Abstract
Heavy metals (HMs) are versatile elements of nature with five times higher atomic weight and density than water. HMs are ubiquitous in nature due to the industrial, domestic, agricultural, medical and technological applications. These are toxic at trace levels and therefore attract more and more interest for their least bioaccumulation and thus high persistence in the environment. Among HMs, arsenic, cadmium, chromium, lead and mercury rank as priority metals that are of public health significance and ecological concern. Interestingly, bacteria have been found as efficient tool for heavy metal degradation as well as resistance. Several bacteria have been reported for the HM accumulation which has been controlled by the metal resistance gene, carried on genome or in plasmid. In nature, Gram-negative bacteria are dependent on plant-derived simple carbon (C) compounds. In HMs abundant flora and fauna, they survive by different cellular mechanisms like metal sorption, mineralization, uptake and accumulation, extracellular precipitation, enzymatic mechanisms for oxidation or reduction to a less toxic form and efflux of heavy metals from the cells to adapt in HM stresses. Hence, here we focus on the mechanism of microbial interaction with these heavy metals which can open the new horizon for the exploitation of Gram-negative bacteria and their gene pool as HM remediator agents, biological indicator and plant growth promoters.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keyword
1 Introduction
The extensive existence of the anthropogenic-based pollutant and effluent are the important stress factors that have been responsible for several diseases all over the biological kingdom of environments. Among them, the metals and metalloids with a density above 5 g−1 cm3, known as heavy metals (HMs), are increasing day by day and are a global threat to living beings and ecological health on earth (Zhou et al. 2015). HMs are high-density nondegradable naturally occurring earth crust compounds, which are much toxic even at a very trivial dosage or concentration. HMs are problematic to environment because of their nonbiodegradability, higher toxicity and bioaccumulation in food chain of living organism. They enter our systems through inhalation, adsorption by cell surface contact within industrial exposure, manufacturing, agriculture and residential settings. HMs are represented by arsenic (As), cadmium (Ca), chromium (Cr), cobalt (Co), lead (Pb), mercury (Hg), nickel (Ni), selenium (Se) and zinc (Zn) which are highly toxic even in trace amounts (Turpeinen et al. 2002; Siddiquee et al. 2015). HMs basically come from naturally as well as anthropogenic sources (Fu and Wang 2011). Some important natural sources of heavy metals are natural rock weathering process, volcanic eruptions, forest fires, sea salt sprays, wind-borne soil particles and biogenic sources. Industrial activities such as leather tanning, energy production, electroplating, oil industries including crude oil and hydrocarbon exploration and utilization, emissions from vehicular traffic gas exhausts, fuel production and downwash from power lines are also the major sources of HMs. However, the growth and development of living beings require some traces of heavy metals like Fe, Cu, Zn, Mo, etc., but the excess of these metals can be harmful for plants as well as concerned food chain for animals (Wintz and Fox 2002). For example, during plant growth, if they accumulate the HMs in more concentration, the plant growth and cellular metabolism, absorption as well as transportation of vital component will be inhibited (Xu and Shi 2000). Robin et al. (2012) have reported the harmful effects of HMs to plant growth and development, which are responsible for various diseases in animal. Studies have revealed that HM pollution is a serious global environmental problem which is adversely affecting the composition and activity of soils and its microbial communities (Xie et al. 2016).
HM pollutants consistently get deposited in the major sink of nature, “the soils”. In the nature, the cycle of elements and metals is managed by soil bacteria, fungi, actinomycetes, algae and other microorganisms, and they are responsible for the decomposition of material elements and nutrient conversion via various biochemical reactions in the soil. Soil microbes are more sensitive to soil conditions than large animals or plants, and hence they serve as an indicator for soil environmental quality (Dian 2018). But the HM pollution can alter the soil microbiota and their activity such as soil enzyme activity, composition of soil microbial community and structure, plant growth, etc. (Sadler et al. 1967; Giller et al. 1998; Rajapaksha et al. 2004; Singh et al. 2017). Gülser and Erdoğan (2008) studied the effects of HM pollution on microbial enzyme activities and basal soil respiration of soils. Mills and Colwell (1977) found that HMs are detrimental to microorganisms even at the low concentrations. Ahmad et al. (2005) reported microbial diversity losses in soil by metal toxicity and which was validated by microcosm test. The study revealed that Pb, Mn and Ni were highly toxic, followed by Cd, Hg, Cr and Cu and Zn were the least. However, the toxicity of HMs was concentration as well as time dependent.
During the long-term exposures and history of HM contamination, several microorganisms have followed the resistance strategies and developed the detoxification or assimilation or bioremediation mechanisms to counter the toxic effects (Azarbad et al. 2016; Tipayno et al. 2018; Yang et al. 2019). The term “resistance to HMs” refers to the mechanism of detoxification of toxic metals by bacteria (Gadd 1992). Khan et al. (2016) isolated the HM-resistant Gram-negative bacteria Salmonella enterica 43Ca and found that the resistance was in order of Pb2+ > Cd2+ > As3+ > Zn2+ > Cr6+ > Cu2+ > Hg2+. The metal resistance mechanisms allow the microbial populations to survive and maintain the functional sustainability of their communities. Moreover, the HM resistance in bacteria allow them to be employed as potential eco-friendly and cost-effective bioremediation tools for HMs which transform the toxic HMs into a less harmful state (Abbas et al. 2014; Ma et al. 2016; Ndeddy and Babalola 2016). Some of the metal resistance factors such as bioaccumulation (Jin et al. 2018), reduction (Nies 1999), biosorption (Quintelas et al. 2011), siderophore production (Singh et al. 2019; Prajakta et al. 2019) and the formation of biofilms (Von Bodman et al. 2003) have been explored for the control of metal pollution with a view to promote mitigation of the environmental impacts.
Gram-negative bacteria are attractive model microorganisms for the laboratory because of their fast growth, easy manipulation, genetic stability in large cultures and well-studied secretion system. Gram-negative bacteria have been studied extensively for the metal resistance. Moreover, Gram-negative microbes and their secreted products are also utilized at commercial scale which has made it more biotechnologically relevant for researchers, who have focused significantly on the homeostasis and resistance of Gram-negative bacteria in the presence of HMs. Therefore, it can open the opportunity for microbial mediated alleviation of HM stress and decreased accumulation of metals in agriculture. Hence, the critical evaluation of mechanistic system of Gram-negative bacteria is needed. So, the present chapter is aimed to display the resistance capabilities and mechanism of Gram-negative bacteria to cope with toxic concentrations of HMs generally considered to be environmental pollutants.
2 HM Resistance in Gram-Negative Bacteria: Molecular and Ecological Prospective
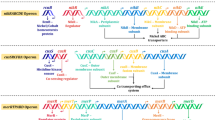
In the course of evolution, bacteria adapted to the increased content of HM ions in places of ore deposits. Its adaptation ensured the appearance in bacteria of various mechanisms for the protection of sensitive components from the action of heavy metal ions such as the type and number of pathways for the transport of metal ions into the cell; the localization of resistance genes on the chromosome, plasmid or transposon; and the role of these ions in normal cell metabolism (Choudhury and Srivastava 2001; Subhashini et al. 2017). Resistance to HMs in Gram-negative bacteria is described here according to previous research. Basically, the protection mechanisms are driven by some fundamental procedures, such as biosorption, intracellular sequestration, extracellular sequestration, extracellular barrier for preventing the metal entry into the microbial cell, methylation of metals and reduction of heavy metal ions by the microbial cell (Gomathy and Sabarinathan 2010; Chandrangsu et al. 2017). An illustration of HM resistance in Gram-negative bacteria is shown in Fig. 7.1.
The extracellular barrier is an important mechanism to prevent these ions from going into the cell. The membrane of the cell, cell wall or capsule can avoid entry of the metal ions within the cell. Different taxonomic groups of bacteria can bind metal ions by polarized groups of the cell wall or capsule (phosphate, carboxyl, hydroxyl and amino groups) (Taniguchi et al. 2000). Sorption is considered as a passive process, and during this process, bacterial cells of dead bacteria bind with the ions of metal. It was shown that bacterial cells killed by heating possessed a similar or higher sorption capacity as viable cells.
A passive sorption of ions of heavy metal was observed in nonviable cells of P. putida at high levels, while some studies revealed that the accumulation of metal ions by viable bacterial cells occurs in two stages – rapid non-specific sorption on the surface of the cell wall and, far along, long-term gathering of these ions of metal in the cytoplasm (Gadd 1990, McEldowney 2000). HM ions can also be bound by bacterial capsular polymers, mainly carboxyl groups of polysaccharides. The ability to bind metal ions by extracellular biopolymers was observed in Marinobacter sp. and Acinetobacter sp. (Bhaskar and Bhosle 2006). Interestingly, in P. aeruginosa biofilm cells had a significantly higher resistance to HMs (Cu, Pb and Zn) (Teitzel and Parsek 2003).
Active transport or, in other words, efflux represents the most extensive system for the resistance of bacteria to metal ions. By active transport, bacteria can remove metal ions from the cell. Efflux systems can be encoded by both chromosomal (Franke et al. 2001) and plasmid genetic determinants (Cervantes and Gutierrz-Corona 1994).
HM resistance in Gram-negative bacteria is mostly contributed by lipopolysaccharide of cell walls, a significant component of the outer membrane. The outer layers of cells probably determine how much of a metal penetrates the cytoplasm. HM resistance in Gram-negative bacteria also relates to secretion system that deliver multiple effector proteins into cells or into the extracellular milieu. Wang et al. (2015) have revealed that T6SS4 of Y. pseudotuberculosis have a prominent role in transporting the zinc ions (Zn2+) from the environment into bacterial cells to mitigate the detrimental hydroxyl radicals induced by multiple stressors and prevent the cells from having oxidative damage. Similarly, an H3-T6SS secreted effector TseF (PA2374) of P. aeruginosa is involved in iron uptake by interacting with outer membrane vesicles (OMVs) and the Pseudomonas quinolone signal (PQS) system (Lin et al. 2017).
2.1 Lead (Pb)
Pb is not known to be of any biological significance but is toxic at very low concentrations (Bruins et al. 2000). However, the bioavailable fraction of Pb(II), to which microbes are exposed, may be rather low (Kotuby-Amacher et al. 1992). In the Micrococcus luteus and Azotobacter sp., the cell wall and several functional macromolecules are involved in binding Pb(II). The ions were revealed to be presented in the cell wall and cell membrane, and the least portion was found in the cytoplasmic fraction (Tornabene and Edwards 1972; Tornabene and Peterson 1975). Jarosławiecka and Piotrowska-Seget (2014) have reviewed Cupriavidus metallidurans CH34 for its unique mechanism combining efflux and lead precipitation. Pb(II) toxicity can alter the conformation of nucleic acids and proteins, inhibit the enzyme activity and disrupt the membrane functions and oxidative phosphorylation, as well as osmotic balance disruption (Bruins et al. 2000). Hasnain et al. (1993) have isolated the Pb(II)-resistant Gram-negative bacteria (P. marginalis, P. vesicularis and Enterobacter sp.) from metal-contaminated soils, industrial wastes and plants growing on metal-contaminated soils.
2.2 Cadmium (Cd)
In the hazardous substance list, Cd is also in the priority list and is classified at the seventh place by the Agency for Toxic Substances and Disease Registry (ATSDR) in 2017 (ATSDR 2017). Several studies have marked Cd as a highly toxic element even at low concentrations (Figueira et al. 2005, Lima et al. 2006, Aksoy et al. 2014, Vinodini et al. 2015). Tremaroli et al. (2009) have reported that the exposure to metals changes bacterial metabolism and altered growth pattern in time-dependent manner (Khan et al. 2016). Fazeli et al. (2010) have found that Gram-negative bacteria were less sensitive to cadmium compared to the Gram-positive bacteria. During the experiment, 23–50 mg kg−1 cadmium (drinking water) was given to healthy mice, and after 45 days intestinal microflora was aseptically collected and bacterial count was performed. In comparison with the control, it was revealed that bacteria of genera Bacillus cereus, Lactobacillus spp., Clostridium spp., E. coli, Klebsiella spp., Pseudomonas spp., Enterococcus spp. and Proteus spp. were presented which could be due to their possible ability to uptake the Cd ions. In E. coli, Cd effects by extending the lag phase of cultures, though normal proliferation was detected at the end of the lag phase (Jaiganesh et al. 2012). Among the microbial plethora, plant-associated soil bacteria are of great interest because of their potentiality in nitrogen fixation, agriculture and industrial production, crop protection and plant-mediated biodegradation (Singh et al. 2016, Singh et al. 2019, Prajakta et al. 2019, Yang et al. 2019). High Cd resistance in soilborne bacterium Enterobacter sp. strain EG16 was found in multimetal-polluted site of Dabao Mountain, Guangdong, China. It tolerates high level of Cd2+ concentrations (MIC, >250 mg). Strain EG16 accumulated the 31% of the total Cd by surface biosorption (Chen et al. 2016). Similarly, Rhizobium sp. strain E20–8, isolated from Pisum sativum root nodules, grown at non-contaminated field in Southern Portugal was reported as Cd tolerant (Matos et al. 2019).
2.3 Mercury (Hg)
In HMs, Hg is in any form poisonous, is persistent in nature and has been ranked third by the US Government Agency (1999) for Toxic Substances and Disease Registry. These most toxic elements or substances are continuing to be dumped and spilled into the soil, water and atmosphere and consumed by living beings (Clifton 2007). Currently, Hg resistance in bacteria is in its fifth decade. Unfortunatelly, the bacterial metal resistant mechanism is leads to transformation of its toxic target at large scale (Barkay et al. 2003). The Gram-negative bacteria consist of a regulatory gene (merR), an operator/promoter region and at least three structural genes merT, merP and merA as Hg-resistant elements (Etesami 2017). Regulatory genes are basically encoded for three components – a membrane transport protein, a Hg2+-binding protein for periplasmic and an enzyme subunit for mercuric reductase and participated as central player for Hg resistance. In Gram-negative bacteria, the mer operons, a fairly high GC content, averaging 61% overall ((Liebert et al. 2000), have been studied extensively for the bacterial Hg resistance. However, mer operons are highly homologous in most of Gram-negative bacteria (Trajanovska et al. 1997). Hg(II) in Gram-negative bacteria competes with MerT’s cytosolic cysteines as a dithiol derivative which is a substrate form for MerA. MerC is also the most common in Gram-negative bacteria, but the presence of MerF and MerE in sequence data is also recorded. Resistance to inorganic and organic mercurial in bacteria drives with MerB. Recently, MerG, a speciously periplasmic protein, has been reported in some Gram-negative bacteria. Moreover, in several Gram-negative bacteria, an additional protein, MerD, appears which antagonizes MerR’s activation of mer operon transcription. Parkhill and Brown (1990) have reported the presence of MerO, an 18-bp hyphenated dyad with 7-bp palindromes flanking a 4-bp AT-rich centre, in most Gram-negative bacteria.
Several studies have revealed the Hg-resistant Gram-negative bacteria. Pérez-Valdespino et al. (2013) isolated the Aeromonas strains from diarrhoea sample and showed that Hg resistance occurs via mercuric ion reduction and indicated the presence of high variable mer operons in Aeromonas. Hg-resistant Aeromonas strains, A. hydrophila, A. caviae, A. veronii, A. aquariorum and A. media were characterized in Mexico (Aguilera-Arreola et al. 2007). Hg-tolerant P. aeruginosa has isolated from hospital sewage of Brazil (Lima de Silva et al. 2012), surface river water of Pole Khan and Pole Petroshimi stations, Iran (Mirzaei et al. 2013). Gram-negative bacteria from water samples were reported to be resistant at 110–200 μg/ml (Shakoori and Muneer 2002, Alam and Imran 2017). Beijerinckia and Azotobacter sp., N2-fixing bacteria, have been reported for their ability to remove Hg (Ray et al. 1989).
2.4 Chromium (Cr)
Cr is also speculated as one of the 17 extremely hazardous chemicals by the US Environmental Protection Agency (USEPA) (Marsh and McInerney 2001). Cr(VI) compounds are not only toxic for humans but also responsible for the alteration of bacterial diversity in soil ecosystem (Turpeinen et al. 2004; Viti 2006). Moreover, the bacterial growth rate declines (Garbisu et al. 1998), or the lag phase is extended with uncoupling of energy (Nepple et al. 2000), as the chromate concentration is progressively increased (Chardin et al. 2002). The toxicity of Cr(VI) in bacteria also affected the morphological symmetry and filamentous growth, altered the gene expression, activated the SOS response to counteract the oxidative stress and resulted in the induction of prophage-related genes (Ackerley et al. 2006). Chourey et al. (2006) have revealed that Cr stress also negatively affected the cell division, DNA metabolism and gene regulation, chemotaxis and protein transport system, biosynthesis and degradation of murein, membrane response and environmental stress protection mechanisms.
Cr resistance in bacteria has been generally observed in chromium-contaminated habitats such as soil, wastewater, industrial effluents, etc. (Pal et al. 2005). Pseudomonas sp., a Gram-negative bacterial strain, was first reported as Cr(VI) resistant and has the ability to reduce Cr(VI) (Romanenko and Korenkov 1977). Cr detoxification was observed by the reduction of Cr(VI) to Cr(III), through Cr(V) and Cr(IV) intermediates through P. aeruginosa (Bopp and Ehrlich 1988a). Interestingly, chromium (III) is less toxic than chromium (VI) (approximately 1000 times) due to their impermeability to cell membrane. Several studies have already been revealed the resistance of Gram-negative bacteria to Cr such as Cr-resistant and Cr-reducing bacteria Serratia marcescens (Campos et al. 2005) Acinetobacter and Ochrobactrum isolated from the activated sludge of a wastewater treatment plant, Portugal (Francisco et al. 2002), E. casseliflavus (Saranraj et al. 2010) E. gallinarum with the ability to reduce the chromate to 100% at a concentration of 200 mg/l (Sayel et al. 2012). The removal of Cr(VI) from aqueous solution using kaolin-supported bacterial biofilms was evaluated by Khyle et al. (2018). They revealed that the adsorptive capacity of Gram-negative E. coli is higher than the Gram-positive S. epidermidis. Thacker and Madamwar (2005) have isolated and identified the Cr-resistant (>300 ppm) Ochrobactrum sp. DM1 from chemical industry sites and speculated 30 kDa inducible protein for chromium reduction.
Cr resistance in bacterial cell was clearly illustrated by Ahemad (2014). He reported that Cr enters into the bacterial cell via sulphate transporter (encoded by chromosomal DNA), which is due to the homology between Cr and sulphate. Bacterial cells resist to chromate toxicity by exorcise the intracellular chromates outside through efflux systems (encoded by plasmid DNA). Aerobic and anaerobic reduction of Cr(VI) ion into Cr(III) ion involves soluble reductase which requires NAD(P) and electron transport pathway by cytochrome b or c along the respiratory chains in the inner membrane, respectively. While, Cr(VI) ion redox cycle produced the Cr(V) ion by the production of reactive oxygen species (ROS) in oxidative stresses.
2.5 Copper (Cu)
Cu is an essential element required in traces for cellular process and participates as component of proteins. But, during changes in cuprous and cupric, it generates the reactive toxic radicals (Ridge et al. 2008). Hiniker et al. (2005) reported that free copper causes the cellular sulfhydryl pool depletion and decreases the cellular viability. Hence, the bacterial system has evolved the mechanism to control the intracellular copper level and save itself from Cu cation toxicity (Waldron and Robinson 2009).
Biocidal action of Cu ions is due to the electrostatic attraction with cell membrane and is affected more in Gram-negative bacteria than Gram-positive bacteria (Vergara-Figueroa et al. 2019). The excess of Cu in Gram-negative bacteria is mainly controlled by the cue regulon which is composed by CueR, included with P1B-1-type ATPase coding genes (a periplasmic multicopper oxidase (MCO)), for sensing the Cu(I) ions’ presence in cytoplasm and small metal chaperones of cytoplasm (Outten et al. 2000, Espariz et al. 2007, Zhang and Rainey 2008). Cu(I) export from cytoplasm to the periplasm is done by ATPase (Rensing et al. 2000), and Cu(I) is converted into a less toxic Cu(II) by oxygen-mediated oxidation governed by MCO (Singh et al. 2004). Majorly, periplasmic Cu homeostasis in aerobic condition is maintained by the above described mechanisms in Gram-negative bacteria. In E. coli, CueR regulon is composed by the ATPase (copA) and the MCO, while in P. fluorescens SBW25 it is by the ATPase and the Cu chaperone-coding genes, respectively (Outten et al. 2001; Zhang and Rainey 2008).
Cu-resistant P. syringae was reported by Cha and Cooksey (1991), which has the ability to produce Cu-inducible proteins CopA, CopB and CopC that are responsible for binding bacterial cell and copper ions. Moreover, the long-term exposure of Cu also stimulates the genetic determinants for Cu adaptation. Five isolates from three Gram-negative genera, Sphingomonas, Stenotrophomonas and Arthrobacter, were procured and all were resistant (3.1–4.7 mM) to Cu (Altimira et al. 2012). The Gram-negative Enterobacteriaceae (E. coli and S. enterica) encoded the homologous Cue-responsive regulon system (Samanovic et al. 2012). Gram-negative strains of E. coli, Enterobacter spp., Klebsiella pneumoniae and Pseudomonas aeruginosa have been isolated from the hospital environment of AGH University of Science and Technology, Kraków, and all were more sensitive to copper (Różańska et al. 2018).
2.6 Arsenic (As)
Arsenic (As) is also often present in the environment and is very toxic for most microorganisms. For the resistance, some microbial strains have genetic determinants, e.g. bacterial plasmids. In Gram-negative bacteria, it encodes specific efflux pumps that have the ability to extrude the As from cell cytoplasm. The efflux pump consists of a two-component ATPase complex (ArsA and ArsB) and is integrated with membrane subunit (Rosen and Liu 2009; Yang et al. 2012). arsBC gene pair is commonly found in Gram-negative bacterial chromosome. Several studies have reported the various genes and gene clusters associated with plasmids as well as chromosomes, for example, As-resistant arsRBC or arsRDABC gene cluster associated with plasmids in E. coli and A. multivorans AIU 301 (Suzuki et al. 1998), ars operon variants in the marine strain P. fluorescens MSP3 (Chen et al. 2016), As genomic island of SinA plasmid in Sinorhizobium sp. and Thiomonas sp. (Freel et al. 2015). The diversity analysis of the arsenic-contaminated old tin mine area in Thailand has procured 262 isolates, and among them, A. koreensis and β-proteobacteria were found as the dominant species of the soil. Interestingly, Areonmit et al. (2010) have revealed that majority of the As-resistant isolates were Gram-negative bacteria. Harmin et al. (2018) have isolated the As-resistant E. asburiae, S. paucimobilis, Pantoea spp., Rhizobium rhizogenes and R. radiobacter (MIC of >1500 mg/L of As).
2.7 Iron (Fe)
In HM resistance, iron uptake system is very significant in bacteria because Fe-mediated gene expression is controlled by the “global” transcriptional regulator fur, which is conserved in many bacterial genera. More than 90 genes in E. coli and 87 in P. aeruginosa are known to be regulated by fur. Studies revealed that Fur also regulates a varied range of metabolic functions in bacteria, such as respiration, chemotaxis, the tricarboxylic acid cycle, glycolysis, amino acid biosynthesis, DNA synthesis and sugar metabolism, protecting the cell from oxidative damage and redox stress conditions. Typically, genes that are involved in Fe uptake are expressed only when Fe is deficient (Guerinot and Yi 1994). Some of the bacteria like B. japonicum is best exemplified to know the regulatory function of fur protein (Singh et al. 2011). In B. japonicum, the symbiont of soybeans, an outer membrane protein that is made in response to Fe starvation is similar to the hydroxamate receptor, FhuA, of E. coli (James et al. 2008). When Fe is replete, Fur, which interacts with Fe, binds to DNA sequences (fur boxes) that overlap the target promoters, repressing their transcription (Guerinot and Yi 1994, Braun et al. 1998). In the absence of Fe, Fur no longer binds, allowing transcription to occur. Fur also affects the transcription of genes concerned with traits as varied as toxin production, superoxide dismutase or acid tolerance (Tsolis et al. 1995). At some promoters, Fur is a positive regulator, in response to the cell’s Fe status (Tsolis et al. 1995). Viable fur alleles have also been isolated in P. aeruginosa, based on the fact that fur mutant strains are manganese resistant (Prince et al. 1993).
3 Biotechnological Prospective of Heavy Metal Resistance
Interactions of microorganisms with HMs are vital for various biotechnological interests. Tolerance to HMs is generally present in bacteria due to the horizontal gene transfer (Ianeva 2009). Naturally occurring microorganisms are capable of reducing and detoxifying heavy metal contamination from industrial effluents. This adaptation occurs due to the development of the cellular protection mechanism system against toxic metal ions of microorganisms. Naturally occurring bacteria such as Gemella sp. and Micrococcus sp. showed biodegradation capacity to metals like cadmium (Cd), chromium (Cr) and lead (Pb) where Hafnia sp. showed resistance to cadmium (Cd) (Marzan et al. 2017). Several studies have revealed that Gram-negative bacteria are likely to be more tolerant to HMs than Gram-positive (Silva et al. 2012). HM resistance in biotechnology has great importance for creating the value-added product by adding metal resistance to a microorganism for the facilitation of biotechnological process, biomining of expensive metals and bioremediation of metal-contaminated environments. Bacteria might be established in the sewage plant or plasmids with a broad host range of replication, and it could be cloned into the bacterial community for economic and social uses. For HM resistance E. coli, Pseudomonas sp., etc. could be a better system to understand how bacteria manage metal homeostasis via several mechanisms and timing of metal sequestration.
References
Abbas HS, Ismail MI, Mostafa MT, Sulaymon HA (2014) Biosorption of heavy metals: a review. J Chem Sci Technol v3:74–102
Ackerley DF, Barak Y, Lynch SV, Curtin J, Matin A (2006) Effects of chromate stress on Escherichia coli K-12. J Bacteriol 188:3371–3381
Aguilera-Arreola MG, Hernández-Rodríguez C, Zúñiga G, Figueras MJ, Garduño RA et al (2007) Virulence potential and genetic diversity of Aeromonas caviae, Aeromonas veronii, and Aeromonas hydrophila clinical strains from Mexico and Spain: a comparative study. Can J Microbiol 53:877–887
Ahemad M (2014) Bacterial mechanisms for Cr(VI) resistance and reduction: an overview and recent advances. Folia Microbiol (Praha) 59(4):321–332
Ahmad I, Hayat S, Ahmad A, Inam A, Samiullah (2005) Effect of heavy metal on survival of certain groups of indigenous soil microbial population. J Appl Sci Environ Manage 9(1):115–121
Aksoy E, Salazar J, Koiwa H (2014) Cadmium determinant 1 is a putative heavy-metal transporter in Arabidopsis thaliana. FASEB J 28(617):4
Alam M, Imran M (2017) Metal tolerance analysis of gram-negative bacteria from hospital effluents of Northern India. J Appl Pharm Sci 7(4):174–180
Altimira F, Yá̃ez C, Bravo G, González M, Rojas L et al (2012) Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of Central Chile. BMC Microbiol 12:193. https://doi.org/10.1186/1471-2180-12-193
Areonmit PJ, Sajjaphan K, Sadowsky MJ (2010) Structure and diversity of arsenic-resistant bacteria in an old tin mine area of Thailand. J Microbiol Biotechnol 20(1):169–178
ATSDR (2017) Substance priority list. Available from: https://www.atsdr.cdc.gov/ SPL
Azarbad H, Van GCAM, Niklińska M, Laskowski R, Röling WFM et al (2016) Resilience of soil microbial communities to metals and additional stressors: DNA-based approaches for assessing stress-on-stress responses. Int J Mol Sci 17:20
Barkay T, Susan M, Miller Anne Summers O (2003) Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev 27(2–3):355–384
Bhaskar PV, Bhosle NB (2006) Bacterial extracellular polymeric substance (EPS): a carrier of heavy metals in the marine food-chain. Environ Int 32(2):191–198
Bopp LH, Ehrlich HL (1988a) Chromate resistance and reduction in Pseudomonas fluorescens strain LB300. Arch Microbiol 150:426–431
Bopp LH, Ehrlich HL (1988b) Chromate resistance and reduction in Pseudomonas fluorescens strain LB300. Arch Microbiol 150:426–431
Braun V (1998) Pumping iron through cell membranes. Science 282:2202–2203
Bruins MR, Kapil S, Oehme FW (2000) Microbial resistance to metals in the environment. Ecotoxicol Environ Saf 45:198–207
Campos VL, Moraga R, Yánez J, Zaror CA, Mondaca MA (2005) Chromate reduction by Serratia marcescens isolated from tannery effluent. Bull Environ Contam Toxicol 75(2):400–406
Cervantes C, Gutierrz-Corona F (1994) Copper resistance mechanisms in bacteria and fungi. FEMS Microbiol Rev 14(2):121–138
Cha JS, Cooksey DA (1991) Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. PNAS U S A 88(20):8915–8919
Chandrangsu P, Rensing C, Helmann JD (2017) Metal homeostasis and resistance in bacteria. Nat Rev Microbiol 15(6):338–350
Chardin B, Dolla A, Chaspoul F, Fardeau ML, Gallice P et al (2002) Bioremediation of chromate: thermodynamic analysis of the effects of Cr(VI) on sulfate-reducing bacteria. Appl Microbiol Biotechnol 60:352–360
Chen J, Yoshinaga M, Garbinski LD, Rosen BP (2016) Synergistic interaction of glyceraldehydes-3-phosphate dehydrogenase and ArsJ, a novel organoarsenical efflux permease, confers arsenate resistance. Mol Microbiol 100:945–953. https://doi.org/10.1111/mmi.13371
Choudhury R, Srivastava S (2001) Zinc resistance mechanisms in bacteria. Curr Sci 81(7):768–775
Chourey K, Thompson MR, Morrell-Falvey J, VerBerkmoes NC, Brown SD et al (2006) Global molecular and morphological effects of 24-h chromium(VI) exposure on Shewanella oneidensis MR-1. Appl Environ Microbiol 72:6331–6344
Clifton JC (2007) Mercury exposure and public health. Pediatr Clin N Am 54(2):237–269
Dian C (2018) Effects of heavy metals on soil microbial community. IOP Conf Ser Earth Environ Sci 113:12009. https://doi.org/10.1088/1755-1315/113/1/012009
Espariz M, Checa SK, Audero ME, Pontel LB, Soncini FC (2007) Dissecting the Salmonella response to copper. Microbiology 153:2989–2997
Etesami H (2017) Bacterial mediated alleviation of heavy metal stress and decreased accumulation of metals in plant tissues. Mechanisms and future prospects. Ecotoxicol Environ Saf 147:175–191
Fazeli M, Hassanzadeh P, Alaei S (2010) Cadmium chloride exhibits a profound toxic effect on bacterial microflora of the mice gastrointestinal tract. Hum Exp Toxicol 30(2):152–159
Figueira EMDAP, Gusmão LAI, Pereira SIA (2005) Cadmium tolerance plasticity in Rhizobium leguminosarum bv. viciae: glutathione as a detoxifying agent. Can J Microbiol 51:7–14
Francisco R, Alpoim MC, Morais PV (2002) Diversity of chromium-resistant and -reducing bacteria in a chromium-contaminated activated sludge. J Appl Microbiol 92(5):837–843
Franke S, Grass G, Nies DH (2001) The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology 147(4):965–972
Freel KC, Krueger MC, Farasin J, Brochier-Armanet C, Barbe V et al (2015) Adaptation in toxic environments: arsenic genomic islands in the bacterial genus Thiomonas. PLoS One 10:e0139011. https://doi.org/10.1371/journal.pone.0139011
Fu F, Wang Q (2011) Removal of heavy metal ions from wastewaters: a review. J Environ Manag 92:407–418
Gadd GM (1990) Heavy metal accumulation by bacteria and other microorganisms. Experientia 46:834–840. https://doi.org/10.1007/BF01935534
Gadd GM (1992) Metals and microorganisms: a problem of definition. FEMS Microbiol Lett 100:197–204
Garbisu C, Alkorta I, Llama MJ, Serra JL (1998) Aerobic chromate reduction by Bacillus subtilis. Biodegradation 9:133–141
Giller KE, Witter E, McGrath SP (1998) Toxicity of heavy metals to microorganisms and microbial processes in agricultural soil: a review. Soil Biol Biochem 30:1389–1414
Gomathy M, Sabarinathan KG (2010) Microbial mechanisms of heavy metal tolerance- a review. Agric Rev 31(2):133–138
Guerinot ML, Yi Y (1994) Iron: nutritious, noxious and not readily available. Plant Physiol 104:815–820
Gülser F, Erdoğan E (2008) The effects of heavy metal pollution on enzyme activities and basal soil respiration of roadside soils. Environ Monit Assess 145(1):127–133
Harmin T, Abdullah S, Idris SRS, Mushrifah A, Nurina B (2018) Arsenic resistance and biosorption by isolated rhizobacteria from the roots of Ludwigia octovalvis. Int J Microbiol 2018:1–10
Hasnain, Yasmin S, Yasmin A (1993) The effects of lead resistant pseudomonads on the growth of Triticum aestivum seedlings under lead stress. Environ Pollut 81:179–184
Hiniker A, Collet JF, Bardwell JC (2005) Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J Biol Chem 280:33785–33791
Ianeva OD (2009) Mechanisms of bacteria resistance to heavy metals. Mikrobiol Z 71(6):54–65
Jaiganesh T, Rani JDV, Girigoswami A (2012) Spectroscopically characterized cadmium sulfide quantum dots lengthening the lag phase of Escherichia coli growth. Spectrochim Acta A 92:29–32
James KJ, Hancock MA, Moreau V, Molina F, Coulton JW (2008) TonB induces conformational changes in surface-exposed loops of FhuA, outer membrane receptor of Escherichia coli. Protein Sci 17:1679–1688
Jarosławiecka A, Piotrowska-Seget Z (2014) Lead resistance in micro-organisms. Microbiol (United Kingdom) 160(1):12–25
Jin Y, Luan Y, Ning Y, Wang L (2018) Effects and mechanisms of microbial remediation of heavy metals in soil: a critical review. Appl Sci 8:1336
Khan Z, Rehman A, Hussain SZ, Nisar MA (2016) Cadmium resistance and uptake by bacterium, Salmonella enterica 43C, isolated from industrial effluent. AMB Exp 6:54. https://doi.org/10.1186/s13568-016-0225-9
Khyle GQ, Bonifacio D, Cybelle MF, Meng-Wei W (2018) Removal of chromium(VI) and zinc(II) from aqueous solution using kaolin-supported bacterial biofilms of Gram-negative E. coli and gram-positive Staphylococcus epidermidis. Sustain Environ Res 28(5):206–213
Kotuby-Amacher J, Gambrell RP, Amacher MC (1992) The distribution and environmental chemistry of lead in soil at an abandoned battery reclamation site. Eng Aspects Metal Waste Manag:1–24
Liebert CA, Watson A, Summers O (2000) The quality of merC, a module of the mer mosaic. J Mol Evol 51:607–622
Lima AIG, Corticeiro SC, Figueira EMDAP (2006) Glutathione-mediated cadmium sequestration in Rhizobium leguminosarum. Enzym Microb Technol 39:763–769
Lima de Silva AA, de Carvalho MA, de Souza SA, Dias PM, Filho d S et al (2012) Heavy metal tolerance (Cr, Ag and Hg) in bacteria isolated from sewage. Brazil J Microbiol 43(4):1620–1631
Lin J, Zhang W, Cheng J, Yang X, Zhu K et al (2017) A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun 8:14888. https://doi.org/10.1038/ncomms14888
Ma Y, Rajkumar M, Zhang C, Freitas H (2016) Beneficial role of bacterial endophytes in heavy metal phytoremediation. J Environ Manag 174:14–25
Marsh TL, McInerney MJ (2001) Relationship of hydrogen bioavailability to chromate reduction in aquifer sediments. Appl Environ Microbiol 67(4):1517–1521
Marzan LW, Hossain M, Mina SA, Akter Y, Chowdhury AMMA (2017) Isolation and biochemical characterization of heavy-metal resistant bacteria from tannery effluent in Chittagong city, Bangladesh: bioremediation viewpoint. Egypt J Aquat Res 43:65–74
Matos D, Sa C, Cardoso P, Pires A, Rocha SM et al (2019) The role of volatiles in Rhizobium tolerance to cadmium: effects of aldehydes and alcohols on growth and biochemical endpoints. Ecotoxicol Environ Saf 186:109759
McEldowney S (2000) The impact of surface attachment on cadmium accumulation by Pseudomonas fluorescens H2. FEMS Microbiol Ecol 33(2):121–128
Mills AL, Colwell RR (1977) Microbiological effects of metal ions in Chesapeake Bay water and sediment. Bull Environ Contam Toxicol 18:99–103
Mirzaei N, Rastegari H, Kargar M (2013) Antibiotic resistance pattern among gram- negative mercury resistant bacteria isolated from contaminated environments, jundishapur. J Microbiol 6(10):e8085
Ndeddy ARJ, Babalola OO (2016) Effect of bacterial inoculation of strains of Pseudomonas aeruginosa, alcaligenes faecalis and Bacillus subtilis on germination, growth and heavy metal (cd, Cr, and Ni) uptake of Brassica juncea. Int J Phytorem 18(2):200–209
Nepple BB, Kessi J, Bachofen R (2000) Chromate reduction by Rhodobacter sphaeroides. J Ind Microbiol Biotechnol 25:198–203
Nies DH (1999) Microbial heavy-metal resistance. Appl Microbiol Biotechnol 51(6):730–750. https://doi.org/10.1007/s002530051457
Outten FW, Outten CE, Hale J, O’Halloran TV (2000) Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, CueR. J Biol Chem 275:31024–31029
Outten FW, Huffman DL, Hale JA, O’Halloran TV (2001) The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem 276:30670–30677
Pal A, Dutta S, Mukherjee PK, Paul AK (2005) Occurrence of heavy metal-resistance in microflora from serpentine soil of Andaman. J Basic Microbiol 45(3):207–218
Parkhill J, Brown NL (1990) Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501, the 19bp spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res 18:5157–5162
Pérez-Valdespino A, Celestino-Mancera M, Villegas-Rodríguez VL, Curiel-Quesada E (2013) Characterization of mercury-resistant clinical Aeromonas species. Brazil J Microbiol 44(4):1279–1283. https://doi.org/10.1590/S1517-83822013000400036
Prajakta BM, Suvarna PP, Singh RP, Rai AR (2019) Potential biocontrol and superlative plant growth promoting activity of indigenous Bacillus mojavensis PB-35(R11) of soybean (Glycine max) rhizosphere. SN Appl Sci 1:1143. https://doi.org/10.1007/s42452-019-1149-1
Prince RW, Cox CD, Vasil ML (1993) Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J Bacteriol 175:2589–2598
Quintelas C, da Silva VB, Silva B, Figueiredo H, Tavares T (2011) Optimization of production of extracellular polymeric substances by Arthrobacter viscosus and their interaction with a 13X zeolite for the biosorption of Cr(VI). Environ Technol 32:1541–1549
Rajapaksha RMCP, Tobor-Kapłon MA, Bååth E (2004) Metal toxicity affects fungal and bacterial activities in soil differently. Appl Environ Microbiol 70(5):2966–2973
Ray S, Gachhui R, Pahan K, Chaudhury J, Mandal A (1989) Detoxification of mercury and organomercurials by nitrogen-fixing soil bacteria. J Biosci 14(2):173–182
Rensing C, Fan B, Sharma R, Mitra B, Rosen BP (2000) CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. PNAS U S A 97:652–656
Ridge PG, Zhang Y, Gladyshev VN (2008) Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS One 3:e1378. https://doi.org/10.1371/journal.pone.0001378
Robin RS, Muduli PR, Vardhan KV, Ganguly D, Abhilash KR et al (2012) Heavy metal contamination and risk assessment in the marine environment of Arabian Sea, along the southwest coast of India. A J Chem 2(4):191–208. https://doi.org/10.5923/j.chemistry.20120204.03
Romanenko VI, Korenkov VN (1977) A pure culture of bacterial cells assimilating chromates and bichromates as hydrogen acceptors when grown under anaerobic conditions. Mikrobiolo 46:414–417
Rosen BP, Liu Z (2009) Transport pathways for arsenic and selenium: a minireview. Environ Int 35:512–515
Różańska A, Chmielarczyk A, Romaniszyn D, Majka G, Bulanda M (2018) Antimicrobial effect of copper alloys on Acinetobacter species isolated from infections and hospital environment. Antimicrob Resist Infect Control 7:10. https://doi.org/10.1186/s13756-018-0300-x
Sadler WR, Trudinger PA, Mineral D (1967) The inhibition of microorganisms by heavy metals. 2(3):158–168. https://doi.org/10.1007/BF00201912
Samanovic MI, Ding C, Thiele DJ, Darwin KH (2012) Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe 11(2):106–115
Saranraj P, Stella D, Reetha D, Mythili K (2010) Bioadsorption of chromium resistant Enterococcus casseliflavus isolated from tannery effluents. J Ecobiotechnol 2(7):17–22
Sayel H, Bahafid W, Joutey NT, Derraz K, Benbrahim KF (2012) Cr (VI) reduction by Enterococcus gallinarum isolated from tannery waste-contaminated soil. Ann Microbiol 62(3):1269–1277
Shakoori AR, Muneer B (2002) Copper-resistant bacteria from industrial effluents and their role in remediation of heavy metals in wastewater. Folia Microbiologica (Praha) 47:43–50
Siddiquee S, Rovina K, Azad SA (2015) Heavy metal contaminants removal from wastewater using the potential filamentous fungi biomass: a review. J Micro Biochem Technol 07(06):384–393
Silva AADL, Agostinho ARDC, Márcia ALDS, Sérgio ALTD, Patrícia MDSF et al (2012) Heavy metal tolerance (Cr, Ag and Hg) in bacteria isolated from sewage. Braz J Microbiol 43(4):1620–1631
Singh SK, Grass G, Rensing C, Montfort WR (2004) Cuprous oxidase activity of CueO from Escherichia coli. J Bacteriol 186:7815–7817
Singh RP, Singh RN, Srivastava AK, Kumar S, Dubey RC et al (2011) Structural analysis and 3D-modelling of fur protein from Bradyrhizobium japonicum. J Appl Sci Environ Sani 6(3):357–366
Singh RP, Manchanda G, Singh RN, Srivastava AK, Dubey RC (2016) Selection of alkalotolerant and symbiotically efficient chickpea nodulating rhizobia from North-West Indo Gangetic Plains. J Basic Microbiol 56:14–25. https://doi.org/10.1002/jobm.201500267
Singh RP, Manchanda G, Li ZF, Rai AR (2017) Insight of proteomics and genomics in environmental bioremediation. In: Bhakta JN (ed) Handbook of research on inventive bioremediation techniques. IGI Global, Hershey. https://doi.org/10.4018/978-1-5225-2325-3
Singh RP, Manchanda G, Maurya IK, Maheshwari NK, Tiwari PK et al (2019) Streptomyces from rotten wheat straw endowed the high plant growth potential traits and agro-active compounds. Biocatal Agric Biotechnol 17:507–513. https://doi.org/10.1016/j.bcab.2019.01.014
Subhashini DV, Singh RP, Manchanda G (2017) OMICS approaches: tools to unravel microbial systems. Directorate of Knowledge Management in Agriculture, Indian Council of Agricultural Research. ISBN: 9788171641703. https://books.google.co.in/books?id=vSaLtAEACAAJ
Suzuki K, Wakao N, Kimura T, Sakka K, Ohmiya K (1998) Expression and regulation ofthe arsenic resistance operon of Acidiphilium multivorum AIU 301 plasmid pKW301 in Escherichia coli. Appl Environ Microbiol 64:411–418
Taniguchi J, Hemmi H, Tanahashi K, Amano N, Nakayama T et al (2000) Zinc biosorption by a zinc-resistant bacterium, Brevibacterium sp. strain HZM-1. Appl Microbiol Biotechnol 54(4):581–588
Teitzel GM, Parsek MR (2003) Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl Environ Microbiol 69(4):2313–2320
Thacker U, Madamwar D (2005) Reduction of toxic chromium and partial localization of chromium reductase activity in bacterial isolate DM1. W J Microbiol Biotechnol 21:891–899
Tipayno SC, Truu J, Samaddar S, Truu M, Preem JK (2018) The bacterial community structure and functional profile in the heavy metal contaminated paddy soils, surrounding a nonferrous smelter in South Korea. Ecol Evol 8:6157–6168. https://doi.org/10.1002/ece3.4170
Tornabene TG, Edwards HW (1972) Microbial uptake of lead. Science 176:1334–1335
Tornabene TG, Peterson SL (1975) Interaction of lead and bacterial lipids. Appl Microbiol 29:680–684
Trajanovska S, Britz ML, Bhave M (1997) Detection of heavy metal ion resistance genes in gram-positive and gram-negative bacteria isolated from a lead-contaminated site. Biodegradation 8:113–124
Tremaroli V, Workentine ML, Weljie AM, Vogel HJ, Ceri H et al (2009) Metabolomic investigation of the bacterial response to a metal challenge. Appl Environ Microbiol 75:719–728
Tsolis RM, Bäumler AJ, Heffron F (1995) Role of Salmonella typhimurium Mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect Immun 63(5):1739–1744
Turpeinen R, Kairesalo T, Haggblom M (2002) Microbial activity community structure in arsenic, chromium and copper contaminated soils. J Environ Microbiol 35(6):998–1002
Turpeinen R, Kairesalo T, Haggblom M (2004) Microbial community structure and activity in arsenic, chromium and copper contaminated soils. FEMS Microbiol Ecol 47:39–50
US Department of Health and Human Services, Public Health Service (1999) Toxicological profile for mercury. US Department of Health and Human Services, Atlanta, pp 1–600
Vergara-Figueroa J, Alejandro-Martín S, Pesenti H, Cerda F, Fernández-Pérez A et al (2019) Obtaining nanoparticles of Chilean natural zeolite and its ion exchange with copper salt (Cu2+) for antibacterial applications. Material 12(13):E2202. https://doi.org/10.3390/ma12132202
Vinodini N, Chatterjee PK, Chatterjee P, Chakraborti S, Nayanatara A et al (2015) Protective role of aqueous leaf extract of Moringa oleifera on blood parameters in cadmium exposed adult wistar albino rats. Int J Curr Res Acad Rev 3:192–199
Viti C (2006) Response of microbial communities to different doses of chromate in soil microcosms. J Appl Soil Ecol 34:125–139
von Bodman SB, Bauer WD, Coplin DL (2003) Quorum sensing in plant-pathogenic bacteria. Annu Rev Phytopathol 41:455–482
Waldron KJ, Robinson NJ (2009) How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol 7:25–35
Wang T, Si M, Song Y, Zhu W, Gao F et al (2015) Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog 11:e1005020. https://doi.org/10.1371/journal.ppat.1005020
Wintz HT, Fox VC (2002) Functional genomics and gene regulation in biometals research. Biochem Soc Trans 30:766–768
Xie Y, Fan J, Zhu W, Amombo E, Lou Y et al (2016) Effect of heavy metals pollution on soil microbial diversity and bermudagrass genetic variation. Front Plant Sci 7:755. https://doi.org/10.3389/fpls.2016.00755
Xu Q, Shi G (2000) The toxic effects of single Cd and interaction of Cd with Zn on some physiological index of [Oenanthe javanica (Blume) DC]. J Nanjing Normal Uni 23(4):97–100
Yang HC, Fu HL, Lin YF, Rosen BP (2012) Pathways of arsenic uptake and efflux. Curr Top Membr 69:325–358
Yang YJ, Singh RP, Lan X, Zhang CS, Sheng DH et al (2019) Synergistic effect of Pseudomonas putida II-2 and Achromobacter sp. QC36 for the effective biodegradation of the herbicide quinclorac. Ecotoxicol Environ Saf. https://doi.org/10.1016/j.ecoenv.2019.109826
Zhang XX, Rainey PB (2008) Regulation of copper homeostasis in Pseudomonas fluorescens SBW25. Environ Microbiol 10:3284–3294
Zhou Y, Xu YB, Xu JX, Zhang XH, Xu SH et al (2015) Combined toxic effects of heavy metals and antibiotics on a Pseudomonas fluorescens strain ZY2 isolated from swine wastewater. Int J Mol Sci 16:2839–2850
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Singh, R.P., Anwar, M.N., Singh, D., Bahuguna, V., Manchanda, G., Yang, Y. (2020). Deciphering the Key Factors for Heavy Metal Resistance in Gram-Negative Bacteria. In: Singh, R., Manchanda, G., Maurya, I., Wei, Y. (eds) Microbial Versatility in Varied Environments. Springer, Singapore. https://doi.org/10.1007/978-981-15-3028-9_7
Download citation
DOI: https://doi.org/10.1007/978-981-15-3028-9_7
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-3027-2
Online ISBN: 978-981-15-3028-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)