nano-biological materials with precise structure and size, DNA nanostructures have wide application prospect in many fields.
5.2.2 The Development of DNA Nanostructures
With the advantages of bottom-up self-assembly strategy, high controllability, and precision, DNA nanotechnology based on DNA components has attracted the attention of related research fields. In 1982, professor Seeman put forwarded that DNA can form specific structure through Watson Crick’s pairing principle and the structure of the single can form in sticky end complex two-dimensional or three-dimensional structure, leading the whole of the development in the field of DNA nanotechnology [16, 17]. After this, the researchers in DNA, for “building materials,” by “bottom-up” constructing method, designed and synthesized DNA, and the DNA of different shapes of various functional nanostructures, DNA nanotechnology has penetrated into many research fields [18,19,20]. In the early days of DNA nanotechnology, the synthesized structure was only a cross and topological structure formed by a number of single DNA strands paired with complementary bases. In 1993, Seeman’s team designed multi-crossing structures that guarantee the mechanical strength of DNA nanostructures, such as multiple crossing sites between the double helix domains that forming rigid planar structures [21]. After all these, a large number of three-dimensional polyhedron structures were synthesized by the method of multichain base pairing, such as tetrahedron. Then, Mao et al. made further improvements, greatly reducing the types of DNA strands needed, reducing costs and experimental errors [22]. Rothemund invented the DNA origami in 2006, and the emergence of the technology makes the production of complex DNA nanostructures ability improved greatly [23]. The DNA origami is a nucleation self-assembly process, and the whole process passes through several nucleation points at one time. Therefore, the complexity of self-assembly of graphic modules generated by DNA origami is greatly increased. Through DNA origami techniques, researchers have constructed a variety of intricate nanopatterns and nanostructures, including smiley faces, dolphins, maps of China, huge rocks, nuts, bridges, flask, stereoscopic vases, pentacle stars, squares, rectangles, triangles, hollow boxes, tetrahedrons, and cubes [24,25,26]. In addition, each staple chain in a template constructed by DNA module or DNA origami can be extended to a specific identifiable sequence, which allows templates constructed by DNA nanotechnology to be further functionalized and widely used in many research fields [27].
With the continuous progress of technology, DNA nanostructures begin to move from the purely basic, structural research to specific applications. In the 552 issue of Nature magazine in 2017, four articles were published in the form of cover articles, which consisted of the mass synthesis of single DNA strand, the construction of large-scale DNA structure by DNA nano-tiles, and the 2D DNA origami patterns made by combining computer programming [28,29,30]. These directions show us the wide application prospect of DNA nanostructure. As it turns out, DNA nanostructure is developing faster and applied in more directions than we thought. When it comes to applications of biological and medicine fields, DNA nanostructures are widely used in the design of biosensors [31,32,33,34]. Considering the chemical nature of DNA nanostructures, multiple oligonucleotide sequences or aptamers are connected to various DNA nanostructures and constructed as biosensors of microRNA or protein, so as to be used in the detection of various diseases. DNA nanostructures also show promising prospects as drug carriers [35, 36]. Through the characteristics of easy editing of DNA, researchers modified drugs or aptamers on the DNA nanostructure to obtain the targeted drug delivery with good biocompatibility. For example, Kim et al. reported for the first time a DNA mirror nanostructure which is self-assembled, with good biocompatibility and greater serum stability [37]. On this basis, after incubating doxorubicin, Kim et al. obtained stronger tumor inhibition effect in vivo and in vitro than the conventional way of drug administration. DNA nanostructures have also made a breakthrough in the field of gene editing, a hot topic in recent years [38, 39]. For example, Gu et al. achieved efficient transfection of CRISPR-cas9 by using the structure of DNA strands, providing a new idea for gene editing. Beyond biomedicine, DNA nanostructures play a role beyond our wildest dreams [40]. Organick published an article in Nature Biotechnology in February 2018, claiming that the writing and reading of binary data in DNA has been realized by taking advantage of the easy editing of DNA and benefiting from the extremely high data density of DNA, which is expected to bring technological innovation in big data storage [41]. It is very clear that with the increasing understanding of DNA nanostructures, interdisciplinary research is beginning to make groundbreaking breakthroughs [42]. These researches and advances have shown us a new material and structure motif with broad application prospect, and it is exerting broad and far-reaching influence in related fields.
5.3 Tetrahedral Framework Nucleic Acids
tFNA is one of the most widely studied DNA nanostructures in recent years. DNA tetrahedral nanomaterials are simple and strong pyramidal 3D structural models, which have high mechanical rigidity and stability, rich in functional modification sites, simple production process, and high yield [43,44,45,46]. In particular, the DNA tetrahedral nanomaterials based on synthetic models, such as Turberfield, have shown promising applications in biological detection, in vivo imaging, gene carrier, and drug delivery [47]. DNA tetrahedral framework nucleic acids have some unique advantages over traditional DNA nanostructures in terms of biocompatibility, relative stability, and ease of editing.
5.3.1 Self-assembly
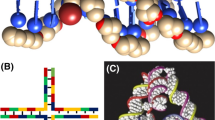
tFNAs are three-dimensional DNA nanostructures with tetrahedral shape, which is formed by the automatic hybridization of each strand through the clever DNA sequence design and the principle of complementary pairing [48,49,50]. Each ssDNA are divided into three segments, the three pieces respectively with other three different pieces ssDNA hybrid tetrahedral structure formation, and each small piece of the two hybrid combinations form tetrahedron has a double helix structure. On the tetrahedron vertex, ssDNA 5′- and 3′- at the end of each intersection, forming a port or on the edge [51]. DNA materials can achieve fine control of material molecules at the nanoscale. All at once, making the two adjacent sides of DNA tetrahedron at a certain angle, so as to ensure the correct formation of the structure of DNA tetrahedron and a certain degree of stability, each single strand of DNA between the adjacent two small fragments contains one base or two which are not paired with other sequences [52, 53]. Furthermore, in order to synthesize a regular tetrahedron of DNA, each ssDNA fragment must contain the same number of bases [54,55,56]. According to the Watson-Crick complementary base pairing principle, the four ss DNA were added to the TM buffer solution in equal quantities, and the four single-stranded DNA could be automatically complementary hybridized into a tetrahedral three-dimensional DNA structure through a one-step annealing operation. The synthesis conditions are as follows: 10 min at 95 °C, after that cooling to 4 °C for 20 min [57,58,59,60].
5.3.2 The Physical, Chemical, and Biological Characteristics
5.3.2.1 The Nanoscale and Editable
The DNA molecule is 2 nm wide; the distance between the two bases is 0.34 nm, and as a result the double helix structure increases by about 3.4 nm per 10 base pairs (bp) [61]. These properties determine the size adaptability of nucleic acid molecules. The size of DNA molecule determines that DNA material is an ideal nanomaterial. tFNA is a three-dimensional tetrahedral structure composed by multiple single-stranded DNA through the principle of complementary pairing of bases. The shape and size of tFNAs can be precisely controlled [62,63,64]. Researches show that the efficiency of the cells uptake of DNA nanostructure was influenced by its size and shape [65]. DNA nanostructures vary in size from nanometer to micron. This flexibility allows us to try a variety of sizes to compare and adjust the efficiency of endocytosis.
DNA molecules have a continuous length of 150 bp. When the double strand length is less than 150 bp, the DNA strand is rigid and not easily bent. According to this characteristic of DNA molecule, its rigidity and flexibility can be controlled by changing the number of bases on the double helix strand. Using this characteristic of DNA materials, researchers can design different rigid and flexible control nanomaterials according to different needs [66]. At present, the length of tFNAs used in most studies is 21 bp, which is rigid.
5.3.2.2 The Ability to Enter Cells
The efficiency of a material’s uptake by cells is not only affected by its charge but also largely related to the geometric properties such as the morphology and size of the material [67, 68]. Many investigations have shown that nanomaterials with a scale of 10–100 nm are relatively easier to be actively absorbed by cells [69]. Recent research has shown that DNA nanostructures can freely enter cells with great efficiency. The cell membrane is the main barrier for DNA oligonucleotides entering into a cell. Since the cell membrane is negatively charged, common DNA single strand and double helix can hardly enter the cell through the membrane. While DNA nanostructures are negatively charged, their unique nanoscale properties allow them to enter cells through endocytosis (including endocytosis and pinocytosis), an energy-dependent active transport process rather than simple diffusion [70]. Fan Chunhai’s team observed the interaction between RAW264.7 macrophages and fluorescently labeled tFNAs by confocal microscopy. After 2 h of culture, strong fluorescence signal was observed in the cytoplasm, which indicated that the DNA tetrahedral nanostructures can be ingested by cells. In the meantime, the single-stranded DNA that synthesis the tetrahedron was incubated with the cell, and only very weak fluorescence signals detected in the cytoplasm illustrated the formation of the tetrahedral structure for effective cells intake is important [71]. Flow cytometry instrument quantitative analysis results also show that compared with single DNA, after forming tetrahedral nanostructures, the amount taken up by cells increased significantly. Moreover, the tetrahedral structure of DNA can effectively resist the degradation of nucleases in biological culture medium and extracellular and inactivated fetal bovine serum. The tetrahedron structure remained intact after 4 hours of incubation. After the cells were incubated for 8 h, the cells and the DNA tetrahedron marked by Cy3 and Cy5 fluorescent markers are still well coincidently aligned, which fully proved that the formation of DNA nanostructures has a good stability. It is also can be used in biomedical research in the field of one of the important characteristics. Besides, Liang et al. found that, unlike the traditional DNA structure, tFNAs can cross the cell membrane to a certain extent and obtain a certain degree of lysosomal escape by modifying the nuclear nucleic acid aptamer [67]. This suggests that tFNAs may have a biological effect on cells that ordinary nucleic acids do not have, and at the same time, it may lead to breakthroughs in targeted drug delivery as a carrier and in biological imaging and intelligent drug transportation in animals. However, research on the movement of DNA nanostructures into cells and where they end up in cells is still in its infancy and controversial.
5.3.2.3 Biocompatibility and Biodegradability
DNA, as a natural biomolecule existing in all biological systems, has a large number of regulatory tool enzymes. Therefore, DNA nanomaterials have good biocompatibility and have no toxicity to organisms [72]. Moreover, DNA nanomaterials have good biodegradability because they it can be degraded by a variety of DNA enzymes. For DNA nanostructures, the shape and size do not influent their biocompatibility, which allows researchers to design the structures according to their needs, greatly expanding their applications in the life sciences [73].
To verify the safety of DNA nanomaterials in vivo, researchers investigated the distribution and metabolic behavior of DNA nanostructures of different shapes in tumor mice [64]. The results showed that DNA nanomaterials had no significant effect on tumor cell growth, apoptosis, and metabolism-related gene expression. After 12 days of injection of nanomaterial, the body weight of tumor mice did not change significantly compared with that of the control group. At the same time, histopathological analysis showed that these nanomaterials did not cause hepatorenal toxicity. All these results fully demonstrate that DNA nanomaterials have good biocompatibility. In addition, DNA of nanomaterials security in vivo and in vitro was evaluated by other researchers at the same time [74]. Preliminary results show that all kinds of DNA nanostructures have little cytotoxicity. DNA nanomaterials are degraded in vivo and excreted through the metabolic system, which does not lead to the accumulation of toxicity of organs, so it has good biological safety. It is important to note that although the size and shape of DNA nanostructures usually will not affect its biocompatibility, research shows that different DNA nanostructures is greatly different from the absorption and distribution in the body. Therefore, in order to achieve the ideal application effect of biology, we need to prepare a variety of DNA nanostructures and compare their distribution and metabolism in the body, so as to select the most optimal nanostructures for further research.
5.3.2.4 High Chemical Reactivity and Multiple Modification Sites
DNA and functional molecules can be coupled in a variety of ways: covalent modification, nucleic acid hybridization, biotin-affinity interactions, and DNA double-stranded embedding [75]. Based on the precise controllability of DNA nanostructures, it is possible to precisely control the position of functional molecules. The modification sites of DNA tetrahedron are abundant and it is a high capacity carrier. According to the modification position of functional groups or molecules in DNA tetrahedron, there are four main modification methods: vertex, capsule, mosaic, and cantilever [76].
Vertex modification refers to the modification of functional molecules on the vertices of DNA tetrahedron [77]. In addition, bioactive molecules or molecular specific recognition sequence can be modified at the vertices of DNA tetrahedron according to the experimental requirements. In order to synthesize such modified DNA tetrahedron, biological active molecules or specific sequences are often modified at the 5′ or 3′ ends of ss DNA, so that the two ends of four single-stranded DNA are joined at the vertex of the DNA tetrahedron. After that the designed four ss DNA are added to the TM buffer solution in equal quantities, and the DNA tetrahedral framework nucleic acid is formed through one-step annealing hybridization. Studies have indicated that the aptamer AS1411 has the effect of inhibiting the proliferation of tumor cells [78]. Li Qianshun et al. modified AS1411 aptamer to the vertex part of DNA tetrahedron. The results of this experimental showed that the DNA tetrahedral framework nucleic acid modified with AS1411 at the vertices could inhibit the growth of tumor cells. Moreover, because of the high selectivity of AS1411, it had almost no adverse effect on the growth of normal cells. This research provides a reference for the use of DNA tetrahedron as the carrier of collaborative therapy to deliver a variety of bioactive molecules.
Capsule modification involves wrapping functionalized molecules in a cage at the center of tetrahedral DNA [79]. The cavity in the center of the DNA tetrahedron can be used to encapsulate some nanoscale materials. For example, the DNA tetrahedral framework nucleic acids wrapped in cytochrome C can regulate the entry of apoptotic enzyme activator (Apaf-1). When Apaf-1 and cytochrome C forms a complex, the complex can initiate the cascade reaction of apoptotic protease. Erben et al. constructed a tetrahedron DNA nanostructure whose central cavity structure can contain functional small molecules [80]. They modified cytochrome C to the 5′ end of an oligonucleotide. By changing the sequence of oligonucleotides, the position (internal or external) of cytochrome C relative to the tetrahedron of DNA is regulated. This design can be used to the regulate function of protein. Zhou et al. prepared larger tetrahedral dendritic macromolecules by using AuNPs-wrapped DNA tetrahedron as monomer [81]. By replacing AuNPs with corresponding antigens, this DNA tetrahedral dendritic macromolecule gold nanoparticle conjugation method has promising applications in cancer treatment and immunotherapy.
Mosaic modification refers to the inlaying of functional biological small molecules or groups in the interior of the double helix structure of DNA tetrahedron by means of conjugation [82]. For example, in order to conduct biological imaging analysis or the analysis of the transportation route of DNA tetrahedral nanomaterials, the method of mosaic functionalization is often used to embed fluorescently labeled biomolecules or dyes in the interior of the double helix structure of DNA tetrahedron. Mosaic modification is widely used in the delivery of small-molecule anticancer drugs. The small-molecule anticancer drugs were embedded in the edge of the DNA tetrahedron, and the anticancer drugs were introduced into the target cells in the largest number by virtue of their ability to cross the cell membrane independently, and the cytotoxicity of DNA tetrahedron is little, so as to effectively improve the utilization rate of drugs and at the same time reduce the negative effects on human body to a greater extent. The amount of free doxorubicin (Dox) entering target cells is relatively small, and it has little cytotoxicity to drug-resistant cells. Dox, combined with DNA tetrahedral framework nucleic acids, can enter target cells in a relatively large number and has great toxicity to drug-resistant cells. Using DNA tetrahedron as carrier to transport Dox into breast cancer cells, which is drug-resistant, can better overcome the problem of drug resistance. In order to embed Dox into the double helix structure of DNA tetrahedron, Kim et al. incubated Dox with tFNAs and filtered unloaded Dox with G25 gel [83]. The experimental results show that DNA tetrahedron, as a drug transport system, can significantly inhibit the proliferation of drug-resistant cells and promote cell apoptosis. In summary, tFNAs, used as a drug carrier to reverse drug resistance of cancer cells in clinic, has a good application prospect.
Cantilever modification means the suspension of functional molecules on the side arms of the DNA tetrahedron. For example, by designing four ss DNA base sequences, the intersection of the 5′ and 3′ ends of the ss DNA is located on the edge of the tetrahedral nanostructure (middle or other non-vertex), where the 5′ or 3′ ends without complementary pairing extend outward for modification of functional molecules or groups [84]. Lee et al. used DNA tetrahedron as the carrier to deliver siRNAs into cells for silencing the expression of tumor-related target genes [85]. At first, they designed six single strands of ssDNA with complementary overhangs at the 3′ ends and then self-assembled the DNA tetrahedron. Among them, each edge of the tetrahedron is suspended in the middle of the uncomplementary paired sequence, which is used to connect the siRNA sequence. Finally, siRNA is immobilized on a cantilever of a DNA tetrahedron into the cell to silence targeted genes.
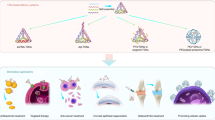
5.3.3 The Application of tFNAs as a Drug Carrier in Biomedicine Fields
As the carrier of drugs (small-molecule drugs, proteins, nucleic acids, etc.), nanomaterials have been the focus of nanobiology research for accurate drug delivery and controlled drug release. As an efficient and customizable carrier, DNA nano-self-assembly structure has the following advantages: good biocompatibility, targeting, controllability of structure, morphology, and surface chemical modification [86]. Anthracyclines commonly refer to small-molecule anticancer drugs that can be inserted into double strands of DNA to block the synthesis of biological macromolecules in living organisms. Nucleic acid drug molecules can be directly connected to DNA self-assembly structures by base complementary pairing. The study shows that DNA tetrahedron as a drug carrier can make nucleic acid drug molecules play a good role in the body [87, 88]. Other studies have reported that some nucleic acid drug molecules, such as nucleic acid aptamers and siRNA, can enter the cell with the help of DNA tetrahedron and play a role on cancer cells [89,90,91]. Functional DNA tetrahedron is widely used in biosensors, drug delivery, and biological imaging, due to its advantages of both DNA tetrahedron and specific functional molecules. Research has fully confirmed that DNA tetrahedron as a carrier can realize accurate drug delivery and controlled release in vivo, which has great application potential in the field of nano-diagnosis and treatment.
5.3.3.1 Transport Small-Molecule Drugs for Cancer Therapy
Cancer is a serious disease that seriously affects the health and life of mankind. Timely and effective treatment after the diagnosis of cancer is very important to improve the survival rate of patients. So far, chemotherapy is still the main treatment for malignant tumors. However, it has shortcomings such as poor solubility, poor cell penetration, poor specificity, large toxic and side effects, and easy to produce multidrug resistance. The cytotoxic effect of traditional chemotherapeutic drugs often causes systemic toxic reaction throughout the body, which reduces the patient’s tolerance and leads to the decrease of therapeutic effect [92]. Therefore, it is urgently needed to construct efficient new drug carriers to achieve efficient targeted drug transport, improve drug efficacy, reduce drug toxicity and side effects, and reverse the drug resistance of tumors. Ever since it was proposed, DNA nanotechnology has attracted wide attention in the field of biological detection and drug transportation, especially in the detection, imaging, and treatment of tumors, because of its structural programmability, good biocompatibility, no obvious cytotoxicity, and immunostimulation [93]. Doxorubicin (Dox) is a first-line antitumor drug, which can inhibit many kinds of tumors. Dox works by embedding bases into DNA, preventing mRNA formation and thus inhibiting DNA and RNA synthesis. Doxorubicin’s mechanism of action is to prevent the formation of mRNA by embedding DNA double strand, thus inhibiting the synthesis of DNA and RNA. It has the strongest inhibitory effect on RNA [94]. It is a cyclically nonspecific drug which has a killing effect on tumor cells in different growth cycles. DNA nanostructure is used as a carrier to carry Dox for antitumor, which is of great significance for improving the efficacy of Dox, reducing side effects and overcoming cell resistance. Using DNA tetrahedron as Dox carrier, Yan et al. obtained Dox-tFNA complex by incubating the synthesized DNA tetrahedron with Dox, and the Dox load efficiency was greatly improved [95]. The results indicate that the Dox-tFNA complex is not only cytotoxicity to human breast cancer cells but also cytotoxicity to Dox-resistant cancer cells. Furthermore, in comparison with free Dox, the killing effect of Dox-tFNAs complex on cancer cells was significantly improved and the clearance efficiency was decreased.
Targeted therapy means that drugs can be combined with targeted tumor cells to inhibit tumor growth. It is one of the important technical paths to overcome the toxicity of traditional chemotherapy system. MUC1, a kind of transmembrane glycoprotein, is overexpressed in a variety of cancer cells. MUC1 also plays a certain role in tumor growth, metabolism and metastasis [96]. In addition, the structure of MUC1 expressed in tumor cells is different from that expressed in normal tissue cells, so MUC1 has become a target molecule with certain tumor characteristics, which provides a biological basis for its application in targeted therapy. Studies have shown that monoclonal antibody against MUC1, vaccine, small-molecule ligand, and other methods can achieve certain initial effects in tumor inhibition experiments in vitro and in vivo [97]. It has been shown that tFNAs can be used as a drug deliverer to load the antitumor drug Dox.
Paclitaxel (PTX) is a diterpenoid alkaloid compound separated and purified from the bark of Taxus cuspidata. It is the most outstanding anticancer drug found in nature, which has been widely used in the therapy of ovarian cancer, breast cancer, lung cancer, and partial head and neck cancer. Due to the rapid proliferation of tumor cells, it can play an antitumor role by inhibiting the mitosis process of tumor cells [98]. PTX, as a new anti-microtubule drug, promotes the polymerization of microtubules, thus preventing the depolymerization of microtubules and disrupting the normal function of cells, thus inhibiting the mitosis of tumor cells and promoting cell apoptosis. Although the clinical treatment is very good, the wide application of PTX is limited by multidrug resistance. Recently, tFNAs have been thought as promising drug nanocarriers. In order to overcome paclitaxel resistance, tFNAs are used to deliver PTX into MDR cells as an efficient vehicle. Xie Xueping et al. incubated synthesized tFNAs and PTX to construct a PTX/tFNA drug transport system [99]. They compared the toxic effects of PTX/tFNA drug delivery system with that of free PTX on lung cancer cells and explored the mechanism by which PTX/tFNA drug delivery system reverse drug resistance. This study shows that PTX/tFNA drug delivery system can inhibit the growth of multidrug-resistant cancer cells significantly because of highly efficient load rate of PTX by the drug system. In addition, this study shows that PTX/tFNA drug delivery system has great potential in revers drug resistance and tFNAs have great application prospects in drug delivery and the treatment of multidrug-resistant cancers.
5.3.3.2 Transport Functional Nucleic Acids
Functional nucleic acid refers to nucleic acid molecules with special functions, including two categories: one is the DNA/RNA with similar properties to antibodies, which can specifically bind to the target molecule, called nucleic acid aptamer; another is nucleic acid molecule with similar catalytic activity to proteases is called DNAzymes [100]. Functional nucleic acid has the advantages of strong binding force, good selectivity, wide range of targets, good biocompatibility, easy synthesis, and easy functional modification. Functional nucleic acids include aptamer, antisense oligonucleotides, small interfering RNA (siRNA), microRNA, and other nucleotide sequences with special functions, which are widely used in the diagnosis and treatment of diseases. However, the unmodified functional nucleic acid molecules are easy to be degraded by various nucleases in the physiological environment, and the carrier needs to be able to protect the integrity of nucleic acid molecules for a long time and extend the circulation time of nucleic acid drugs in the body, so as to ensure that the nucleic acid drug molecules reach the target cells and play a role [101]. DNA tetrahedron and nucleic acid drug molecule homology is a good nucleic acid drug carrier. Since both the carrier and the drug are nucleic acids, it is convenient to carry the drug molecules by nucleic acid hybridization or embedding [102, 103].
5.3.3.2.1 Antisense Oligodeoxynucleotide
Antisense oligonucleotides (AONs) are nucleic acid molecules that inhibit the expression of a target gene by specifically binding to its DNA or mRNA in a sequence. By binding with specific target sequences of mRNA or DNA, antisense oligonucleotides can block mRNA transcription and translation, regulate the information transfer from gene to protein, and inhibit protein expression. The data show that oligonucleotides designed with 15–20 base sequences can specifically target any specific single gene in the human genome [104]. Since 1978, when Zamecnik and Stephenson et al. first reported that antisense oligonucleotides could inhibit the replication of chicken sarcoma virus, a large number of studies have confirmed the ability of antisense oligonucleotides to specifically inhibit gene expression, and their great potential in disease treatment has been gradually recognized [105]. Antisense oligonucleotides targeting specific oncogenes, growth factors and their receptors, and signaling pathways have been widely used as a new tool for in vivo and in vitro studies and as a promising drug for the therapy of diseases such as diabetes, asthma, cancer, and viral infections, attracting great interest [106]. However, antisense oligonucleotides are rapidly degraded when they enter the cell without modification, and the degradation rate is greatly reduced after modification or carrier binding. At present, most carriers are cationic particles and liposomes, which are cytotoxic. tFNA is considered to be the most ideal nano-drug carrier because of its editable, easy modification, and biocompatibility. Zhang Xiaolin et al. designed antisense oligonucleotides targeting the c-met gene and constructed antisense oligonucleotide-DNA tetrahedral drug delivery system for the treatment of cancer. The results showed that DNA tetrahedron could successfully transport antisense oligonucleotides into cells and antisense oligonucleotides could bind to the target gene mRNA, inhibit the expression of related proteins, and finally achieve the purpose of inhibiting tumor cell proliferation and promoting cell apoptosis [107]. In addition, Keum et al. constructed DNA tetrahedrons with antisense characteristics by using five oligonucleotides, which can bind to targeted mRNA and block the expression of some specific genes after entering cells [90]. The results indicated that, in comparison with linear DNA, DNA tetrahedrons show higher ability of cell uptake and gene silencing.
5.3.3.2.2 Antisense Peptide Nucleic Acid
Antisense peptide nucleic acid (asPNA) is a kind of synthetic DNA analogue, which has higher water solubility, stability, and base specificity compared with nucleic acid. The antisense peptide nucleic acid and double strand of DNA can form a triplet structure, which can block gene transcription and translation and inhibit gene replication by inhibiting the extension of DNA primers [108]. Antisense peptide nucleic acid can downregulate or inhibit expression of the target genes in gene replication, transcription, and translation, so as to achieve the goal of disease treatment. Antisense peptide nucleic acid, as third-generation gene therapy agent, has been widely used in vitro to inhibit the proliferation of tumor cells and the treatment of bacterial or viral infections. However, without the help of any drug delivery system, asPNA has a hard time crossing cell membranes [109, 110]. The further improvement of the intake of asPNA and the ability of asPNA to enter the nucleus has been a key issue affecting its wide clinical application. The traditional drug delivery system has many disadvantages, such as protease sensitivity, insufficient bioavailability, poor effective targeting, lack of cell specificity, immunogenicity to animal cells, and potential cytotoxicity. It is difficult for the unmodified synthetic antisense peptide nucleic acid to pass through the bacterial cell wall and be absorbed by the bacteria [111]. Therefore, bacterial cell wall is the biggest obstacles to the development of antisense peptide nucleic acid as a therapeutic antimicrobial agent. The cell penetrating peptide covalently connects with the antisense nucleic acid to facilitate the asPNA to penetrate the bacterial cell wall and transfer it into the cell. Although they are very effective carriers of asPNA, they can’t specifically identify bacterial cell walls and can show growth inhibition at high doses. In Zhang Yuxin’s study, a complex asPNA-tFNAs drug delivery system was constructed. In this study, with tFNAs as the carrier, asPNA with high specificity, high affinity, and specific targeting of ftsZ gene was transported into bacteria, in order to inhibit the expression of this gene and achieve the purpose of bacteria inhibition [112]. Guided by the principle of base complementary pairing, asPNA replaces part of the tFNAs sequence without changing the original structure, shape, size, or excellent carrier properties of the DNA tetrahedron. The results showed that the asPNA-tFNA vector system could penetrate the bacterial cell wall and carry antisense peptide nucleic acid targeting specific genes to block the expression of the specific genes, thus effectively inhibiting the growth and proliferation of MRSA bacteria. John has designed a self-assembling DNA tetrahedral framework nucleic acid as the drug carrier and incorporated a targeted asPNA in its structure to penetrating the cell wall of bacteria. This research show that the minimum inhibitory concentration (MIC) was reduced when the asPNA is carried by the DNA tetrahedron, contrasting with no reduction in MIC when PNA4 is used alone [109]. The drug delivery system has the ability to penetrate the bacterial cell wall and deliver the targeted synthesis of asPNA, which has a wide range of optimization selection and potential application.
5.3.3.2.3 Aptamer
Aptamer is a small fragment oligonucleotide sequence or short polypeptide selected in vitro, which can bind with the corresponding ligand and has the advantages of high affinity and strong specificity. The appearance of aptamers provides a new research platform for chemical biology and biomedical science to identify nucleic acid aptamers quickly and efficiently [113]. In 1990, Tuerk and Ellingtong were the first to screen the specific RNA aptamers of phage T4DNA polymerase and organic dye, respectively [114, 115]. Since the concept of the aptamer was put forward in the 1990s, researchers have been devoting themselves to the study of the aptamer and found that the aptamer has many advantages. First of all, adapters have the advantages of short detection cycle, low detection limit, high affinity and strong specificity, which make adapters have larger surface area and a large number of receptor binding sites.The aptamer can combine with the target material based on van der Waals forces, hydrogen bonds and hydrophobic effects to form three dimensional structures such as helix, hairpin, stem ring, convex ring, clover and pseudo structure. Secondly, compared with antibodies, the selected aptamers are easy to be synthesized in vitro in large quantities, with good repeatability and high stability, and are easy to be stored [116]. Since then, aptamers have been widely used in cell imaging, drug development, disease treatment, and microbial detection. Researchers have developed a variety of anticancer drugs, and nanomaterials are widely used for drug delivery, but they have the disadvantages of poor biocompatibility, unable to deliver in a targeted manner and harmful to other cells. The aptamer can specifically bind to the target, and the nanomaterial-specific aptamer complex can realize the targeted drug delivery in the cell. Liu et al. constructed an icosahedral nanomaterial, which, after binding with the specific aptamer DNA, can carry doxorubicin and deliver it to the lesion location in a directional manner, specifically leading to the death of epithelial cancer cells [117].
AS1411 is a guanine-rich aptamer, which can form G-4 chain structure, can specifically bind to nuclides, and has many special biological activities. Nucleolinins are highly expressed on the nucleus and the surface of the tumor cell membrane, and AS1411 can enter the nucleus by the shuttle action of nucleolinins in the cell. At the same time, AS1411 can inhibit DNA replication; make the cells stay in the S phase, thus inhibiting cell proliferation; and promote cell apoptosis by interfering with the binding of nuclides and bcl-2 [118]. AS1411 has a great prospect in the treatment and diagnosis of cancer. Because of its good biocompatibility, good stability and modifiability, and simple synthesis method, DNA tetrahedron has a great prospect in drug loading, tumor therapy, and other fields. DNA tetrahedron can be used as nanometer drug carrier material with good biocompatibility [119]. Li Qianshun et al. modified AS1411 aptamer at the vertex of DNA tetrahedron to study the specific recognition effect of the drug delivery system targeting tumor cells [120]. The results showed that Apt-tFNA complex could enter McF-7 cells in large quantities and accumulate in the nucleus. However, relatively few Apt-tFNAs can enter into normal cells L929, suggesting that AS1411-modified tFNAs are an effective drug delivery vector targeting tumors. However, the amount of Apt-tFNA complex entering normal cell L929 was relatively small. These results suggest that tFNAs modified with AS1411 is an effective drug delivery vector targeting tumors cells. Bermudez et al. studied the effect of DNA tetrahedron modified with AS1411 aptamer on HeLa cells. The results showed that AS1411-tFNA drug delivery system was more easily absorbed than the AS1411 aptamer alone and it could inhibit the proliferation of HeLa cells significantly for up to 24 h [121]. However, AS1411-tFNA complex had little effect on the growth of noncancerous cervical cells. This result demonstrated that the AS1411 aptamer suspended on the tetrahedron of DNA is more easily recognized by the receptor and inhibits the growth of cancer cells through specific uptake and absorption.
5.3.3.2.4 siRNA
RNA interference (RNAi) refers to the phenomenon of gene silencing caused by degradation of target mRNA or inhibition of translation. The advantages of specificity, efficiency, and stability of RNA interference technology make it a hot topic in mechanism research in biomedical field in recent years, and it is widely used in cancer mechanism research and cancer therapeutic drug research [122, 123]. Single-stranded or double-stranded RNA containing 21–23 bases, namely, small stranded interfering RNAs (siRNAs), can effectively and specifically block the expression of homologous genes in vivo through RNA interference, promote homologous mRNA degradation, and induce cells to show the phenotype of specific gene deletion. siRNAs combine with proteins, such as eLF2c, Gemin3, Gemin4, and so on, forming the RNA-induced silencing complex (RISC). RISC can specifically recognize and degrade target mRNAs under the antisense chain of siRNAs [124,125,126]. Binding of siRNAs to target mRNA sites is a highly sequentially specific process, following the principle of complementary pairing of the Watson-Crick bases. Wilkins et al. found the regulatory program of RNA interference on Caenorhabditis elegans genes, and they found that RNA interference is the innate antiviral immune defense mechanism of Caenorhabditis elegans, and it is a conserved sequence-specific post-transcription gene silencing mechanism in the evolutionary process [127]. The discovery of RNA interference opens the door to the application of siRNAs therapy. siRNAs have shown great advantages and potential in basic research in molecular biology and in the treatment of viral infections, tumors, and genetic diseases. Tumors are often malignant growths of cells caused by overexpression of proto-oncogenes or loss of control of tumor suppressor genes. siRNAs can be used to specifically block the expression of these genes in vivo for therapeutic purposes. The development of RNA interference technology is of great significance for elucidating signal transduction pathways and discovering new drug targets, but their safe and effective introduction methods and methods of stable expression in vivo, as well as the expression of target gene downregulation at mRNA and protein levels caused by RNA interference effect need to be further studied [128]. The short time in vivo and short half-life in plasma of siRNA make its application limited. The lack of safe and effective vectors restricts the application of siRNA. Anderson et al. introduced siRNA into live nude mouse tumor model by using DNA tetrahedral nanomaterials as the carrier to inhibit the expression of target genes for tumor therapy [85]. The team suspended the siRNA on an arm of the DNA tetrahedron by means of base complementary pairing, which transported the DNA tetrahedron modified with the siRNA to the lesion site through the ligand connection with the cancer cell receptor. The results show that DNA nanostructure can improve the stability of RNA molecules, thus significantly improving the efficiency of RNA interference. Besides, Kim et al. used DNA tetrahedron as the transporter of siRNA to silence some genes in cancer cells, which modified folic acid molecule on the tetrahedron to promote the process of RNA interference [129]. In addition, blood circulation time of DNA tetrahedron modified by siRNA is longer than that of free siRNA. Since gene silencing can be carried out by transferring siRNA through tFNAs, this drug delivery system can not only silence tumor target genes by transferring siRNA but also be used to treat other gene-related diseases.
5.3.3.2.5 Num, Dpt.
CpG oligonucleotide sequences are widely found in bacterial genomes and a small amount in mammalian genomes, which are active agents to induce innate immunity and acquired immune response. It is regarded as the signal of pathogen invading the immune system and is recognized by toll-like receptor 9 to induce the immune response [130]. CpG oligonucleotide sequences can be used as a potential therapeutic DNA and immune adjuvant in the adjuvant treatment of infectious diseases, cancer, allergies, and asthma. However, unmodified CpG oligonucleotide sequences is generally easy to be degraded by nuclease, with low cell uptake rate, requiring high dose and repeated administration, which severely limits the application of CpG [131]. Since natural CpG sequences are easily degraded by nucleases, the development of nontoxic nanocarriers with high transport efficiency is of great significance to improve the stability and clinical treatment effect of CpG. In recent years, the development of DNA nanotechnology to solve the problem of nucleic acid drug delivery provides a new tool. A large number of research reports have shown that nanomaterial-loaded CpG nucleic acid drug has high activity, low toxicity and good biocompatibility, and is expected to become a new immunotherapy drug for the prevention and treatment of related diseases [132,133,134]. Fan Chunhai’s research group first used DNA tetrahedral nanostructures as CpG carriers. The DNA tetrahedral nanostructures in this experiment were assembled using four single-stranded DNA strands through a simple annealing reaction [135]. These four vertices of the tetrahedron extend 1–4 CpG oligonucleotide sequences. The cell uptake of CpG-tetrahedron was detected by fluorescence imaging and flow cytometry. The results showed that single-stranded CpG DNA was difficult to be absorbed by cells, but CpG-tetrahedron could be widely consumed by cells, proving that the tetrahedron structure plays an important role in promoting cell uptake. In addition, a series of experiments have demonstrated that the tetrahedral nanostructure can enhance the stability of CpG DNA inside and outside the cell. Moreover, they used CpG-modified tFNAs to study the immune activation of macrophages (RAW 264.7). Inflammatory cytokines stimulated by CpG-modified tFNAs, such as TNF-α, IL-6, and IL-12, were much more numerous than single-stranded CpG sequences. This study indicated that tFNAs can increase CpG load, thus inducing immune response more obviously, suggesting that tFNAs can be used as a good carrier for immunotherapy.
5.3.3.3 Transport Peptides and Proteins
Peptides and proteins, including vaccines, immunoglobulin, and enzymes, can be used as drugs for the prevention, treatment, and diagnosis of diseases. Peptides and proteins are endogenous substances of human body or developed for biological regulators in vivo [136]. Peptides and proteins act by promoting or inhibiting physiological and biochemical processes in the human body or in bacterial viruses. Advantages of peptides and proteins include low side effects, high potency, and strong pertinence, and they will not accumulate in the body and cause poison [137]. Polypeptides and protein-based drugs are the most active and the fastest developing drugs in the field of pharmaceutical research and development, and they are among the most promising industries of the twenty-first century. However, there are also many problems in the application of peptides and proteins drugs. For example, the structure of protein drugs is unstable, and it may be inactivated due to a variety of complex chemical degradation and physical changes in the in vivo and in vitro environment [138]. In addition, protein drugs have many disadvantages, such as short half-life, high clearance rate, large molecular weight, poor transmittance, vulnerability to the destruction of enzymes, bacteria and body fluids, and low bioavailability of non-injectable drugs. Studies have shown that peptides and proteins can bind to DNA nanostructures and enter cells [136, 139, 140]. Yan’s team reported on the construction of nanoscale vaccines using DNA tetrahedrons. By the specific binding of biotin-avidin, the biotin-modified DNA tetrahedron loaded a model antigen of streptomycin into mice [141]. The tetrahedral antigen complex can induce a strong antibody response in mice continuously and steadily, while the tetrahedral or antigen alone does not stimulate any immune response. Catabolite activator protein (CAP) is an important transcription factor that regulates over 100 genes. Recently, Turberfield’s team provided a template for DNA-protein assembly by designing CAP recognition sites on the side chains of DNA tetrahedrons and assembling caps into tetrahedrons in a non-covalent manner [142]. Zhao et al. found a fast and efficient method for enzyme inclusion: first, attach a single enzyme molecule to half of the DNA cage, and then combine the two half-cage structures to successfully load the enzyme into the DNA cage cavity [143]. They also recorded glucose oxidase (GOX) and HRP in a DNA cage at a ratio of 1:1. As a carrier of proteins, DNA nanocages (such as tFNAs) can protect proteins from the influences of external environment (such as the biodegradation of proteins by enzymes), to protect the activity of proteins and improve the stability of proteins.
5.3.3.4 Transport Multiple Drug Molecules
When two or more drugs are used together, if they act in the same direction, the effect of mutual enhancement is called synergy. Based on the synergistic effect, the therapeutic effect can be enhanced when the DNA tetrahedron co-transports multiple drug molecules which act in the same direction [144]. In many cases, the actual treatment of a disease requires the synergistic action of multiple drugs for composite therapy, which requires the nanomaterial delivery system to simultaneously deliver multiple drug molecules and maintain their respective activity. Because DNA tetrahedral modification sites are abundant, it can carry different drug molecules at the same time. To enhance the effect of killing tumor cells and drug targeting, Dai Phuendong et al. conjugated MUC1 nucleic acid aptamer (Apt) with DNA tetrahedron to construct a targeted doxorubicin delivery system [145]. This study also evaluated whether the vector system could bind to MUC1-positive tumor cells specifically and be cytotoxic to MUC1-positive tumor cells. MUC1 aptamer and DNA tetrahedron can be used to construct the drug carrier with targeted action. Since MUC1 aptamer can specifically identify tumor cells, it improves the targeting effect of DOX and the killing effect on tumor cells. The results showed that Apt-TDN-Dox drug carrier system could produce significant cytotoxicity to MUC1-positive cancer cells, which is more effective than the free Dox. In addition, Liu et al. used DNA tetrahedron to co-transport CpG and streptavidin. CpG sequence can be used as an adjuvant to enhance the efficacy of vaccine due to its strong immune stimulation activity, so the antigen-CpG-DNA tetrahedron complex can continuously induce a stronger immune response [146]. Therefore, the coordinated delivery strategy is of great significance in reducing the dosage of drugs and improving the therapeutic effect, which is worthy of further exploration by researchers.
5.3.4 The Current Problems of tFNAs as a Drug Carrier in Biomedicine Fields We Faced
The simple chemical synthesis connects functional molecules (inorganic small molecules, biological macromolecules, inorganic nanoparticles, etc.) to the DNA tetrahedron to form multifunctional DNA composites. These composites have shown great potential in the field of nano-diagnosis and treatment. However, there are still some problems in the application of DNA tetrahedron to living system [147,148,149]. For example, high cationic concentrations are required for DNA self-assembly structures to maintain structural stability; DNA’s ability to self-assemble structures across cell membranes is limited; After DNA self-assembly structure enters biological system, it is easily degraded by various biological enzymes in serum. These problems restrict its application to some extent. Therefore, it is very important to improve the stability of DNA self-assembly structure and improve kinetic performance and cellular uptake efficiency of DNA self-assembly structure.
5.3.4.1 Improves the Stability of DNA Tetrahedron In Vivo
As mentioned above, we believe that DNA tetrahedron has great application prospects in the biomedical field, including the use of DNA tetrahedron alone to regulate the biological behavior of cells, and the use of DNA tetrahedron as a carrier to achieve targeted therapy, which requires DNA tetrahedron to have certain stability in the systemic circulation and to avoid immune recognition to some extent [150]. The application of DNA tetrahedron to living system requires special consideration of its stability at low magnesium ion concentration, 37 °C, and nuclease conditions. Fetal bovine serum is a mixture used in cell and tissue culture usually. It contains a variety of biological enzymes that have a strong degradation effect on nucleic acid molecules. However, Bermudez’s team confirmed that the degradation rate of DNA tetrahedral structure in FBS was only 1/3 of that of a single strand [151]. They believed that this was mainly because the folded DNA structure affected the binding of enzymes to DNA strands, thus preventing the degradation of DNA strands by biological enzymes. In a solution containing 80% mouse serum, tFNAs were completely degraded after 12 h. However, in in vivo circulation, there are more adverse factors, including enzyme degradation, protein absorption and conditioning, RES clearance, etc., which put forward higher requirements for the stability of tFNA’s in vivo circulation. We need to use various methods to hide the surface features of DNA nanostructures in order to inhibit nonspecific clearance in vivo. The researchers added a variety of polymers, cationic particles, and so on to the DNA structure to coat the nucleic acid, thus achieving better stability and drug carrying capacity. Among the methods, polyetherimide (PEI) is a classic method. With proper PEI and nucleic acid ratio, the preparation can be completed after incubation at room temperature [152]. The synthesized complex has high transfection efficiency in the gene transfer field and can promote the lysosome escape of nucleic acid through proton sponge effect. However, due to the cationic characteristics of PEI, the biocompatibility is poor, in recent years, the academic community has also proposed a variety of PEI modification or replacement materials, so as to improve the cytotoxicity of PEI.
In addition, DNA chain modification can also inhibit the degradation of DNA by nucleases. Sleiman’s team modified a C12-alkyl chain on the DNA strand, using the complex to connect the DNA cubes to human serum albumin (HSA) to form a nanoscale superstructure that remained stable in the serum for 22 h, while DNA cubes without HSA remained stable in the serum for only a few minutes [153]. Dietz et al. studied the influence of temperature on the stability of DNA self-assembly structure, and they found that the self-assembly structure of DNA constructed by multilayer origami has a strong stability, which can be stable at more than 50 °C, and multilayer origami can effectively inhibit the degradation of nucleases [154]. These results suggest that DNA dense accumulation, chemical modification of DNA strands, and end protection are conducive to the good stability of DNA nanomaterials in the medium containing enzymes and at high temperature. In addition, the use of virus liposome garment at the end of the shell, such as package structure of DNA self-assembly, also can improve the DNA stability of nanomaterials in the physiological environment. Due to the complex internal environment of the organism, it has not been determined whether DNA nanostructures can maintain stability for a long time under the condition of low ion concentration in vivo. At present, the research of intelligent drug delivery is limited to in vitro, not involving cell and in vivo applications. How to develop and construct DNA nanomaterials that can cope with the complex biological environment, effectively complete the loading, transportation and unloading of macromolecules, and truly realize the biological motors in nature, so as to truly apply DNA nanomaterials to clinical medicine, still needs further exploration and solution by researchers.
5.3.4.2 Improves the Cell Uptake Efficiency of DNA Tetrahedron
Turberfield team first explored cells can ingest DNA self-assembly structure in without the help of transfection reagents [155]. They modified Cy3 and Cy5 fluorescence on the two edges of the DNA tetrahedron, respectively, and the result confirmed that the DNA tetrahedron could enter the cell without transfection reagent through fluorescence imaging. The fluorescence resonance energy transfer between Cy3 and Cy5 showed that the tetrahedron entering the cell still maintained a good integrity. Later, Mirkin’s team constructed “spherical nucleic acid” nanostructures by densifying DNA strands on nanoparticles, which could also enter cells efficiently without transfection agents, suggesting that three-dimensional DNA nanostructures were more conducive to cell uptake than single strands [156]. Studies have shown that the DNA tetrahedron enters the cell through receptor-mediated endocytosis and then enters the lysosome of the cell, which can remain stable in the cell for at least 8 h. Use the function of target molecules can enhance cell active uptake of DNA nanostructures. Mao et al. increased the uptake of DNA nanostructures by modifying folic acid on DNA nanostructures to make folate receptors highly expressed on the surface of tumor cells [157]. Nucleic acid aptamers can be directly connected to DNA nanomaterials through DNA synthesis, which can not only improve the uptake efficiency of DNA nanomaterials but also have specificity. “Hidden” DNA nanostructures are also an effective way to promote cell uptake of DNA nanomaterials. Perrault and Shih used viral capsid proteins and liposomes to wrap the DNA octahedron, which reduced its immunogenicity by 2 times and increased its biological effect by 17 times [158, 159]. In summary, compared with linear DNA chains, the symmetry, size, and functional modification of three-dimensional DNA nanostructures have obvious effects on the uptake efficiency of cells. Moreover, there is no definitive answer to the question of how DNA nanostructures interact with cells. The study of cellular uptake of DNA nanostructures is crucial for the subsequent biomedical applications of DNA nanostructures.
5.3.4.3 Improve the Synthetic Yield and Reduce the Synthesis Cost of DNA Tetrahedron
At present, using DNA nanostructure as drug carrier also has some disadvantages, such as low yield and high cost. First, although the construction of a rich and diverse nanostructure has been achieved, the mechanism of DNA assembly is still not very clear, and the thermodynamics and dynamics of the assembly process are still unclear [160]. Because of the unclear assembly mechanism, the mismatch problem in the assembly process is difficult to solve, and it is also difficult to get a stable high yield. Therefore, it is still a difficult problem to explore the assembly mechanism and obtain high yield and stable assembly. Secondly, DNA nanomaterials need to be purified. To solve this problem, the purification methods such as gel electrophoresis, high-performance liquid chromatography, and density gradient centrifugation were developed to improve the purity of the product to a certain extent [161]. In the end, although DNA nanotechnology has been applied to biomedicine, electronic science, and other fields, the high preparation cost of DNA nanostructure restricts the practical application of DNA nanotechnology [162]. Compared with other nanomaterials that can be prepared on a large scale, the synthesis cost of nanomaterials constructed from DNA molecules is still high. Therefore, the development of cheaper and more convenient means of DNA amplification is another challenge to be tackled. In recent years, with the development of DNA synthesis technology, the cost and price of commercial synthesis are rapidly decreasing [159, 163]. It is believed that the cost of synthesizing DNA nanostructures on a large scale commercially will fall to a level that medical applications can afford in the near future.
5.4 Conclusion
This paper reviews the concept of DNA nanomaterials and the development of DNA nanotechnology. In this paper, the synthesis and physicochemical characteristics of DNA tetrahedron, which is the most widely used and most studied of DNA nanomaterials, as well as the research progress of DNA tetrahedron for intelligent drug delivery, are also described in detail. By loading small molecules and protein drugs into DNA tetrahedron, the drug resistance of some drugs was reversed, and the problems of easy degradation, poor water solubility, and short retention time of some drug molecules were solved. Translocation of functional nucleic acid molecules by DNA tetrahedron is a new concept of cross-domain generation. Based on the compatibility and homology between drug molecules and carriers, DNA tetrahedron provides an incomparable platform for the transport of nucleic acid drugs. The intelligent drug delivery system based on DNA tetrahedron realizes the controlled release of drug molecules at the targeted sites, significantly improves the efficacy, and reduces the side effects. Functional DNA tetrahedron nanomaterials have the advantages of simple self-assembly, high yield, stable structure, good biocompatibility, and not easy degradation. They show a wide range of applications and good prospects in biosensors, separation analysis, biological imaging, and drug delivery.
At present, the application of DNA tetrahedral framework nucleic acids as drug carriers still faces many challenges. First, in the field of diagnosis and treatment of disease, at present, mice are still the main subjects of in vivo experiments on DNA tetrahedral framework nucleic acids, which still face many challenges nanostructures as drug transporters is one of the main targets of future research. Secondly, the exact mechanism by which cells take up the tetrahedron of DNA is still unclear. Recent studies have shown that the size, shape, charge, and cell type of DNA tetrahedral framework nucleic acids affect their cellular uptake, intracellular transport, and eventual destination. At present, little is known about the final transformation of DNA tetrahedron in cells, which is an important question for future research. In addition, the main challenges are to further improve the yield and the stability of DNA tetrahedral framework nucleic acids and explore the cellular uptake mechanism.
Despite many problems, DNA nanotechnology is still an invention with great potential. We have reason to believe that the development of DNA nanotechnology will bring new light to human beings in DNA chips, nano-devices, biomedicine, and other aspects and will certainly promote the progress of related fields. With the development of related researches, DNA nanostructures will have a broader development prospect in the application of drug transporters and intelligent drug carriers. DNA nanotechnology is still evolving as a cross-cutting science, drawing strength from fields such as physical chemistry, optics, and electronics. We have reason to believe that eventually it will show up in the biomedical field and be widely used.
References
Ding T, Yang J, Pan V, Zhao N, Lu Z, Ke Y, Zhang C. DNA nanotechnology assisted nanopore-based analysis. Nucl Acids Res. 2020; https://doi.org/10.1093/nar/gkaa095.
Seeman NC. DNA nanotechnology at 40. Nano Lett. 2020;20(3):1477–8.
Seeman NC. Nucleic acid junctions and lattices. J Theoret Biol. 1982;99(2):237–47.
Fan C, Li Q. Advances in DNA nanotechnology. Small. 2019;15(26):e1902586. https://doi.org/10.1002/smll.201902586.
Huo S, Li H, Boersma AJ, Herrmann A. DNA nanotechnology enters cell membranes. Adv Sci (Weinh). 2019;6(10):1900043.
Li S, Tian T, Zhang T. Advances in biological applications of self-assembled DNA tetrahedral nanostructures. Mater Today. 2018;8(11):S1369–7021.
Zhao D, Liu M, Li Q. Tetrahedral DNA nanostructure promote endothelia cells proliferation, migration and angiogenesis via Notch signaling pathway. ACS Appl Mater Interfaces. 2018;10(44):37911–8.
Lin S, Zhang Q, Zhang T. Tetrahedral DNA nanomaterial regulates the biological behaviors of adipose-derived stem cells via DNA methylation on Dlg3. ACS Appl Mater Interfaces. 2018;10(38):32017–25.
Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171(4361):964–7.
Ma VPY, Salaita K. DNA nanotechnology as an emerging tool to study mechanotransduction in living systems. Small. 2019;15(26):e1900961. https://doi.org/10.1002/smll.201900961.
Kimna C, Lieleg O. Engineering an orchestrated release avalanche from hydrogels using DNA-nanotechnology. J Control Release. 2019;304:19–28.
Seeman NC, Fan C, Wang S, Schanze K, Fernandez L. Forum on translational DNA nanotechnology. ACS Appl Mater Interfaces. 2019;11(15):13833–4.
Hu Y, Niemeyer CM. From DNA nanotechnology to material systems engineering. Adv Mater Weinheim. 2019;31(26):e1806294. https://doi.org/10.1002/adma.201806294.
Madsen M, Gothelf KV. Chemistries for DNA nanotechnology. Chem. Rev. 2019;119(10):6384–458.
Yang J, Meng Z, Liu Q, Shimada Y, Olsthoorn RCL, Spaink HP, Herrmann A, Kros A. Performing DNA nanotechnology operations on a zebrafish. Chem Sci. 2018;9(36):7271–6.
Shen H, Wang Y, Wang J, Li Z, Yuan Q. Emerging biomimetic applications of DNA nanotechnology. ACS Appl Mater Interfaces. 2019;11(15):13859–73.
Chen T, Ren L, Liu X, Zhou M, Li L, Xu J, Zhu X. DNA nanotechnology for cancer diagnosis and therapy. Int J Mol Sci. 2018;19(6) https://doi.org/10.3390/ijms19061671.
Chidchob P, Sleiman HF. Recent advances in DNA nanotechnology. Curr Opin Chem Biol. 2018;46:63–70.
Ke Y, Castro C, Choi JH. Structural DNA nanotechnology: artificial nanostructures for biomedical research. Annu Rev Biomed Eng. 2018;20:375–401.
Ye D, Zuo X, Fan C. DNA nanotechnology-enabled interfacial engineering for biosensor development. Annu Rev Anal Chem (Palo Alto Calif). 2018;11(1):171–95.
Zhang S, Fu TJ, Seeman NC. Symmetric immobile DNA branched junctions. Biochemistry. 1993;32(32):8062–7.
He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452(7184):198–201.
Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440(7082):297–302.
Hu Q, Li H, Wang L, Gu H, Fan C. DNA nanotechnology-enabled drug delivery systems. Chem. Rev. 2019;119(10):6459–506.
Nummelin S, Kommeri J, Kostiainen MA, Linko V. Evolution of structural DNA nanotechnology. Adv. Mater. Weinheim. 2018;30(24):e1703721. https://doi.org/10.1002/adma.201703721.
Chandrasekaran AR, Rusling DA. Triplex-forming oligonucleotides: a third strand for DNA nanotechnology. Nucl Acids Res. 2018;46(3):1021–37.
Sun W. DNA nanotechnology: DNA robots that sort cargoes. Nat Nanotechnol. 2017;12(12):1120–7.
Niemeyer CM. DNA nanotechnology: On-command molecular Trojans. Nat Nanotechnol. 2017;12(12):1117–9.
Adam V, Prusty DK, Centola M, Škugor M, Hannam JS, Valero J, Klöckner B, Famulok M. Expanding the toolbox of photoswitches for DNA nanotechnology using Arylazopyrazoles. Chemistry. 2018;24(5):1062–6.
Zhou C, Duan X, Liu N. DNA-nanotechnology-enabled chiral plasmonics: from static to dynamic. Acc. Chem. Res. 2017;50(12):2906–14.
Yata T, Takahashi Y, Tan M, Nakatsuji H, Ohtsuki S, Murakami T, Imahori H, Umeki Y, Shiomi T, Takakura Y, Nishikawa M. DNA nanotechnology-based composite-type gold nanoparticle-immunostimulatory DNA hydrogel for tumor photothermal immunotherapy. Biomaterials. 2017;146:136–45.
Howorka S. DNA nanotechnology: bringing lipid bilayers into shape. Nat Chem. 2017;9(7):611–3.
Kotani S, Hughes WL. Multi-arm junctions for dynamic DNA nanotechnology. J. Am. Chem. Soc. 2017;139(18):6363–8.
Mathur D, Medintz IL. Analyzing DNA nanotechnology: a call to arms for the analytical chemistry community. Anal. Chem. 2017;89(5):2646–63.
Pastore C. DNA nanotechnology: a nucleosome clamp. Nat Nanotechnol. 2017;12(1):4–8.
Cox AJ, Bengtson HN, Rohde KH, Kolpashchikov DM. DNA nanotechnology for nucleic acid analysis: multifunctional molecular DNA machine for RNA detection. Chem. Commun. (Camb.). 2016;52(99):14318–21.
Kim KR, Kim HY, Lee YD, Ha JS, Kang JH, Jeong H, Bang D, Ko YT, Kim S, Lee H, Ahn DR. Self-assembled mirror DNA nanostructures for tumor-specific delivery of anticancer drugs. J Control Release. 2016;243:121–31.
Wu S, Wang P, Xiao C, Li Z, Yang B, Fu J, Chen J, Wan N, Ma C, Li M, Yang X, Zhan Y. A quick-responsive DNA nanotechnology device for bio-molecular homeostasis regulation. Sci Rep (2016);6:31379–82
Kumar V, Palazzolo S, Bayda S, Corona G, Toffoli G, Rizzolio F. DNA nanotechnology for cancer therapy. Theranostics. 2016;6(5):710–25.
Gu Y, Gao J, Cao M, Dong C, Lian J, Huang L, Cai J, Xu Z. Construction of a series of episomal plasmids and their application in the development of an efficient CRISPR/Cas9 system in Pichia pastoris. World J. Microbiol. Biotechnol. 2019;35(6):79–86.
Organick L, Ang SD, Chen YJ, Lopez R, Yekhanin S, Makarychev K, Racz MZ, Kamath G, Gopalan P, Nguyen B, Takahashi CN, Newman S, Parker HY, Rashtchian C, Stewart K, Gupta G, Carlson R, Mulligan J, Carmean D, Seelig G, Ceze L, Strauss K. Erratum: Random access in large-scale DNA data storage. Nat. Biotechnol. 2018;36(7):660–8.
Keyser UF. Enhancing nanopore sensing with DNA nanotechnology. Nat Nanotechnol. 2016;11(2):106–8.
Mao C, Pan W, Shao XR. the clearance effect of tetrahedral DNA nanostructures on senescent human dermal fibroblasts. ACS Appl Mater Inter. 2019;11(2):1942–50.
Shao X, Ma W, Xie X. Neuroprotective effect of tetrahedral DNA nanostructures in a cell model of Alzheimer’s disease. ACS Appl Mater Inter. 2018;10(28):23682–92.
Ma W, Xie X, Shao X. Tetrahedral DNA nanostructures facilitate neural stem cell migration via activating RHOA/ROCK2 signaling pathway. Cell Prolifer. 2018;51(6):e12503.
Ma W, Shao X, Zhao D. Self-assembled tetrahedral DNA nanostructures promote neural stem cell proliferation and neuronal differentiation. ACS Appl Mater Inter. 2018;10(9):7892–900.
Goodman RP, Berry RM, Turberfield AJ. The single-step synthesis of a DNA tetrahedron. Chem. Commun. (Camb.). 2004;(12):1372–3.
Zhang Q, Lin S, Shi S. Anti-inflammatory and anti-oxidative effects of tetrahedral DNA nanostructures via the modulation of macrophage responses. ACS Appl Mater Inter. 2018;10(4):3421–30.
Shi S, Lin S, Li Y. Effects of tetrahedral DNA nanostructures on autophagy in chondrocytes. Chem Communicat (Camb). 2018;54(11):1327–30.
Zhou M, Liu N, Shi S. Effect of tetrahedral DNA nanostructures on proliferation and Osteo/odontogenic differentiation of dental pulp stem cells via activation of the notch signaling pathway. Nanomedicine. 2018;14(4):1227–36.
Shao X, Lin S, Peng Q. Tetrahedral DNA nanostructure: a potential promoter for cartilage tissue regeneration via regulating chondrocyte phenotype and proliferation. Small. 2017;13(12):1602770.
Tian T, Zhang T, Zhou T. Synthesis of an Ethyleneimine/tetrahedral DNA nanostructure complex and its potential application as a multi-functional delivery vehicle. Nanoscale. 2017;9(46):18402–12.
Shi S, Lin S, Shao X. Modulation of chondrocyte motility by tetrahedral DNA nanostructures. Cell Prolifer. 2017;50:e12368.
Shi S, Peng Q, Shao X. Self-assembled tetrahedral DNA nanostructures promote adipose-derived stem cell migration via lncRNA XLOC 010623 and RHOA/ROCK2 signal pathway. ACS Appl Mater Inter. 2016;8(30):19353–63.
Wiraja C, Zhu Y, Lio D. Framework nucleic acids as programmable carrier for transdermal drug delivery. Nature Commun. 2019;10(1):1147–52.
Han X, Jiang Y, Li S. Multivalent aptamer-modified tetrahedral DNA nanocage demonstrates high selectivity and safety for anti-tumor therapy. Nanoscale. 2018;11(1):339–47.
Wang D, Chai Y. Lattice-Like DNA tetrahedron nanostructure as scaffold to locate GOx and HRP enzymes for highly efficient enzyme cascade reaction. ACS Appl Mater & Inter. 2020;12(2):2871–7.
He L, Lu D, Liang H. mRNA-initiated, three-dimensional DNA amplifier able to function inside living cells. J Am Chem Soc. 2018;140(1):258–63.
Ke YG, Ong LL, Shih WM. Three-dimensional structures self-assembled from DNA bricks. Science. 2012;338(6111):1177–83.
Zheng H, Lang Y, Yu J. Affinity binding of aptamers to agarose with DNA tetrahedron for removal of hepatitis B virus surface antigen. Colloids Surfaces B-Biointerfaces. 2019;178:80–6.
Reuter M, Dryden DTF. The kinetics of YOYO-1 intercalation into single molecules of double-stranded DNA. Biochem. Biophys. Res. Commun. 2010;403(2):225–9.
Wang J, Wang DX. Three-dimensional DNA nanostructures to improve the hyperbranched hybridization chain reaction. Chem Sci. 2019;10(42):9758–67.
Zhu C, Yang J, Zheng J, Chen S, Huang F, Yang R. Triplex-functionalized DNA tetrahedral nanoprobe for imaging of intracellular pH and tumor-related messenger RNA. Anal. Chem. 2019;91(24):15599–607.
Li M, Ding H, Lin M, Yin F, Song L, Mao X, Li F, Ge Z, Wang L, Zuo X, Ma Y, Fan C. DNA framework-programmed cell capture via topology-engineered receptor-ligand interactions. J. Am. Chem. Soc. 2019;141(47):18910–5.
Hu J, Liu MH, Zhang CY. Construction of tetrahedral DNA-quantum dot nanostructure with the integration of multistep förster resonance energy transfer for multiplex enzymes assay. ACS Nano. 2019;13(6):7191–201.
Xu Y, Jiang S, Simmons CR, Narayanan RP, Zhang F, Aziz AM, Yan H, Stephanopoulos N. Tunable nanoscale cages from self-assembling DNA and protein building blocks. ACS Nano. 2019;13(3):3545–54.
Tian R, Ning W, Chen M, Zhang C, Li Q, Bai J. High performance electrochemical biosensor based on 3D nitrogen-doped reduced graphene oxide electrode and tetrahedral DNA nanostructure. Talanta. 2019;194:273–81.
Ding H, Li J, Chen N, Hu X, Yang X, Guo L, Li Q, Zuo X, Wang L, Ma Y, Fan C. DNA nanostructure-programmed like-charge attraction at the cell-membrane interface. ACS Cent Sci. 2018;4(10):1344–51.
Zhu G, Xu Z, Yang Y, Dai X, Yan LT. Hierarchical crystals formed from DNA-functionalized Janus nanoparticles. ACS Nano. 2018;12(9):9467–75.
He L, Lu D, Liang H, Xie S, Zhang X, Liu Q, Yuan Q, Tan W. mRNA-Initiated, three-dimensional DNA amplifier able to function inside living cells. J. Am. Chem. Soc. 2018;140(1):258–63.
Zhang Y, Cui Z, Kong H, Xia K, Pan L, Li J, Sun Y, Shi J, Wang L, Zhu Y, Fan C. One-Shot immunomodulatory nanodiamond agents for cancer immunotherapy. Adv. Mater. Weinheim. 2016;28(14):2699–708.
Li N, Wang M, Gao X, Yu Z, Pan W, Wang H, Tang B. A DNA tetrahedron nanoprobe with controlled distance of dyes for multiple detection in living cells and in vivo. Anal. Chem. 2017;89(12):6670–7.
Qu X, Yang F, Chen H, Li J, Zhang H, Zhang G, Li L, Wang L, Song S, Tian Y, Pei H. Bubble-mediated ultrasensitive multiplex detection of metal ions in three-dimensional DNA nanostructure-encoded microchannels. ACS Appl Mater Interfaces. 2017;9(19):16026–34.
Schlichthaerle T, Strauss MT, Schueder F, Woehrstein JB, Jungmann R. DNA nanotechnology and fluorescence applications. Curr. Opin. Biotechnol. 2016;39:41–7.
Angell C, Xie S, Zhang L, Chen Y. DNA nanotechnology for precise control over drug delivery and gene therapy. Small. 2016;12(9):1117–32.
Estrich NA, Hernandez-Garcia A, de Vries R, LaBean TH. Engineered Diblock polypeptides improve DNA and gold solubility during molecular assembly. ACS Nano. 2017;11(1):831–42.
Abdel-Rahman LH, Abu-Dief AM, El-Khatib RM, Abdel-Fatah SM. Sonochemical synthesis, DNA binding, antimicrobial evaluation and in vitro anticancer activity of three new nano-sized Cu(II), Co(II) and Ni(II) chelates based on tri-dentate NOO imine ligands as precursors for metal oxides. J. Photochem. Photobiol. B, Biol. 2016;162:298–308.
Sadowski JP, Calvert CR, Zhang DY, Pierce NA, Yin P. Developmental self-assembly of a DNA tetrahedron. ACS Nano. 2014;8(4):3251–9.
Matthies M, Agarwal NP, Schmidt TL. Design and synthesis of triangulated DNA origami trusses. Nano Lett. 2016;16(3):2108–13.
Erben CM, Goodman RP, Turberfield AJ. Single-molecule protein encapsulation in a rigid DNA cage. Angew. Chem. Int. Ed. Engl. 2006;45(44):7414–7.
Chen D, Sun D, Wang Z, Qin W, Chen L, Zhou L, Zhang Y. A DNA nanostructured aptasensor for the sensitive electrochemical detection of HepG2 cells based on multibranched hybridization chain reaction amplification strategy. Biosens Bioelectron. 2018;117:416–21.
Chen Q, Liu H, Lee W, Sun Y, Zhu D, Pei H, Fan C, Fan X. Self-assembled DNA tetrahedral optofluidic lasers with precise and tunable gain control. Lab Chip. 2013;13(17):3351–4.
Lee CS, Kim TW, Oh DE, Bae SO, Ryu J, Kong H, Jeon H, Seo HK, Jeon S, Kim TH. In vivo and in vitro anticancer activity of doxorubicin-loaded DNA-AuNP nanocarrier for the ovarian cancer treatment. Cancers (Basel). 2020;12(3) https://doi.org/10.3390/cancers12030634.
Yan W, Xu L, Xu C, Ma W, Kuang H, Wang L, Kotov NA. Self-assembly of chiral nanoparticle pyramids with strong R/S optical activity. J. Am. Chem. Soc. 2012;134(36):15114–21.
Lee H, Lytton-Jean AKR, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7(6):389–93.
Liang L, Li J, Li Q. Single particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angewandte Chemie Int Ed English. 2014;53(30):7745–50.
Lin M, Wang J, Zhou G. Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angewandte Chemie Int Ed English. 2015;54(7):2151–5.
Li Y, Yan T, Chang W. Fabricating an intelligent cell-like nano-prodrug via hierarchical self-assembly based on the DNA skeleton for suppressing lung metastasis of breast cancer. Biomater Sci. 2019;7(9):3652–61.
Zhang L, Abdullah R, Hu X. Engineering of bioinspired, size-controllable, self-degradable cancer-targeting DNA nanoflowers via the incorporation of an artificial sandwich base. J Am Chem Soc. 2019;141:4282–90.
Keum JW, Ahn J, Bermudez H. Design, assembly, and activity of antisense DNA nanostructures. Small. 2011;7(24):3529–35.
Thubagere AJ, Li W, Johnson RF. A cargo-sorting DNA robot. Science. 2017;357(6356):1095–6.
Chen G, Liu D, He CB. Enzymatic synthesis of periodic DNA nanoribbons for intracellular pH sensing and gene silencing. J Am Chem Soc. 2015;137(11):3844–51.
Sun WJ, Jiang TY, Lu Y. Cocoon-like self-degradable DNA nano-clew for anticancer drug delivery. J Am Chem Soc. 2014;136(42):14722–5.
Tacar O, Sriamornsak P, Dass CR. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J Pharm Pharmacol. 2013;65(2):157–70.
Yan J, Chen J, Zhang N, Yang Y, Zhu W, Li L, He B. Mitochondria-targeted tetrahedral DNA nanostructures for doxorubicin delivery and enhancement of apoptosis. J Mater Chem B. 2020;8(3):492–503.
Wei YP, Zhang YW, Mao CJ. A silver nanoparticle-assisted signal amplification electrochemiluminescence biosensor for highly sensitive detection of mucin 1. J Mater Chem B. 2020; https://doi.org/10.1039/c9tb02773d.
Han X, Jiang Y, Li S, Zhang Y, Ma X, Wu Z, Wu Z, Qi X. Multivalent aptamer-modified tetrahedral DNA nanocage demonstrates high selectivity and safety for anti-tumor therapy. Nanoscale. 2018;11(1):339–47.
Zhang YM, Yu Q, Liu Y. Reply to comment on “Photo-controlled reversible microtubule assembly mediated by paclitaxel-modified cyclodextrin”. Angew Chem Int Ed Engl. 2020; https://doi.org/10.1002/anie.202000894.
Xie X, Shao X, Ma W. Overcoming drug-resistant lung cancer by paclitaxel loaded tetrahedral DNA nanostructures. Nanoscale. 2018;10(12):5457–65.
Liu J, Song L, Liu S. A DNA-based nanocarrier for efficient gene delivery and combined cancer therapy. Nano Lett. 2018;18(6):3328–34.
Lin S, Zhang Q. Antioxidative and angiogenesis-promoting effects of tetrahedral framework nucleic acids in diabetic wound healing with activation of the Akt/Nrf2/HO-1 pathway. ACS Appl Mater Interf. 2020; https://doi.org/10.1021/acsami.0c00874.
Thomsen RP, Malle MG. A large size-selective DNA nanopore with sensing applications. Nat Commun. 2019;10:5655–60.
Li BL, Setyawati MI, Chen L. Directing assembly and disassembly of 2D MoS2 nanosheets with DNA for drug delivery. ACS Appl Mater Interf. 2017;9(18):15286–96.
Kim J, Jeon S, Kang SJ, Kim KR, Thai H, Bao D, Lee S, Kim S, Lee YS, Dae-Ro A. Lung-targeted delivery of TGF-β antisense oligonucleotides to treat pulmonary fibrosis. J Control Release. 2020; https://doi.org/10.1016/j.jconrel.2020.03.016.
Zamecnik PC, Stephenson ML. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci USA. 1978;75(1):280–4.
McCabe KM, Hsieh J, Thomas DG, Molusky MM, Tascau L, Feranil JB, Qiang L, Ferrante AW, Tall AR. Antisense oligonucleotide treatment produces a type I interferon response that protects against diet-induced obesity. Mol Metab. 2020;34:146–56.
Zhang X, Liu N, Zhou M, Zhang T, Tian T, Li S, Tang Z, Lin Y, Cai X. DNA nanorobot delivers antisense oligonucleotides silencing c-met gene expression for cancer therapy. J Biomed Nanotechnol. 2019;15(9):1948–59.
Lee HT, Kim SK, Yoon JW. Antisense peptide nucleic acids as a potential anti-infective agent. Microbiology. 2019;57(6):423–30.
Readman JB, Dickson G, Coldham NG. Tetrahedral DNA nanoparticle vector for intracellular delivery of targeted peptide nucleic acid antisense agents to restore antibiotic sensitivity in cefotaxime-resistant Escherichia coli. Nucl Acid Ther. 2017;27(3):176–81.
Ouyang X, Li J, Liu H. Rolling circle amplification-based DNA origami nanostructures for intracellular delivery of immunostimulatory drugs. Small. 2013;9(18):3082–7.
Mercurio S, Cauteruccio S, Manenti R, Candiani S, Scarì G, Licandro E, Pennati R. Ciona intestinalis exploring miR-9 involvement in neural development using peptide nucleic acids. Int J Mol Sci. 2020;21(6) https://doi.org/10.3390/ijms21062001.
Zhang Y, Ma W, Zhu Y. Inhibiting methicillin-resistant staphylococcus aureus by tetrahedral DNA nanostructure-enabled antisense peptide nucleic acid delivery. Nano Lett. 2018;18(9):5652–9.
Zhu G, Zheng J, Song E. A Self-assembled, aptamer tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc Natl Acad Sci USA. 2013;110(20):7998–8003.
Tuerk C, Eddy S, Parma D, Gold L. Autogenous translational operator recognized by bacteriophage T4 DNA polymerase. J. Mol. Biol. 1990;213(4):749–61.
Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–10. https://doi.org/10.1126/science.2200121.
Quirico L, Orso F, Esposito CL, Bertone S, Coppo R, Conti L, Catuogno S, Cavallo F, de Franciscis V, Taverna D. Axl-148b chimeric aptamers inhibit breast cancer and melanoma progression. Int. J. Biol. Sci. 2020;16(7):1238–51.
Liu M, Ma W, Li Q. Aptamer-targeted DNA nanostructures with doxorubicin to treat protein tyrosine kinase 7-positive tumours. Cell Prolifer. 2019;52(1):e12511.
He XY, Ren XH, Peng Y, Zhang JP, Ai SL, Liu BY, Xu C, Cheng SX. Aptamer/peptide-functionalized genome-editing system for effective immune restoration through reversal of PD-L1-mediated cancer immunosuppression. Adv Mater Weinheim. 2020:e2000208.
Kardani A, Yaghoobi H, Alibakhshi A, Khatami M. Inhibition of miR-155 in MCF-7 breast cancer cell line by gold nanoparticles functionalized with antagomir and AS1411 aptamer. J Cell Physiol. 2020; https://doi.org/10.1002/jcp.29584.
Li Q, Zhao D, Shao X. Aptamer-modified tetrahedral DNA nanostructure for tumor-targeted drug delivery. ACS Appl Mater Interf. 2017;9(42):36695–701.
Charoenphol P, Bermudez H. Aptamer-targeted DNA nanostructures for therapeutic delivery. Mol. Pharm. 2014;11(5):1721–5.
Lan B, Wu J, Li N, Pan C, Yan L, Yang C, Zhang L, Yang L, Ren M. Hyperbranched cationic polysaccharide derivatives for efficient siRNA delivery and diabetic wound healing enhancement. Int J Biol Macromol. 2020; https://doi.org/10.1016/j.ijbiomac.2020.03.164.
Menanteau-Ledouble S, Schachner O, Lawrence ML, El-Matbouli M. Effects of siRNA silencing on the susceptibility of the fish cell line CHSE-214 to Yersinia ruckeri. Vet. Res. 2020;51(1):45–9.
Ren K, Zhang Y, Zhang X. In Situ SiRNA assembly in living cells for gene therapy with MicroRNA triggered cascade reactions templated by nucleic acids. ACS Nano. 2018;12(11):10797–7806.
Hoffmann M, Hersch N, Merkel R. Changing the way of entrance: highly efficient transfer of mRNA and siRNA via Fusogenic nano-carriers. J Biomed Nanotechnol. 2019;15(1):170–83.
Kim KR, Jegal H. A self-assembled DNA tetrahedron as a carrier for in vivo liver-specific delivery of siRNA. Biomater Sci. 2020;8(2):586–90.
Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436(7053):1044–7.
Xu C, Liu W, Hu Y, Li W, Di W. Bioinspired tumor-homing nanoplatform for co-delivery of paclitaxel and siRNA-E7 to HPV-related cervical malignancies for synergistic therapy. Theranostics. 2020;10(7):3325–39.
Kim KR, Jegal H, Kim J. A self-assembled DNA tetrahedron as a carrier for in vivo liver-specific delivery of siRNA. Biomater Sci. 2020;8:586–90.
Tsai MH, Chuang CC, Chen CC, Yen HJ, Cheng KM, Chen XA, Shyu HF, Lee CY, Young JJ, Kau JH. Nanoparticles assembled from fucoidan and trimethylchitosan as anthrax vaccine adjuvant: In vitro and in vivo efficacy in comparison to CpG. Carbohydr Polym. 2020;236:116041–7.
Yu W, Sun J, Liu F, Yu S, Xu Z, Wang F, Liu X. Enhanced immunostimulatory activity of cytosine-phosphate-guanosine immunomodulator by the assembly of polymer DNA wires and spheres. ACS Appl Mater Interfaces. 2020;12(15):17167–76.
Kim TH, Park J, Kim D. Anti-bacterial effect of CpG-DNA involves enhancement of the complement systems. Int J Mol Sci. 2019;20(14):3397.
Mendoza A, Pflueger J, Lister R. Capture of a functionally active methyl-CpG binding domain by an arthropod retrotransposon family. Genome Res. 2019;29(8):1277–86.
Sun R, Xiang T, Tang J, Peng W, Luo J, Li L, Qiu Z, Tan Y, Ye L, Zhang M, Ren G, Tao Q. 19q13 KRAB zinc-finger protein ZNF471 activates MAPK10/JNK3 signaling but is frequently silenced by promoter CpG methylation in esophageal cancer. Theranostics. 2020;10(5):2243–59.
Pei H, Zuo X, Zhu D, Huang Q, Fan C. Functional DNA nanostructures for theranostic applications. Acc. Chem. Res. 2014;47(2):550–9.
Schiffels D, Szalai VA, Liddle JA. Molecular precision at micrometer length scales: hierarchical assembly of DNA-protein nanostructures. ACS Nano. 2017;11(7):6623–9.
Setyawati MI, Kutty RV, Leong DT. DNA nanostructures carrying stoichiometrically definable antibodies. Small. 2016;12(40):5601–11.
Xie X, Zhang Y, Ma W, Shao X, Zhan Y, Mao C, Zhu B, Zhou Y, Zhao H, Cai X. Potent anti-angiogenesis and anti-tumour activity of pegaptanib-loaded tetrahedral DNA nanostructure. Cell Prolif. 2019;52(5):e12662.
Ma J, Xue L, Zhang M, Li C, Xiang Y, Liu P, Li G. An electrochemical sensor for Oct4 detection in human tissue based on target-induced steric hindrance effect on a tetrahedral DNA nanostructure. Biosens Bioelectron. 2019;127:194–9.
Xu Y, Jiang S, Simmons CR. Tunable nanoscale cages from self-assembling DNA and protein building blocks. ACS Nano. 2019;13:3545–54.
Ke Y, Sharma J, Liu M, Jahn K, Liu Y, Yan H. Scaffolded DNA origami of a DNA tetrahedron molecular container. Nano Lett. 2009;9(6):2445–7.
Crawford R, Erben CM, Periz J, Hall LM, Brown T, Turberfield AJ, Kapanidis AN. Non-covalent single transcription factor encapsulation inside a DNA cage. Angew. Chem. Int. Ed. Engl. 2013;52(8):2284–8.
Zhao Z, Fu J, Dhakal S, Johnson-Buck A, Liu M, Zhang T, Woodbury NW, Liu Y, Walter NG, Yan H. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat Commun. 2016;7:10619–22.
Huang Y, Huang W, Chan L. A multifunctional DNA origami as carrier of metal complexes to achieve enhanced tumoral delivery and nullified systemic toxicity. Biomaterials. 2016;103:183–96.
Dai B, Hu Y, Duan J, Yang XD. Aptamer-guided DNA tetrahedron as a novel targeted drug delivery system for MUC1-expressing breast cancer cells in vitro. Oncotarget. 2016;7(25):38257–69.
Liu X, Xu Y, Yu T, Clifford C, Liu Y, Yan H, Chang Y. A DNA nanostructure platform for directed assembly of synthetic vaccines. Nano Lett. 2012;12(8):4254–9.
Schlapak R, Danzberger J, Armitage D, Morgan D, Ebner A, Hinterdorfer P, Pollheimer P, Gruber HJ, Schäffler F, Howorka S. Nanoscale DNA tetrahedra improve biomolecular recognition on patterned surfaces. Small. 2012;8(1):89–97.
Simmons CR, Schmitt D, Wei X, Han D, Volosin AM, Ladd DM, Seo DK, Liu Y, Yan H. Size-selective incorporation of DNA nanocages into nanoporous antimony-doped tin oxide materials. ACS Nano. 2011;5(7):6060–8.
Zhao H, Wang M. Simultaneous fluorescent detection of multiplexed miRNA of liver cancer based on DNA tetrahedron nanotags. Talanta. 2020;210:120677.
Wei B, Dai MJ, Yin P. Complex shapes self-assembled from single-stranded DNA tiles. Nature. 2012;485(7400):623–6.
Keum JW, Bermudez H. Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem Commun (Camb). 2009;(45):7036–8.
Tian T, Zhang T, Zhou T, Lin S, Shi S, Lin Y. Synthesis of an ethyleneimine/tetrahedral DNA nanostructure complex and its potential application as a multi-functional delivery vehicle. Nanoscale. 2017;9(46):18402–12.
Lacroix A, Edwardson TGW, Hancock MA, Dore MD, Sleiman HF. Development of DNA nanostructures for high-affinity binding to human serum albumin. J Am Chem Soc. 2017;139(21):7355–62.
Castro CE, Kilchherr F, Kim DN, Shiao EL, Wauer T, Wortmann P, Bathe M, Dietz H. A primer to scaffolded DNA origami. Nat. Methods. 2011;8(3):221–9.
Walsh AS, Yin HF, Erben CM, Wood MJA, Turberfield AJ. DNA cage delivery to mammalian cells. ACS Nano. 2011;5(7):5427–32.
Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312(5776):1027–30.
Ko SH, Chen Y, Shu D, Guo P, Mao C. Reversible switching of pRNA activity on the DNA packaging motor of bacteriophage phi29. J. Am. Chem. Soc. 2008;130(52):17684–7.
Perrault SD, Shih WM. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano. 2014;8(5):5132–40.
Goodman RP, Schap IAT, Tardin CF. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310(5754):1661–5.
Bergamini C, Angelini P, Rhoden KJ. A practical approach for the detection of DNA nanostructures in single live human cells by fluorescence microscopy. Methods. 2014;67(2):185–92.
Samanta A, Banerjee S, Liu Y. DNA nanotechnology for nanophotonic applications. Nanoscale. 2015;7(6):2210–20.
Ma W, Zhan Y, Zhang Y. An intelligent DNA nanorobot with enhanced protein lysosomal degradation of HER2. Nano Lett. 2019;19(7):4505–17.
Chandrasekaran AR, Punnoose JA, Zhou L, Dey P, Dey BK, Halvorsen K. DNA nanotechnology approaches for microRNA detection and diagnosis. Nucl Acids Res. 2019;47(20):10489–505.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Zhang, X., Lin, Y. (2021). The Application and Problems of Tetrahedral Framework Nucleic Acids as a Drug Carrier in Biomedicine Fields. In: Lin, Y., Zhou, R. (eds) Advances in Nanomaterials-based Cell Biology Research. Springer, Singapore. https://doi.org/10.1007/978-981-16-2666-1_5
Download citation
DOI: https://doi.org/10.1007/978-981-16-2666-1_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-2665-4
Online ISBN: 978-981-16-2666-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)