Abstract
Neovascular age-related macular degeneration (nAMD) and polypoidal choroidal vasculopathy (PCV) have some shared risk factors and clinical manifestation, but there are also some different features. Genetic variants are an important risk factor for both conditions. In this chapter, we reported an updated meta-analysis comparing the genetic variants between PCV and nAMD. Totally 57 SNPs in 20 genes were investigated. Among them, 11 SNPs in ARMS2-HTRA1 and rs77466370 in FGD6 showed significant differences between PCV and nAMD, but the other SNPs had similar distribution between PCV and nAMD, including variants in CFH, VEGF, C2, CFB. These results suggest that PCV and nAMD shares the majority of genetic components, but the variants that distribute differently between these two conditions may explain the pathogenic and clinical difference of PCV and nAMD.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Age-related macular degeneration
- Polypoidal choroidal vasculopathy
- Meta-analysis
- Single nucleotide polymorphism
- Genetic association
8.1 Introduction
Age-related macular degeneration (AMD) is a leading cause of blindness and central vision impairment and blindness in elderly patients [1]. There are two types of AMD, dry and wet (exudative or neovascular) AMD (nAMD), which is characterized by atrophy of retinal pigment epithelium (RPE) and choroidal neovascularization respectively. The clinical manifestations of nAMD include choroidal neovascularization, subretinal fluid, hemorrhage, exudation, and fibrosis. Polypoidal choroidal vasculopathy (PCV) is characterized by the branching vascular network of the choroid and polyp-like aneurysmal dilations of its terminals [2]. Clinically, PCV is manifested as serosanguineous detachments of the pigmented epithelium and exudative changes that can recur in several episodes.
It is still controversy whether PCV presents a subtype of nAMD or a distinct disease. PCV and nAMD have some shared characters but there are also some different features, including risk factors, clinical manifestations, natural course, and response to treatment.
Both PCV and nAMD are commonly seen in elderly patients. However, PCV presents a younger age than nAMD [3, 4]. Although both PCV and nAMD occur in any race, PCV is known to be more prevalent in pigmented ethnicity while nAMD has a high prevalence in European than in Asian [5]. Smoking is a proven risk factors for both PCV and nAMD, while female gender is a protective factor for both conditions [3, 6]. Diabetes was found to be more prevalent in nAMD than in PCV patients [4].
Clinically, Both PCV and nAMD present as exudation or hemorrhage at the macular region. But there are also different characters between them. nAMD is predominantly located at the fovea or parafoveal region, while PCV may involve perifoveal, peripapillary, or even peripheral retina. The histological feature of PCV is majorly polypoidal enlargement of the terminal of the choroidal vessel. nAMD is characterized by choroidal neovascularization above or underneath the RPE. The choroidal thickness of nAMD is usually thin but that of PCV is usually thick. The natural history of PCV is multiple, recurrent episodes while nAMD is a progressive disease. Although both disorders can be treated using photodynamic therapy or anti-vascular endothelial growth factor (VEGF) antibody, nAMD responses better to anti-VEGF therapy and PCV responses better to photodynamic therapy [7].
Genetic studies of AMD have identified susceptibility single-nucleotide polymorphisms (SNPs) in multiple genes, including rs1061170 in complement factor H (CFH), rs10490924 in age-related maculopathy susceptibility 2 (ARMS2), and rs11200638 in high-temperature requirement factor H (HTRA1) [8, 9]. In 2016, the International AMD Genomics Consortium reported 34 loci associated with AMD [10]. Due to the similarities between nAMD and PCV, major gene SNPs for nAMD have also been evaluated in PCV. The CFH SNP rs1061170 was not found to be associated with PCV [11], while an adjacent SNP rs800292 was significantly associated [11,12,13]. Both rs10490924 and rs11200638 at the ARMS2-HTRA1 locus were associated with PCV [11, 12, 14, 15]. In 2012, we published a meta-analysis investigating genetic associations of PCV with SNPs in the ARMS2, HTRA1, CFH, and complement component 2 (C2) genes. The results also showed that one SNP, rs10490924, in ARMS2 showed a significant difference between PCV and AMD [16]. In 2015, we reported the updated meta-analysis of the association of genetic variants with PCV, which found 31 polymorphisms in 10 genes/loci (including ARMS2, HTRA1, CFH, C2, CFB, RDBP, SKIV2L, CETP, 8p21, and 4q12) were significantly associated with PCV. Twelve polymorphisms at the ARMS2-HTRA1 locus showed significant differences between PCV and nAMD. There are many new articles investigating these topics since the publication of the latest meta-analysis. In this chapter, we further updated our meta-analysis comparing the genetic association profiles between PCV and nAMD.
8.2 Methods of Meta-Analysis
A systematic literature search was performed using EMBASE, PubMed, Web of Science, and Chinese Biomedical Literature Database. The search used the terms (polypoidal choroidal vasculopathy or PCV) and (gene or genetic or polymorphism or variant or SNP or DNA). We retrieved all related records published from February 1, 2015, and September 27, 2018, and then added the articles published before Feb 2015 that were included in our previous meta-analysis. The reference lists of all eligible studies, reviews, and meta-analyses were also screened to prevent that any relevant studies were omitted.
The retried records were reviewed by two independent reviewers (L.M. and X. L.) and any inconsistency was resolved by discussion with another reviewer (H.C.). The following criteria were used when assessing the records [1]. case-control studies, cohort studies, or population-based studies that evaluated the difference of gene variants between PCV and nAMD; and [2] allele or genotype counts and/or frequencies being presented or able to be calculated from the data in the study. For those reports published by the same study group on the same gene markers, only the latest study was included. Case reports, animal studies, reviews, conference abstracts, comments, articles without sufficient data, or published in language other than English were excluded.
The data from included studies were extracted by the two independent reviewers (L.M. and Z.L.) and any inconsistency was resolved by discussion with another reviewer (H.C.). If there were several cohorts in the same article, they were treated as independent study. The following information from each record was extracted: first author, year of publication, the ethnicity of study subjects, study design, genotyping method, and sample size, demographics, allele, and genotype distribution in PCV and nAMD.
The distribution of genetic variants between PCV and nAMD from all included studies were pooled. Three genetic models were used, including allelic, dominant, and recessive models. The effect size was assessed using a summary odds ratio (OR) and its 95% confidence intervals (CIs) of each SNP. The software, Review Manager software (RevMan, version 5.3.5, The Cochrane Collaboration, Copenhagen, Denmark) was used for statistical analysis. The I2 statistic was adopted to assess the heterogeneity among the studies. The I2 values correspond with no (<25%), low (25%–50%), moderate (50%–75%), and high heterogeneity (≥75%). If the I2 value was ≥50%, the fixed effects model was used in the meta-analysis, otherwise, the random effects model was used. A summary P value <0.05 was considered statistically significant. We performed a sensitivity analysis by omitting one study at a time and calculating the pooled ORs for the remaining studies. Funnel plots were constructed to assess potential publication bias.
8.3 Results of Updated Meta-Analysis
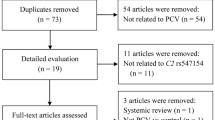
Our literature search yielded a total of 1315 reports published between February 1, 2015, and September 27, 2018, from EMBASE, PubMed, Web of Science and Chinese Biomedical Literature Database. Out of these, 502 articles were excluded due to duplicates. After assessing the titles and abstracts, a further 606 reports with unrelated topics were omitted. For the remaining 107 studies, the full-texts were retrieved and reviewed. Another 89 reports were excluded, among which 62 studies were on AMD but not PCV, 2 were reviews, 23 were non-genetic studies, and 1 was a case report. Finally, 18 articles were eligible for the meta-analysis. A further 66 studies published before 2015 that were used in our previous meta-analysis were added. However, 19 of these studies were excluded because they only studied in PCV patients. Thus, a total of 65 studies were included in the meta-analysis. Figure 8.1 shows the flowchart of literature inclusion and exclusion with the specification of reasons and Table 8.1 shows the characters of the included studies.
In these 65 studies , both PCV and nAMD were assessed for associations with a total of 57 SNPs in 20 genes or loci (i.e., ARMS2, HTRA1, CFH, VEGF-A, C2, CFB, SKIV2L, CETP, 8p21, 4q12, ELN, LIPC, LPL, FGD6, ABCA1, ABCG1, PGF, TLR3, LOXL1, and PEDF; Table 8.1). In total, 11 SNPs at the ARMS2-HTRA1 locus and 1 in FGD6 showed significant differences between PCV and nAMD (Tables 8.2 and 8.3). There was no significant difference between PCV and nAMD in the remaining 45 SNPs (Table 8.4).
There are 12 studies tested the most-investigated SNP, ARMS2 rs10490924, involving 2361 PCV and 2138 nAMD patients (Table 8.2) [3, 12, 15, 22, 26, 27, 35, 36, 38, 43, 44, 78]. The frequency of the T allele was significantly lower in PCV than in nAMD (summary OR 0.69; 95% CI 0.63–0.75; P = 5.50 × 10−16; Table 8.2 and Fig. 8.2). The association was also statistically significant in both dominant and recessive models (OR = 0.64, P = 8.80 × 10−8 and OR = 0.62, P = 1.47 × 10−13 respectively; Table 8.3 and Fig. 8.2). The results of the sensitivity analysis found that the association remains significant after omitting any single included cohorts (data not shown). And there was no asymmetry on the funnel plots (Fig. 8.5). There are 8 other SNPs in ARMS2, namely rs3750848, rs36212731, rs36212732, rs36212733, rs3750846, rs10664316, c.372_815del443ins54 and rs2672587, were evaluated in 2 to 3 cohorts, and also showed significant differences between PCV and nAMD (ORs values between 0.48 and 0.71, P values between 7.19 × 10−9 and 0.05; Table 8.2).
There are seven studies tested the HTRA1 SNP rs11200638 in 1362 PCV and 1364 nAMD patients [3, 14, 18, 43, 44, 57, 73]. The A allele frequency was lower in PCV compared to nAMD, with a summary OR of 0.75 (95% CI, 0.67–0.84; P = 2.14 × 10−5; Table 8.2 and Fig. 8.3). The association was also statistically significant in both dominant and recessive models (OR = 0.67, P = 0.006 and OR = 0.70, P = 9.87 × 10−6 respectively, Table 8.3 and Fig. 8.3). The results of the sensitivity analysis found that the association remain significant after omitting any single included cohorts (data not shown). And there was no asymmetry on the funnel plots (Fig. 8.5). Another HTRA1 SNP, rs2672587, was also evaluated in two cohorts, and showed significant differences between PCV and nAMD (G allele; OR, 1.41; 95% CI, 1.07–1.85; P = 0.01; Table 8.2).
The SNP rs77466370 in FGD6 was studied in 3318 PCV and 2457 nAMD patients from five cohorts. The summary OR for the T allele was 1.86 (95% CI, 1.48–2.35; P = 1.29 × 10−7; Table 8.2 and Fig. 8.4). The association was statistically significant in the dominant model but no in the recessive model (OR = 1.89, P = 1.52 × 10−7 and OR = 2.19, P = 0.27 respectively; Table 8.3 and Fig. 8.4). The results of the sensitivity analysis found that the association remain significant after omitting any single included cohorts (data not shown). And there was no asymmetry on the funnel plots (Fig. 8.5) .
8.4 Discussion
Genetic variants are important risk factors for both nAMD and PCV. This updated systematic review and meta-analysis compared the distribution of genetic variants between nAMD and PCV. The results showed that 57 SNPs in 20 genes had been investigated in both PCV and nAMD in the same cohorts. The pooled outcomes showed 11 SNPs at the ARMS2-HTRA1 locus and 1 SNP in FGD6 had significant differences between PCV and nAMD. The results are robust because the sensitivity test found consistency when omitting any included studies. There was no publication bias found on the funnel plots. There was no significant difference between PCV and nAMD in the remaining 45 SNPs in CFH, VEGF, C2, CFB, SKIV2L, CETP, 8p21, 4q12, ELN, LIPC, LPL, FGD6, ABCA1, ABCG1, PGF, TLR3, LOXL1, and PEDF.
The similarity and difference of PCV and nAMD attracted the interest in investigating the genetic susceptibility between them. There was a large sample size study investigated the genetic variants of 34 AMD loci for PCV and nAMD in East Asians [75]. The results showed that PCV and tAMD were highly correlated (rg = 0.69, P = 4.68 × 10−3) in genetic variants. Weaker association for PCV compared to nAMD was found at ARMS2-HTRA1 and KMT2E-SRPK2. The different association of ARMS2-HTRA1 variants between PCV and nAMD was confirmed in this meta-analysis. But KMT2E-SRPK2 was investigated in only one study and therefore no meta-analysis was performed. In 2016, an article using exome sequencing identified a rare variant, rs77466370, in FGD6 was significantly associated with PCV (OR = 2.12) but not with CNV (OR = 1.13) [70]. Our meta-analysis confirmed that most genetic polymorphisms were distributed similarly between nAMD and PCV, But also some polymorphisms had a statistically significant difference between PCV and nAMD. These results suggest that PCV and nAMD have shared the majority of genetic background, while the differences of ARMS2-HTRA1 locus and FGD6 variants may be correlated with the differences in the pathologic and clinical manifestations of PCV and nAMD. The molecular mechanisms underlying their differences in pathogenesis remain to be further investigated.
ARMS2-HTRA1 locus located at chromosome 10q26. It was one of the most strong associated locus with AMD [79, 80]. There are many SNPs in this locus and they are in strong linkage disequilibrium. ARMS2 was expressed in the mitochondria of the outer segment of photoreceptors [81]. The function of ARMS2 was suggested to be associated with loss of function of RPE [81]. HTRA1 can inhibit transforming growth factor-βin chronic inflammation [82]. In the HTRA1 transgenic mice model, retinal pigment epithelium atrophy, photoreceptor degeneration, and grape-cluster structure in choroidal vasculature were reported, which is similar to the PCV phenotype [83]. However, our meta-analysis found that the effect size of ARMS2-HTRA1 locus was weaker in PCV compared to nAMD. Further functional studies are needed to elucidate the role of ARMS2-HTRA1 locus in PCV/nAMD.
FGD6 located at chromosome 12q22. FGD6 expresses in all human tissue but has a higher level of expression in retina and choroid, especially in retinal microvascular endothelial cells. Rs77466370, c.986A > G (p.Lys329Arg), is a rare variant with the minor allele frequency of 0.02–0.03 in normal subjects. FGD6-Arg329 has a different pattern of intracellular localization from FGD6-Lys329. In vitro, FGD6 could promote endothelial cells tube formation, furthermore, FGD6-Arg329 promoted more abnormal vessel development in the mouse retina than FGD6-Lys329 [70]. These functional studies support the role of FGD6 in the pathogenesis of PCV.
8.5 Summary
In summary, we pooled the results 57 SNPs in 20 genes that had been investigated in both PCV and nAMD in the same studies. Among them, 11 SNPs at the ARMS2-HTRA1 locus and rs77466370 in FGD6 showed significant differences between PCV and nAMD, but the other SNPs had similar distribution between PCV and nAMD. Our results suggest that PCV and nAMD have shared the majority of genetic components, but the variants distributed differently between these two conditions may explain the pathogenic and clinical differences of PCV and nAMD.
References
Mitchell P, Liew G, Gopinath B, Wong TY. Age-related macular degeneration. Lancet. 2018;392:1147–59. https://doi.org/10.1016/S0140-6736(18)31550-2.
Wong CW, et al. Age-related macular degeneration and polypoidal choroidal vasculopathy in Asians. Prog Retin Eye Res. 2016;53:107–39. https://doi.org/10.1016/j.preteyeres.2016.04.002.
Woo SJ, et al. Analysis of genetic and environmental risk factors and their interactions in Korean patients with age-related macular degeneration. PLoS One. 2015;10:e0132771. https://doi.org/10.1371/journal.pone.0132771.
Sakurada Y, Yoneyama S, Imasawa M, Iijima H. Systemic risk factors associated with polypoidal choroidal vasculopathy and neovascular age-related macular degeneration. Retina. 2013;33:841–5. https://doi.org/10.1097/IAE.0b013e31826ffe9d.
Yannuzzi LA, et al. Polypoidal choroidal vasculopathy and neovascularized age-related macular degeneration. Arch Ophthalmol. 1999;117:1503–10.
Kikuchi M, et al. Elevated C-reactive protein levels in patients with polypoidal choroidal vasculopathy and patients with neovascular age-related macular degeneration. Ophthalmology. 2007;114:1722–7. https://doi.org/10.1016/j.ophtha.2006.12.021.
Laude A, et al. Polypoidal choroidal vasculopathy and neovascular age-related macular degeneration: same or different disease? Prog Retin Eye Res. 2010;29:19–29. https://doi.org/10.1016/j.preteyeres.2009.10.001.
Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–4. https://doi.org/10.1126/science.1110189.
Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–9. https://doi.org/10.1126/science.1109557.
Arakawa S, et al. Genome-wide association study identifies two susceptibility loci for exudative age-related macular degeneration in the Japanese population. Nat Genet. 2011;43:1001–4. https://doi.org/10.1038/ng.938.
Lee KY, et al. Association analysis of CFH, C2, BF, and HTRA1 gene polymorphisms in Chinese patients with polypoidal choroidal vasculopathy. Invest Ophthalmol Vis Sci. 2008;49:2613–9. https://doi.org/10.1167/iovs.07-0860.
Hayashi H, et al. CFH and ARMS2 variations in age-related macular degeneration, polypoidal choroidal vasculopathy, and retinal angiomatous proliferation. Invest Ophthalmol Vis Sci. 2010;51:5914–9. https://doi.org/10.1167/iovs.10-5554.
Tanaka K, et al. Associations of complement factor H (CFH) and age-related maculopathy susceptibility 2 (ARMS2) genotypes with subtypes of polypoidal choroidal vasculopathy. Invest Ophthalmol Vis Sci. 2011;52:7441–4. https://doi.org/10.1167/iovs.11-7546.
Gotoh N, et al. Haplotype analysis of the ARMS2/HTRA1 region in Japanese patients with typical neovascular age-related macular degeneration or polypoidal choroidal vasculopathy. Jpn J Ophthalmol. 2010;54:609–14.
Lima LH, et al. Three major loci involved in age-related macular degeneration are also associated with polypoidal choroidal vasculopathy. Ophthalmology. 2010;117:1567–70. https://doi.org/10.1016/j.ophtha.2009.12.018.
Chen H, et al. Genetic associations in polypoidal choroidal vasculopathy: a systematic review and meta-analysis. Mol Vis. 2012;18:816–29.
Gotoh N, et al. Apolipoprotein E polymorphisms in Japanese patients with polypoidal choroidal vasculopathy and exudative age-related macular degeneration. Am J Ophthalmol. 2004;138:567–73.
Kondo N, Honda S, Ishibashi K, Tsukahara Y, Negi A. LOC387715/HTRA1 variants in polypoidal choroidal vasculopathy and age-related macular degeneration in a Japanese population. Am J Ophthalmol. 2007;144:608–12. https://doi.org/10.1016/j.ajo.2007.06.003.
Gotoh N, et al. Correlation between CFH Y402H and HTRA1 rs11200638 genotype to typical exudative age-related macular degeneration and polypoidal choroidal vasculopathy phenotype in the Japanese population. Clin Exp Ophthalmol. 2008;36:437–42.
Kondo N, Honda S, Ishibashi K, Tsukahara Y, Negi A. Elastin gene polymorphisms in neovascular age-related macular degeneration and polypoidal choroidal vasculopathy. Investig Ophthalmol Vis Sci. 2008;49:1101–5.
Bessho H, Kondo N, Honda S, Kuno SI, Negi A. Coding variant Met72Thr in the PEDF gene and risk of neovascular age-related macular degeneration and polypoidal choroidal vasculopathy. Mol Vis. 2009;15:1107–14.
Goto A, et al. Genetic analysis of typical wet-type age-related macular degeneration and polypoidal choroidal vasculopathy in Japanese population. J Ocul Biol Dis Infor. 2009;2:164–75. https://doi.org/10.1007/s12177-009-9047-1.
Gotoh N, et al. ARMS2 (LOC387715) variants in Japanese patients with exudative age-related macular degeneration and polypoidal choroidal vasculopathy. Am J Ophthalmol. 2009;147:1037–1041.e1032.
Kondo N, Bessho H, Honda S, Negi A. SOD2 gene polymorphisms in neovascular age-related macular degeneration and polypoidal choroidal vasculopathy. Mol Vis. 2009;15:1819–26.
Hayashi H, et al. CFH and ARMS2 variations in age-related macular degeneration, polypoidal choroidal vasculopathy, and retinal angiomatous proliferation. Investig Ophthalmol Vis Sci. 2010;51:5914–9.
Bessho H, Honda S, Kondo N, Negi A. The association of age-related maculopathy susceptibility 2 polymorphisms with phenotype in typical neovascular age-related macular degeneration and polypoidal choroidal vasculopathy. Mol Vis. 2011;17:977–82.
Fuse N, et al. Polymorphisms in ARMS2 (LOC387715) and LOXL1 genes in the Japanese with age-related macular degeneration. Am J Ophthalmol. 2011;151:550–6. https://doi.org/10.1016/j.ajo.2010.08.048.
Lima LH, et al. Elastin rs2301995 polymorphism is not associated with polypoidal choroidal vasculopathy in caucasians. Ophthalmic Genet. 2011;32:80–2. https://doi.org/10.3109/13816810.2010.544362.
Nakata I, et al. Association between the SERPING1 gene and age-related macular degeneration and polypoidal choroidal vasculopathy in Japanese. PLoS One. 2011; https://doi.org/10.1371/journal.pone.0019108.
Sng CCA, et al. Toll-like receptor 3 polymorphism rs3775291 is not associated with choroidal neovascularization or polypoidal choroidal vasculopathy in Chinese subjects. Ophthalmic Res. 2011;45:191–6. https://doi.org/10.1159/000321387.
Yamashiro K, et al. Association of elastin gene polymorphism to age-related macular degeneration and polypoidal choroidal vasculopathy. Invest Ophthalmol Vis Sci. 2011;52:8780–4. https://doi.org/10.1167/iovs.11-8205.
Zhang X, et al. Association of genetic variation on chromosome 9p21 with polypoidal choroidal vasculopathy and neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52:8063–7. https://doi.org/10.1167/iovs.11-7820.
Wu K, et al. Lack of association with PEDF Met72Thr variant in neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in a Han Chinese population. Curr Eye Res. 2012;37:68–72. https://doi.org/10.3109/02713683.2011.618289.
Sakurada Y, Mabuchi F, Yoneyama S, Kubota T, Iijima H. Polymorphisms in ARMS2 (LOC387715) and LOXL1 GENES in the Japanese with age-related macular degeneration. Am J Ophthalmol. 2011;152:499.
Tanaka K, et al. Analysis of candidate genes for age-related macular degeneration subtypes in the Japanese population. Mol Vis. 2011;17:2751–8.
Yanagisawa S, et al. Difference between age-related macular degeneration and polypoidal choroidal vasculopathy in the hereditary contribution of the A69S variant of the age-related maculopathy susceptibility 2 gene (ARMS2). Mol Vis. 2011;17:3574–82.
Bessho H, et al. The association of CD36 variants with polypoidal choroidal vasculopathy compared to typical neovascular age-related macular degeneration. Mol Vis. 2012;18:121–7.
Liang XY, et al. Differentiation of exudative age-related macular degeneration and polypoidal choroidal vasculopathy in the ARMS2/HTRA1 locus. Invest Ophthalmol Vis Sci. 2012;53:3175–82. https://doi.org/10.1167/iovs.11-8135.
Nakata I, et al. Association of genetic variants on 8p21 and 4q12 with age-related macular degeneration in Asian populations. Invest Ophthalmol Vis Sci. 2012;53:6576–81. https://doi.org/10.1167/iovs.12-10219.
Nakata I, et al. Significance of C2/CFB variants in age-related macular degeneration and polypoidal choroidal vasculopathy in a Japanese population. Invest Ophthalmol Vis Sci. 2012;53:794–8. https://doi.org/10.1167/iovs.11-8468.
Nishiguchi KM, et al. C9-R95X polymorphism in patients with neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2012;53:508–12. https://doi.org/10.1167/iovs.11-8425.
Zeng R, et al. An rs9621532 variant near the TIMP3 gene is not associated with neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in a Chinese Han population. Ophthalmic Genet. 2012;33:139–43. https://doi.org/10.3109/13816810.2011.643440.
Zuo C, et al. COL1A2 polymorphic markers confer an increased risk of neovascular age-related macular degeneration in a Han Chinese population. Mol Vis. 2012;18:1787–93.
Cheng Y, et al. Genetic and functional dissection of ARMS2 in age-related macular degeneration and polypoidal choroidal vasculopathy. PLoS One. 2013;8:e53665. https://doi.org/10.1371/journal.pone.0053665.
Guo J, et al. TOMM40 rs2075650 polymorphism shows no association with neovascular age-related macular degeneration or polypoidal choroidal vasculopathy in a Chinese population. Mol Vis. 2013;19:2050–7.
Liu K, et al. Associations of the C2-CFB-RDBP-SKIV2L locus with age-related macular degeneration and polypoidal choroidal vasculopathy. Ophthalmology. 2013;120:837–43. https://doi.org/10.1016/j.ophtha.2012.10.003.
Su Y, et al. Three variants of or near VEGF-A gene are not associated with neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in a Han Chinese population. Ophthalmic Genet. 2013; https://doi.org/10.3109/13816810.2013.858753.
Sun Y, et al. TNFRSF10A-LOC389641 rs13278062 but not REST-C4orf14-POLR2B-IGFBP7 rs1713985 was found associated with age-related macular degeneration in a Chinese population. Invest Ophthalmol Vis Sci. 2013;54:8199–203. https://doi.org/10.1167/iovs.13-12867.
Zhang X, et al. Different impact of high-density lipoprotein-related genetic variants on polypoidal choroidal vasculopathy and neovascular age-related macular degeneration in a Chinese Han population. Exp Eye Res. 2013;108:16–22. https://doi.org/10.1016/j.exer.2012.12.005.
Cheng Y, et al. Toll-like receptor 3 polymorphism is not associated with neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in the Chinese. Genet Mol Res GMR. 2014;13:302–9. https://doi.org/10.4238/2014.January.17.15.
Huang L, et al. Different hereditary contribution of the CFH gene between polypoidal choroidal vasculopathy and age-related macular degeneration in Chinese Han people. Invest Ophthalmol Vis Sci. 2014;55:2534–8. https://doi.org/10.1167/iovs.13-13437.
Huang L, et al. rs4711751 and rs1999930 are not associated with neovascular age-related macular degeneration or polypoidal choroidal vasculopathy in the Chinese population. Ophthalmic Res. 2014;52:102–6. https://doi.org/10.1159/000362763.
Hata M, et al. Two-year visual outcome of ranibizumab in typical neovascular age-related macular degeneration and polypoidal choroidal vasculopathy. Graefes Arch Clin Exp Ophthalmol. 2014; https://doi.org/10.1007/s00417-014-2688-1.
Ji Y, et al. Association of rs6982567 near GDF6 with neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in a Han Chinese Cohort. BMC Ophthalmol. 2014;14:140.
Li F, et al. ABCA1 rs1883025 polymorphism shows no association with neovascular age-related macular degeneration or polypoidal choroidal vasculopathy in a northern Chinese population. Ophthalmic Res. 2014;51:210–5. https://doi.org/10.1159/000357978.
Liang XY, et al. FPR1 interacts with CFH, HTRA1 and smoking in exudative age-related macular degeneration and polypoidal choroidal vasculopathy. Eye (Lond). 2014;28:1502–10. https://doi.org/10.1038/eye.2014.226.
Liu K, et al. Genes in the high-density lipoprotein metabolic pathway in age-related macular degeneration and polypoidal choroidal vasculopathy. Ophthalmology. 2014;121:911–6. https://doi.org/10.1016/j.ophtha.2013.10.042.
Liu K, et al. Gender specific association of a complement component 3 polymorphism with polypoidal choroidal vasculopathy. Sci Rep. 2014;4:7018.
Park DH, Shin JP, Kim IT. Association of plasma malondialdehyde with ARMS2 genetic variants and phenotypes in polypoidal choroidal vasculopathy and age-related macular degeneration. Retina. 2014;34:1167–76.
Tanaka K, et al. Associations of complement factor B and complement component 2 genotypes with subtypes of polypoidal choroidal vasculopathy. BMC Ophthalmol. 2014;14:83. https://doi.org/10.1186/1471-2415-14-83.
Yang F, et al. Complement factor I polymorphism is not associated with neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in a Chinese population. Ophthalmologica. 2014; https://doi.org/10.1159/000358241.
Yoneyama S, et al. Genetic and clinical factors associated with reticular pseudodrusen in exudative age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2014; https://doi.org/10.1007/s00417-014-2601-y.
Yoneyama S, et al. Genetic variants in the SKIV2L gene in exudative age-related macular degeneration in the Japanese population. Ophthalmic Genet. 2014;2014:1–5. https://doi.org/10.3109/13816810.2014.921313.
Zeng R, Zhang X, Wu K, Su Y, Wen F. MMP9 gene polymorphism is not associated with polypoidal choroidal vasculopathy and neovascular age-related macular degeneration in a Chinese Han population. Ophthalmic Genet. 2014;35:235–40. https://doi.org/10.3109/13816810.2014.952832.
Huang L, et al. Gene-gene interaction of CFH, ARMS2, and ARMS2/HTRA1 on the risk of neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in Chinese population. Eye. 2015;29:691–8. https://doi.org/10.1038/eye.2015.32.
Jia Chen L, et al. Association of the vascular endothelial growth factor genes with age-related macular degeneration and polypoidal choroidal vasculopathy. Investig Ophthalmol Vis Sci. 2015;56:786.
Jin E, et al. Evidence of a novel gene HERPUD1 in polypoidal choroidal vasculopathy. Int J Clin Exp Pathol. 2015;8:13928–44.
Meng Q, et al. Effect of high-density lipoprotein metabolic pathway gene variations and risk factors on neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in China. PLoS One. 2015; https://doi.org/10.1371/journal.pone.0143924.
Yu Y, et al. COL8A1 rs13095226 polymorphism shows no association with neovascular age-related macular degeneration or polypoidal choroidal vasculopathy in Chinese subjects. Int J Clin Exp Pathol. 2015;8:11635–40.
Huang L, et al. A missense variant in FGD6 confers increased risk of polypoidal choroidal vasculopathy. Nat Genet. 2016;48:640–7. https://doi.org/10.1038/ng.3546.
Ma L, et al. Association of ABCG1 with neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in Chinese and Japanese. Invest Ophthalmol Vis Sci. 2016;57:5758–63. https://doi.org/10.1167/iovs.16-20175.
Ng TK, et al. HTRA1 promoter variant differentiates polypoidal choroidal vasculopathy from exudative age-related macular degeneration. Sci Rep. 2016; https://doi.org/10.1038/srep28639.
Ye Z, et al. Associations of 6p21.3 region with age-related macular degeneration and polypoidal choroidal vasculopathy. Sci Rep. 2016;6:20914. https://doi.org/10.1038/srep20914.
Zuo C, et al. ENOS polymorphisms in neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in a Chinese Han population. Ophthalmic Genet. 2016;37:394–9. https://doi.org/10.3109/13816810.2015.1107598.
Fan Q, et al. Shared genetic variants for polypoidal choroidal vasculopathy and typical neovascular age-related macular degeneration in east Asians. J Hum Genet. 2017;62:1049–55. https://doi.org/10.1038/jhg.2017.83.
Ma L, et al. Identification of ANGPT2 as a new gene for neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in the Chinese and Japanese populations. Invest Ophthalmol Vis Sci. 2017; https://doi.org/10.1167/iovs.16-20575.
Wen X, et al. Association of IGFN1 variant with polypoidal choroidal vasculopathy. J Gene Med. 2018; https://doi.org/10.1002/jgm.3007.
Yoneyama S, et al. Genetic variants in the SKIV2L gene in exudative age-related macular degeneration in the Japanese population. Ophthalmic Genet. 2014;35:151–5. https://doi.org/10.3109/13816810.2014.921313.
Dewan A, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–92. https://doi.org/10.1126/science.1133807.
Yang Z, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–3. https://doi.org/10.1126/science.1133811.
Kanda A, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci USA. 2007;104:16227–32. https://doi.org/10.1073/pnas.0703933104.
Oka C, et al. HtrA1 serine protease inhibits signaling mediated by Tgfβ family proteins. Development. 2004;131:1041–53. https://doi.org/10.1242/dev.00999.
Jones A, et al. Increased expression of multifunctional serine protease, HTRA1, in retinal pigment epithelium induces polypoidal choroidal vasculopathy in mice. Proc Natl Acad Sci USA. 2011;108:14578–83. https://doi.org/10.1073/pnas.1102853108.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Chen, H., Ma, L., Liao, X., Chen, L.J., Pang, C.P. (2021). Differential Genotypes in Age-Related Macular Degeneration and Polypoidal Choroidal Vasculopathy: A Updated Meta-Analysis. In: Prakash, G., Iwata, T. (eds) Advances in Vision Research, Volume III. Essentials in Ophthalmology. Springer, Singapore. https://doi.org/10.1007/978-981-15-9184-6_8
Download citation
DOI: https://doi.org/10.1007/978-981-15-9184-6_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-9183-9
Online ISBN: 978-981-15-9184-6
eBook Packages: MedicineMedicine (R0)