Abstract
Considering both incidence and mortality rates of gastric cancer (GC) being low affected, the disease remains a frequent source of cancer deaths globally. The prediction of GC is based on its staging, hence detecting at an early stage is crucial for a long life that diagnosed at the later. Identifying the cause and treating it will help the patients to sustain a better prognosis. Both genetic and non-genetic factors play an essential role in causing GC. Besides, viral infection by Helicobacter pylori has been proven to cause GC. Identifying and characterizing molecular biomarkers, epigenetic alterations, long non-coding RNAs, circulating tumor DNA and RNA, abnormal methylation with the help of advanced techniques such as microarray profiling, high-throughput techniques, endoscopy, screening body fluids, quantitative PCR, and the advanced next-generation sequencing increase the source of detecting and identifying the gastric cancer at the earliest. However, certain drugs have been administrated to treat early gastric cancer. This chapter reviews in detail the information regarding prognostic and noninvasive biomarkers for the early detection of gastric cancer.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Gastrointestinal cancer (GC) is rated the fifth most common cancer diagnosed and the third principal cause of mortalities among all cancers globally (Bray et al. 2018). Over the past 10 years, the incidence of GC has publicized tremendous reduction, but the five-year survival index showed patients of GC progression to advance stage from early stages (Luo and Li 2019). Treating cancer on an early-onset might reduce the risk and would be a relief to the global burden of the disease so, early detection of any cancer increases the chance of survival on successful prognosis and treatment. Usually, diagnosis or detection of cancer involves two primary mechanisms: (1) education to prop up early diagnosis, and (2) screening. Many programs have been instituted for better cancer outcomes, one such program established in the year 2010 was Be Clear on Cancer (BCOC). Diagnosis using different techniques like endoscopy and biopsy have been in force, but these techniques lack detecting the disease at the earliest, whereas screening for molecular markers aid in revealing early gastric cancer (EGC).
Research on cancer by identifying and characterizing molecular biomarkers, tumor markers, and genetic alteration such as bulky addition or loss of chromosomal, single nucleotide polymorphisms, epigenetics, mutational alteration, histone protein modification, abnormal DNA methylation, over-expression of miRNA, circulating tumor RNA and DNA, and lncRNAs (long non-coding RNAs) has been on forth to detect cancer at the earliest (Cancer Genome Atlas Research Network and Analysis Working Group 2014). In recent times, the application of high-throughput technologies has taken new approaches into molecular pathogenesis, ensuing a novel classification of gastric carcinoma in support of their genomic characterization. Cancer genome atlas has classified GC into four subtypes basing on the targeted material: (a) Epstein–Barr virus-infected tumors, (b) Microsatellite instability tumors, (c) Genomically stable tumor, and (d) Chromosomally unstable cancer (Cristescu et al. 2015). Another new classification was given by the Asian Cancer Research Group, which is microsatellite stable and instability cancer (Patel et al. 2017).
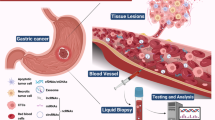
Helicobacter pylori, a Gram-negative, microaerophilic bacterium commonly resite in the human stomach. Recent studies confirmed the risk of gastric carcinoma by H. pylori infection that causes mild to severe gastritis that carries on for a lifetime if not treated with antimicrobial drugs (Correa 1995). The anticipation of dietary intake and screenings reduces the risk of gastric cancer. However, intervention studies such as annihilation through chemoprevention trials have revealed possible strategies. So, understanding the interrelationship of disease and the factors can help researchers and scientists to drive towards novel approaches in the field of reducing the disease progression (Yoon et al. 2011). This chapter aims to discuss the recent advances for early diagnosis of gastric cancer (Fig. 8.1).
2 Screening of GC at the Earliest
Gastroscopy is money-making for the gastroenterologist nowadays; however, the technique is not affordable by the patient since it is expensive. Besides this drawback, it also holds a few complications. Screening procedures that are economically priced, noninvasive and apt for the general population have been required (Tan and Fielding 2006). The progress of high-tech techniques results in molecular markers capable of identifying the disease at the earliest, calculate the disease outcome, and aid admittance for proper therapy.
2.1 Possible Metabolic Biomarkers for GC Metastasis
Metastasis, in general, spreads disease from one organ to another, either adjacent or distant organs. Pathophysiology of the disease confirms the deaths of gastric cancer are primarily a result of metastasis, which can identify metabolic markers. Studies on the metabolomics of human engraft models illustrated principal mechanisms of GC metastasis and probable biomarkers for early diagnosis (Chen et al. 2010). Of the 30 metabolites identified, glutamine was the major metabolite that showed 1.71-fold diminution in expression in the metastatic group rather than non-metastatic. Likewise, praline the upregulated metabolite showed 2.45 times increased expression (Chen et al. 2010). Few studies reported significant variation between the cancerous and non-cancerous group for metabolite composition. Metabolite such as proline, leucine, serine, malic, and lactic acid are known to play an essential role in metastasis of GC (Fig. 8.2). The biomarkers derivative of these metabolites pave pathways that can be support for treating GC at the onset (Hu et al. 2011). Many techniques, namely NMR spectroscopy, liquid and gas chromatography (GC), GC-MS (mass spectrophotometer), capillary electrophoresis-MS, and Fourier spectroscopy are widely used in metabolomics analysis (Jayavelu and Bar 2014).
2.2 Acid Suppression Therapy
Usually, the symptoms EGC are impossible to differentiate compared to benign; such patients are treated with acid suppression drugs, as well as H+ pump inhibitors or hydrogen blockers before gastroscopy. Comparative studies revealed H+ pump inhibitors were healing malignant stomach ulcers with 4 weeks of onset, whereas biopsy and endoscopy are required after acid suppression (Corrigan et al. 1997; Taylor et al. 1978; Wayman et al. 2000).
2.3 Endoscopy Techniques
Modern advances in endoscopic tools have enhanced the sensitivity of identifying EGC. The techniques follow three main steps: (a) detecting suspicious lesions, (b) characterizing the lesions, and finally (c) an accurate diagnosis. Different techniques, namely magnifying and chromoendoscopy, modern high-resolution virtual chromoendoscopy, confocal laser end microscopy, and flexible spectral imaging, have been in use.
Chromoendoscopy, in combination with indigo carmine, is able for recognition and handling targeted biopsies of abnormal areas of the gastric mucosa so that the dye augments tissue abnormality through a high magnification image (Sasako 1997). Magnifying endoscopy assesses gastric lesions at microvascular construction that provides an option of envisaging the histological nature of cancer. Conversely, magnifying endoscopy does not investigate the whole of the gastric mucosa. Another novel technique using infrared light gives deep tissue incursion using an infrared video endoscope (Mataki et al. 2003). Likewise, light-induced fluorescence endoscopy, equipment-based image enhanced endoscopy and endoscopy ultrasonography gave promising results. Endoscopic test plays a vital role in early detection of cancer but the accuracy of detection primarily depends on the endoscopist so that they have a pure knowledge to determine lesions. Combining to or more techniques may increase the scope of better diagnosis so developing minimal invasive endoscopic methods is a challenge to the researchers.
2.4 Serological Test
Quite a few tests using blood samples have been examined to institute aptness as screening tools to detect patients with GC. The tests include screening for pepsinogen, gastrin 17, and Helicobacter pylori antibody. Pepsinogen, a pepsin precursor, be present in two forms, pepsinogen I (PG I) and II (PG II). The difference between the two is their secretion. PG I is secreted mainly by corpus cells where PG II is secreted in cells of the antrum, corpus, Brunner’s gland of the duodenum. The ratio of PG I and PG II is concentrated due to the overproduction of PG II in the antrum, duodenum, and corpus cells (Kikuchi et al. 2000). A study by Kitahara and colleagues revealed the significance of pepsinogen screening is competent in diagnosing GC in patients of atrophic gastritis. The process includes a mixture of PG I and PG I/PG II ratio as an endpoint. However, the test does not apply to mild atrophic gastritis (Kitahara et al. 1999).
As already discussed, H. pylori bacteria play a role in infecting the individuals causing gastric cancer. The antibody of this species acts as a marker in screening dystrophy in patients below 45 years. The test showed a 97% sensitivity and 87% specificity for GC (Sobala et al. 1991). Dissimilarity findings by Whiting and Co. reported reduced activity of H. pylori antibody in patients above 40 years and over 30% had missed diagnosis (Whiting et al. 1998). A study in the year 2005, screening 9293 healthy Japanese with pepsinogen and H. pylori antibody showed promising results and concluded the duo be exclusive screening biomarkers (Watabe et al. 2005).
Gastrin 17, a form of gastrin secreted in G cells, was found in the antrum. In patients of atrophic gastritis, the thrashing of antral G cells resulted in reduced gastrin levels. G-17 subsequently screened these levels as an indicator (Sipponen et al. 1990). Research findings by Sipponen et al. confirmed the use of serology biomarkers, namely pepsinogen, gastrin, and H. pylori antibody together efficiently detected diverse models of gastritis with sensitivity and specificity percentage of 89% and 93%, correspondingly.
2.5 Tumor Biomarkers
For early diagnosis and detection of gastric cancer clinically, tumor marks such as carcinoembryonic antigen (CEA), carbohydrate antigen (CA), and alpha-fetoprotein (AFP) were used (Tsai et al. 2016a). However, the specificity and sensitivity of these biomarkers are stated to be reduced (Tsai et al. 2016a; Tong et al. 2016). Many study evidence explained the improved expression level of oncogenes in gastric cancer where they excite cell cycle and tumor cells growth and also by inhibiting apoptosis. Several genes were identified that showed a positive response in identifying gastric cancer initially. The genes, namely xeroderma pigmentosum group (xpg), stc1, iftim1 (interferon-induced transmembrane protein 1), matrix metalloproteinase 9 (MMP9), and pituitary tumor transforming gene 1 were known to determine GC (Kanda and Kodera 2015).
xpg/ercc5 (xeroderma pigmentosum group G/excision repair cross-complementing group 5) an enzyme from the nucleotide excision repair system is said to involve in revamping DNA lesions that are a result of genomic instability. The expression of ercc5 was significantly reported to have progressed results towards GC gastritis and also coupled with tumor development (Deng et al. 2014). By microarray profiling, the gene ifitm1 was detected in upregulating tumor cell lines and gastric cancer tissues. The gene ifitm1 also plays a vital role in implicating invasions and transfer GC cells to enhance inflammatory response that is a part of tumor progression (Lee et al. 2012). The gene MMP-9 an enzyme plays a role in tumor growth expansion, metastasis, and invasion in gastric carcinoma (Zheng et al. 2006).
Microarray profiling discovered few overexpressed genes such as KRT17, COL10A1, KIAA1199, SPP1, IL11, S100A2, and MMP3 that are related to tumor progression. Out of these, the more used candidate markers were KRT17 and COL10A1 that had enhanced expression for EGC (Chivu et al. 2010). Likely, tumor suppressor genes were also used for early detection as they presented reduced expression in GC patients that resulted in hastened cell growth, declined inhibition of oncogene expression, and the development of cell growth.
A remarkable biomarker gastrokine 1 (GKN1) is extensively expressed in the surface lumen of gastric tissue that indulges in upholding mucosal integrity in the stomach but is not found in GC (Altieri et al. 2017). The gene also acts as a tumor suppressor and modulates apoptosis signaling in GC. These factors and it is lower expression consider the gene as an indicator of increased risk of gastric carcinogenesis (Watanabe et al. 2009).
2.6 Circulating Tumor Cells
Cancer cells freed from a general tumor or the metastatic sites circulate in the blood that is defined as circulating tumor cells (CTCs). The cells are usually sensed by epithelial cell adhesion molecule and cytokeratins. CTCs as diagnostic marker is commonly present in the blood of GC patients.
This gives predictive information after surgical and chemotherapic activities (Haber and Velculescu 2014). CTCs were initially illustrated as expressing epithelial cell markers. EpCAm, cytokeratin (CK): CK8, CK18, and CK19 are CTCs showing an adverse effect for CD45 (Allard et al. 2004). Studies showed the occurrence of CTCs in circulating tumor microemboli representing poor prognosis and controlling disease progression (Chinen et al. 2017).
The soaring heterogeneity of CTCs provoked researchers to expand various methodologies to augment, isolate, and itemize them basing on specific phenotypic and molecular characterization. In general, there are two methodologies used for the isolation and enumeration of CTCs. One is the biological method CellSearch platform for enumeration and the other be physical method, namely Food and Drug Administration (FDA) for clinical purposes. These methods detect EpCAm, CK8, CK18, and CK19 excluding CD45. Using cell-size and phenotype-based systems, as centrifugal microfluidic system based on fluid-assisted separation technique (FAST), or Cascaded Inertial Focusing Microfluidic device, coupled with detection of an extended panel of markers might identify a different subpopulation of CTCs with higher efficiency (Kang et al. 2017; Abdulla et al. 2018). Another technique, immunostaining-fluorescence in situ hybridization (iFISH) platform, claimed to be more sensitive than the CellSearch™ to detect and characterize CTCs in advanced GC patients.
2.7 Circulating Tumor DNA (CtDNA) as a Biomarker
CtDNA investigation developed the liquid biopsy to detect traces of tumor molecular moving body fluids and confer a deeper approach into the cancer heterogeneity, early detection of biological markers, finding therapeutic agents, instances assessment of healing response, and potential resistance and prediction. In general, ctDNA corresponds to only parts of the cell-free circulating DNA (cfDNA) that is noticeably amplified in an advanced stage of the disease (Bettegowda et al. 2014). Studies also showed traces of ctDNA in plasma samples of EGC patients (Alix-Panabières and Pantel 2016; Sumbal et al. 2018). The levels of ctDNA were interrelated with that of vascular invasions, peritoneal repetition, and diagnosis (Fang et al. 2016). In EGC patients, competence for diagnosis was reported with cfDNA consisting of rassfla and apc promoter hypermethylation (Balgkouranidou et al. 2015). Advanced biological techniques such as multiplex MS SNP genotyping, RT quantitative PCR, digital droplet PCR, next-generation sequencing, and advanced nuclear quantification technology were in use to analyze ctDNA in GC individuals to detect the disease at the most basic (Shoda et al. 2015, 2017; Kato et al. 2018).
2.8 CircRNAs
Circular RNAs (CircRNAs) the latest division of non-coding RNA that appears as a closed-loop without ends 5′ and 3′ (Memczak et al. 2013). The presence of CircRNAs in RNA virus was initially reported but, studies found stable and preserved CircRNAs sequence in almost all eukaryotes that organize gene expression by miRNAs connection through microarray profiling and high-throughput RNA sequencing (Chen 2016). The role of CircRNAs was noted in many diseases especially as tumor growth and metastasis (Li et al. 2015). Several CircRNAs are discovered and shown to express in gastric tissues. Of all the types, hsa_circ_0000026, hsa_circRNA_400071, hsa_circRNA_000543, and hsa_circRNA_001959 are reported to have expressed in multiples in GC. hsa_circ_0000026 explained expression of downregulation whereas the other showed differential gene expression (Sui et al. 2017; Huang et al. 2017). Few genes of CirRNA namely cd44, cxxc5, myh9, and malat1suggested to have a role in growth and tumorigenesis. The overall findings discussed here unlocked the path for plasma circRNA profiling that aims to detect definite diagnostic and prognostic circular RNA markers for early gastric cancer individuals.
2.9 LncRNAS Transcriptomes Marker
Long non-coding RNA (LncRNA) are transcripts of 200 nucleotides long with nix or partial perspective towards protein-coding. LncRNAS is labeled as a transcriptomes marker due to its regulation in transcription, translation, cellular differentiation, cell cycle processes, and gene expression (Wang et al. 2015). The factor of its high stable condition while moving in body fluids and also their altitude in tumor tissues made the marker useful to diagnose GC patients at the earliest (Shi et al. 2016; Bolha et al. 2017). A study by Cao and his colleagues revealed 88 differential LncRNAs where 71 showed upregulation and 74 downregulation (Cao et al. 2013). Zhou et al. suggested the use of lncRNA and H19 as potential biomarkers to detect and monitor GC especially for early screening. Besides the discussed markers, lncRNA PVT1candidate, TINCR, CCAT2, AOC4P, BANCR, CUDR, LSINCT-5, PTENP1, and LINC00857 also acts as a possible marker for GC diagnosis (Zhou et al. 2015; Yuan et al. 2016; Zhang et al. 2017; Dong et al. 2015).
2.10 Epigenetic Alteration-Methylation
Few definite genes such as p16nk4a, tcf4, DNA repair (hmlh1 and mgmt), cell growth/differentiation (hoxd10, hai-2/spint2, ndrg2), transcriptional regulation (hltf, pax6, znf545, runx3), cell adhesion/invasion/migration (cdh1, cdh4, apc, flnc, lox, timp3, tsp1), apoptosis (bnip3, xiap, bnip3, bcl2, cacna2d3, dapk, gpx3, pcdh10, pcdh17, casp8, xaf1), angiogenesis (thbs-1 and p73), STAT pathway (socs-1), Ras pathway (rassf1a, rassf2, hdab2ip, rkip), Wnt pathway (dkk-3, ctnnb1), in addition to in multidrug resistance genes (mdr1, gstp1) are reported to regulate in GC persons (Qu et al. 2013; Kazmi et al. 2018). The association of these gene show varied results such as highly methylated in dysplasia and EGC whereas few show lower methylation in advanced stage (Watanabe et al. 2009).
2.11 MiRNAs as a Diagnostic and Prognostic Marker
MicroRNA (miRNA) small non-coding RNA of 19–25 nucleotides long regulates in epigenetic mechanisms such as proliferation, differentiation, cellular processes, and apoptosis. These RNAs are functioned as oncogenes or tumor suppressors basing on the targeted gene (Guimarães et al. 2018). Measuring the serum levels and peripheral blood mononuclear cells show miRNA 21 is overexpressed in gastric patients with a sensitivity of 90%. Few other types of CA199 and CEA reported only 50% specificity (Wu et al. 2015). Gene miR-376c and arid4a are shown to upregulate and downregulate in tissue, plasma, and urine of GC patients (Hung et al. 2017). To date, more than 2500 miRNA genes have been distinguished to express in GC patients. Different studies investigation concluded the presence of various miRNAs, namely miR-196a and 196b, miR-501-3p, miR-143-3p, miR-451a, miR-146a, miR-16, miR-25, miR-92a, miR-451, and miR-486-5p, miR-200a-3p, miR-296-5p, miR-132-3p, miR-485-3p, and miR-22-5p, miR10b-5p, miR132-3p, miR185-5p, and miR195-5p function as noninvasive biomarkers, upregulated and downregulated in gastric cancer persons (Tsai et al. 2016b; Jiang et al. 2017; Zeng et al. 2012; Wang et al. 2018; Zhou et al. 2017).
3 Conclusion
Globally, gastric cancer death rate has increased ten times compared to other cancers. The majority of cancer is diagnosed at the final stage as the disease shows no symptoms at the initial stage due to this treating the patient at the earliest is limited. The detection of biomarkers for the disease diagnosis and prognosis aids in curing the disease at the early stage so, studies are directed towards identification and validation of noninvasive markers, cost-effective, highly stable, specific, and sensitive to the GC patients. Few markers mainly ctDNA, ctRNA, lncRNAs, circRNAs, and miRNAs were discovered and reported promising results for early diagnosis of gastric cancer. However, still, strategies have been to plan to get improved and enriched techniques to detect the disease at the earliest.
References
Abdulla A, Liu W, Gholamipour-Shirazi A, Sun J, Ding X (2018) High-throughput isolation of circulating tumor cells using cascaded inertial focusing microfluidic channel. Anal Chem 90(7):4397–4405
Alix-Panabières C, Pantel K (2016) Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov 6(5):479–491
Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C et al (2004) Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 10(20):6897–6904
Altieri F, Di Stadio CS, Federico A, Miselli G, De Palma M, Rippa E, Arcari P (2017) Epigenetic alterations of gastrokine 1 gene expression in gastric cancer. Oncotarget 8(10):16899
Balgkouranidou I, Matthaios D, Karayiannakis A, Bolanaki H, Michailidis P, Xenidis N et al (2015) Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients. Mutat Res 778:46–51
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al (2014) Detection of circulating tumor DNA in early-and late-stage human malignancies. Sci Transl Med 6(224):224ra24–224ra24
Bolha L, Ravnik-Glavač M, Glavač D (2017) Long noncoding RNAs as biomarkers in cancer. Dis Markers 2017:7243968
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Cancer Genome Atlas Research Network, & Analysis Working Group (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513:202–209
Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY (2013) Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol 19(23):3658
Chen LL (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17(4):205
Chen JL, Tang HQ, Hu JD, Fan J, Hong J, Gu JZ (2010) Metabolomics of gastric cancer metastasis detected by gas chromatography and mass spectrometry. World J Gastroenterol 16(46):5874
Chinen LTD, Abdallah EA, Braun AC, de Campos BDCT, Flores P, Corassa M et al (2017) Circulating tumor cells as cancer biomarkers in the clinic. In: Isolation and molecular characterization of circulating tumor cells. Springer, Cham, pp 1–41
Chivu ME, Necula LG, Dragu D, Badea L, Dima SO, Tudor S et al (2010) Identification of potential biomarkers for early and advanced gastric adenocarcinoma detection. Hepatogastroenterology 57(104):1453–1464
Correa P (1995) Helicobacter pylori and gastric carcinogenesis. Am J Surg Pathol 19:S37–S43
Corrigan NT, O’Riordain MG, McEntee G (1997) Modern anti-ulcer regimens and gastric carcinoma-is there a delay in definitive treatment. Br J Surg Suppl 84:62–63
Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS et al (2015) Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 21(5):449
Deng N, Liu JW, Sun LP, Xu Q, Duan ZP, Dong NN, Yuan Y (2014) Expression of XPG protein in the development, progression and prognosis of gastric cancer. PLoS One 9(9):e108704
Dong L, Qi P, Xu MD, Ni SJ, Huang D, Xu QH et al (2015) Circulating CUDR, LSINCT-5 and PTENP 1 long noncoding RNA s in sera distinguish patients with gastric cancer from healthy controls. Int J Cancer 137(5):1128–1135
Fang WL, Lan YT, Huang KH, Liu CA, Hung YP, Lin CH et al (2016) Clinical significance of circulating plasma DNA in gastric cancer. Int J Cancer 138(12):2974–2983
Guimarães CTU, Martins NNF, da Silva Oliveira KC, Almeida CM, Pinheiro TM, Gigek CO, Calcagno DQ (2018) Liquid biopsy provides new insights into gastric cancer. Oncotarget 9(19):15144
Haber DA, Velculescu VE (2014) Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 4(6):650–661
Hu JD, Tang HQ, Zhang Q, Fan J, Hong J, Gu JZ, Chen JL (2011) Prediction of gastric cancer metastasis through urinary metabolomic investigation using GC/MS. World J Gastroenterol 17(6):727
Huang YS, Jie N, Zou KJ, Weng Y (2017) Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep 16(3):2469–2476
Hung PS, Chen CY, Chen WT, Kuo CY, Fang WL, Huang KH et al (2017) miR-376c promotes carcinogenesis and serves as a plasma marker for gastric carcinoma. PLoS One 12(5):e0177346
Jayavelu ND, Bar NS (2014) Metabolomic studies of human gastric cancer. World J Gastroenterol 20(25):8092
Jiang X, Wang W, Yang Y, Du L, Yang X, Wang L et al (2017) Identification of circulating microRNA signatures as potential noninvasive biomarkers for prediction and prognosis of lymph node metastasis in gastric cancer. Oncotarget 8(39):65132
Kanda M, Kodera Y (2015) Recent advances in the molecular diagnostics of gastric cancer. World J Gastroenterol 21(34):9838
Kang HM, Kim GH, Jeon HK, Kim DH, Jeon TY, Park DY et al (2017) Circulating tumor cells detected by lab-on-a-disc: role in early diagnosis of gastric cancer. PLoS One 12(6):e0180251
Kato S, Okamura R, Baumgartner JM, Patel H, Leichman L, Kelly K et al (2018) Analysis of circulating tumor DNA and clinical correlates in patients with esophageal, gastroesophageal junction, and gastric adenocarcinoma. Clin Cancer Res 24(24):6248–6256
Kazmi HR, Kumari S, Tiwari S, Khanna A, Narayan G (2018) Epigenetic mechanisms and events in gastric cancer-emerging novel biomarkers. Pathol Oncol Res 24(4):757–770
Kikuchi S, Kurosawa M, Sakiyama T, Tenjin H, Miki K, Wada O, Inaba Y (2000) Long-term effect of Helicobacter pylori infection on serum pepsinogens. Jpn J Cancer Res 91(5):471–476
Kitahara F, Kobayashi K, Sato T, Kojima Y, Araki T, Fujino MA (1999) Accuracy of screening for gastric cancer using serum pepsinogen concentrations. Gut 44(5):693–697
Lee J, Goh SH, Song N, Hwang JA, Nam S, Choi IJ et al (2012) Overexpression of IFITM1 has clinicopathologic effects on gastric cancer and is regulated by an epigenetic mechanism. Am J Pathol 181(1):43–52
Li P, Chen S, Chen H, Mo X, Li T, Shao Y et al (2015) Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 444:132–136
Luo M, Li L (2019) Clinical utility of miniprobe endoscopic ultrasonography for prediction of invasion depth of early gastric cancer: a meta-analysis of the diagnostic test from PRISMA guideline. Medicine 98(6):e14430
Mataki N, Nagao S, Kawaguchi A, Matsuzaki K, Miyazaki J, Kitagawa Y et al (2003) Clinical usefulness of a new infrared video endoscope system for diagnosis of early stage gastric cancer. Gastrointest Endosc 57(3):336–342
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338
Patel TN, Roy S, Ravi R (2017) Gastric cancer and related epigenetic alterations. Ecancermedicalscience 11:714
Qu Y, Dang S, Hou P (2013) Gene methylation in gastric cancer. Clin Chim Acta 424:53–65
Sasako M (1997) Radical surgery. Gastric cancer
Shi T, Gao G, Cao Y (2016) Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers 2016:9085195
Shoda K, Masuda K, Ichikawa D, Arita T, Miyakami Y, Watanabe M et al (2015) HER2 amplification detected in the circulating DNA of patients with gastric cancer: a retrospective pilot study. Gastric Cancer 18(4):698–710
Shoda K, Ichikawa D, Fujita Y, Masuda K, Hiramoto H, Hamada J et al (2017) Clinical utility of circulating cell-free Epstein–Barr virus DNA in patients with gastric cancer. Oncotarget 8(17):28796
Sipponen P, Valle J, Varis K, Kekki M, Ihamäki T, Siurala M (1990) Fasting levels of serum gastrin in different functional and morphologic states of the antrofundal mucosa: an analysis of 860 subjects. Scand J Gastroenterol 25(5):513–519
Sobala GM, Pentith JA, Axon ATR, Dixon MF, Crabtree JE, Rathbone BJ et al (1991) Screening dyspepsia by serology to Helicobacter pylori. Lancet 338(8759):94–96
Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J et al (2017) Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol Rep 37(3):1804–1814
Sumbal S, Javed A, Afroze B, Zulfiqar HF, Javed F, Noreen S, Ijaz B (2018) Circulating tumor DNA in blood: future genomic biomarkers for cancer detection. Exp Hematol 65:17–28
Tan YK, Fielding JW (2006) Early diagnosis of early gastric cancer. Eur J Gastroenterol Hepatol 18(8):821–829
Taylor R, Lovell D, Menzies-Gow N, La Brooy SJ, Misiewicz JJ (1978) Misleading response of malignant gastric ulcers to cimetidine. Lancet 311(8066):686–688
Tong W, Ye F, He L, Cui L, Cui M, Hu Y et al (2016) Serum biomarker panels for diagnosis of gastric cancer. Onco Targets Ther 9:2455
Tsai MM, Wang CS, Tsai CY, Huang HW, Chi HC, Lin YH et al (2016a) Potential diagnostic, prognostic and therapeutic targets of microRNAs in human gastric cancer. Int J Mol Sci 17(6):945
Tsai MM, Wang CS, Tsai CY, Huang CG, Lee KF, Huang HW et al (2016b) Circulating microRNA-196a/b are novel biomarkers associated with metastatic gastric cancer. Eur J Cancer 64:137–148
Wang J, Song YX, Wang ZN (2015) Non-coding RNAs in gastric cancer. Gene 560(1):1–8
Wang J, Zhang H, Zhou X, Wang T, Zhang J, Zhu W et al (2018) Five serum-based miRNAs were identified as potential diagnostic biomarkers in gastric cardia adenocarcinoma. Cancer Biomark 23(2):193–203
Watabe H, Mitsushima T, Yamaji Y, Okamoto M, Wada R, Kokubo T et al (2005) Predicting the development of gastric cancer from combining Helicobacter pylori antibodies and serum pepsinogen status: a prospective endoscopic cohort study. Gut 54(6):764–768
Watanabe Y, Kim HS, Castoro RJ, Chung W, Estecio MR, Kondo K et al (2009) Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes. Gastroenterology 136(7):2149–2158
Wayman J, Hayes N, Raimes SA, Griffin SM (2000) Prescription of proton pump inhibitors before endoscopy: a potential cause of missed diagnosis of early gastric cancers. Arch Fam Med 9(4):385
Whiting JL, Hallissey MT, Fielding JWL, Dunn J (1998) Screening for gastric cancer by Helicobacter pylori serology: a retrospective study. Br J Surg 85(3):408–411
Wu J, Li G, Wang Z, Yao Y, Chen R, Pu X, Wang J (2015) Circulating MicroRNA-21 is a potential diagnostic biomarker in gastric cancer. Dis Markers 2015:435656
Yoon H, Kim N, Lee HS, Shin CM, Park YS, Lee DH et al (2011) Helicobacter pylori-negative gastric cancer in South Korea: incidence and clinicopathologic characteristics. Helicobacter 16(5):382–388
Yuan CL, Li H, Zhu L, Liu Z, Zhou J, Shu Y (2016) Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma 63(3):442–449
Zeng C, Rothfuss J, Zhang J, Chu W, Vangveravong S, Tu Z et al (2012) Sigma-2 ligands induce tumour cell death by multiple signalling pathways. Br J Cancer 106(4):693–701
Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y et al (2017) Genome-wide lncRNA microarray profiling identifies novel circulating lncRNAs for detection of gastric cancer. Theranostics 7(1):213
Zheng H, Takahashi H, Murai Y, Cui Z, Nomoto K, Niwa H et al (2006) Expressions of MMP-2, MMP-9 and VEGF are closely linked to growth, invasion, metastasis and angiogenesis of gastric carcinoma. Anticancer Res 26(5A):3579–3583
Zhou X, Yin C, Dang Y, Ye F, Zhang G (2015) Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep 5:11516
Zhou X, Wen W, Zhu J, Huang Z, Zhang L, Zhang H et al (2017) A six-microRNA signature in plasma was identified as a potential biomarker in diagnosis of esophageal squamous cell carcinoma. Oncotarget 8(21):34468
Acknowledgments
The authors are grateful to authorities of Andhra University Visakhapatnam, India, and Krishna University, Machilipatnam, for providing necessary facilities to carry out the research work and for extending constant support.
Conflict of Interest
The authors declare that there is no potential conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Sudhakar, P., Sanapala, P., Naidu, B.P. (2020). Overview of Early Detection of Gastrointestinal Cancer. In: Veera Bramhachari, P., Neelapu, N. (eds) Recent Advancements in Biomarkers and Early Detection of Gastrointestinal Cancers . Diagnostics and Therapeutic Advances in GI Malignancies. Springer, Singapore. https://doi.org/10.1007/978-981-15-4431-6_8
Download citation
DOI: https://doi.org/10.1007/978-981-15-4431-6_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-4430-9
Online ISBN: 978-981-15-4431-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)