Abstract
Pseudomonas aeruginosa is a prevalent pathogenic species reported with severe nosocomial infections. P. aeruginosa infections gain significant concern due to the greater morbidity and mortality coupled with the development of multidrug resistance phenomenon. Furthermore, they are potentially participating in the development of infections to different organs of the human and possibly evade from the host immune defence mechanism. P. aeruginosa involves in the productions several virulence factors which makes the bacterium remains as potential life-threatening pathogen. Additionally, biofilm formation behaviours of P. aeruginosa offer windows to evade different adverse environmental conditions and initiation of adaptation process according to the stress condition and deliver highly resistance strain. The current antibiotics used to combat the Pseudomonas infection are gradually losing their efficacy in controlling the bacterial infections due to the development of the resistance mechanism within the bacterial system. The different resistance mechanisms including intrinsic, acquired and adaptive resistance are supporting the P. aeruginosa for the development of high resistance against several antimicrobial agents. Recently, several alternative therapeutic options were reported such as interference in quorum sensing of P. aeruginosa, phage therapy and development of antimicrobial peptides alternative therapeutic options against antibiotics with limited incidence of resistance development. This chapter overviews the pathogenesis and drug resistance development in P. aeruginosa and also covers the recently available therapeutic alternatives to combat Pseudomonas infection.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
13.1 Introduction
The genus Pseudomonas belongs to the Pseudomonadaceae family, which is composed of a set of Gram-negative rods and flagellated bacteria. Pseudomonas species are ubiquitous in nature inhabiting different habitats including air, soil, and water ecosystems (Streeter and Katouli 2016). These habituation behaviours of Pseudomonas species are due to their versatile metabolic pathways, which subsequently lead to the development of adaptive behaviours against diverse ecological niches. They are capable in utilizing different substrates as carbon source for the synthesis of energy. Moreover, they could subsist in minimum nutrient condition, grow at temperature up to 42 °C and able to respire under anaerobic condition by employing nitrogen as terminal electron acceptor (Kung et al. 2010). They are widely regarded as opportunistic pathogens due to their ability to infect plants, animals and humans. These Pseudomonas strains are capable of producing several pigments including pyoverdine and pyocyanin, which help other way to detect their growth on the agar plates (Gellatly and Hancock 2013). Among the Pseudomonas strains, P. aeruginosa was found to be frequently associated strain in infections associated with humans. However, it is widely observed as opportunistic pathogen, which occasionally infects the normal hosts but capable to develop severe infections in immunocompromised individuals (Klockgether and Tümmler 2017). Furthermore, P. aeruginosa could able to cause wide spectrum of infections when the host fails to maintain its physiological condition including epithelial barriers, neutrophil production and mucociliary layer. Occasionally, even the contaminated medical devices are remains as a reason for the P. aeruginosa infections in humans (Salter 2015). The genome of P. aeruginosa was extensively characterized; initially it was investigated on PAO1 strain which was isolated from the wound infection. It was reported that P. aeruginosa holding a genome size approximately 6.3 Mb and holding 5570 predicted open reading frames (ORFs), which is comparatively larger when compared to other bacterial species (Silby et al. 2011). Due to the excess genome size of P. aeruginosa, they could harbour different genetic materials required for several biological processes including metabolism and efflux of different organic matters as well as putative chemotaxis system (Battle et al. 2009). These remarkable properties of P. aeruginosa facilitate the adaptive mechanism to the versatile environmental conditions. However, some of other bacterial genomes are also reported with larger genome size which occurred due to the duplication of genes unlike the genome of P. aeruginosa where it is composed of greater genetic and functional diversity (Lee et al. 2006). Interestingly, different isolates of P. aeruginosa, i.e. the different strains of P. aeruginosa isolated from different environmental conditions showed greater similarity in their genomes. For example, when the DNA of P. aeruginosa isolated from the soil sample was hybridized with the PAO1 microarray, the result showed the soil isolated shared 89 to 98% of genome similarity with the pathogenic strain (Wolfgang et al. 2003). These results indicated that P. aeruginosa possesses a highly conserved core set of the genetic material responsible for most of the virulence traits. The comparative study between the environmental isolates and the clinical isolates showed genome similarity among different genes responsible for the production virulence factors including pyocyanin, a type III system, lipase, protease, and rhamnolipid (Wiehlmann et al. 2007).
P. aeruginosa causes several infections in humans, including pneumonia that may lead to death when it was encountered by immune-compromised or cystic fibrosis (CF) individuals. P. aeruginosa is capable of producing both acute and chronic diseases, a person with CF arises life-long chronic P. aeruginosa disease that results in eventual death of the individual (Lorè et al. 2012). The disease CF is a genetic disorder where an individual is born with structurally normal lungs, but promotes slow progression of pulmonary malfunctioning with continuous susceptibility to chronic infections. These conditions provoke the development of bronchiectasis resulting in complete failure of the respiratory system, which subsequently leads to death (Bhagirath et al. 2016). An individual with CF is highly susceptible to different pathogenic microorganisms including P. aeruginosa, Staphylococcus aureus, Burkholderia cepacia and Haemophilus influenzae (da Silva Filho et al. 2013). The pulmonary infections in the individual with CF would be developing from their early stage of disease. The overall incidence states that pathogenic microorganisms P. aeruginosa and S. aureus are the common pathogens which are routinely isolated from the patients with CF. The epidemiological investigations evidencing that the rout of entry and transmission pathogen mainly by a direct contact in some cases it is occurring even through contaminated environment (Taccetti et al. 2008). Nevertheless, P. aeruginosa causes several other diseases in humans in which almost all the diseases are developed by different infectious stages including attachment, colonization, local invasion and dissemination to establish systemic infections. This bacterium is also capable of causing infections to major tissues of humans including dermal, ocular, cardiac, respiratory epithelial and transitional epithelial tissues of urinary tract (Kariminik et al. 2017; Azam and Khan 2019). The pathogenesis pattern of the P. aeruginosa are shown as multifactorial which occurs as results of expression of several genes and attacking multiple target on the host that makes P. aeruginosa infection remains as an potential threat in worldwide (Valentini et al. 2018). Although some of the virulence factors of the P. aeruginosa are known for a decade, some of the other significant virulence traits are recently identified owing to the emergence of whole-genome sequencing technology. As discussed earlier both the clinical and the environmental isolates of P. aeruginosa hold the genetic information for most of the virulence factors. However, the pathogenicity and type of infection are possibly altered by the host environment and state of its immune system, as a matter of fact that P. aeruginosa is highly capable to adapt to different lifestyles of the host (Huang et al. 2011).
Several therapeutic options are available and were practised to control the P. aeruginosa infection. However, still these infections remain a major threat leading to greater mortality due to the emergence of the multidrug-resistant strains. There have been several reports elucidating the risk factors associated with the emergence of multidrug-resistant strains. For instance, multidrug-resistant tuberculosis is found to be a serious concern worldwide. World Health Organization (WHO) and government global nations combined programme formulated several measures to control the disease associated with multidrug-resistant Mycobacterium tuberculosis. However, the success rate of this programme found to be very low as compared to the target aimed by the WHO 2015, 75–90% (Kibret et al. 2017). Unsurprisingly, some of the P. aeruginosa strains also showed resistance against third-generation antibiotics including carbapenem and cephalosporins suggesting for the treatment of multidrug-resistant phenomenon (Azam and Khan 2019). Furthermore, the recent antimicrobial agents prescribed for the treatment of pulmonary infections were result in disordering the normal function of the lungs, failure of respiratory system and premature death of the individual with CF. In case of individuals with P. aeruginosa infection, the quality and the life span are directly dependent on the success rate of the treatment process in clearing the initial infection (Stefani et al. 2017). The pathogens reside on the side or organ of infection are showing considerable variation, owing to intensive antimicrobial agent pressure and modification in the antimicrobial agent regimens. Due to these unfavourable conditions on the site of infection, the pathogens are pressured to acquired resistance genes or pretend as new pathogens (Pena et al. 2015; Nguyen et al. 2018). P. aeruginosa develops resistance to antimicrobial agents by altering several biological behaviours including reducing the membrane permeability, activation of efflux systems, synthesis of the enzyme which could able to degrade the antimicrobial agent and potentially modify the structure of the target (Bassetti et al. 2018). Interestingly, P. aeruginosa could develop resistance chromosomally (intrinsic resistance) as well as acquiring the resistance genes from the neighbouring microbes in the environment (Wagner et al. 2016). Till date, the reports on resistance development in P. aeruginosa to at least three antimicrobial agents from different classes of antibiotics including aminoglycosides, antipseudomonal penicillins, cephalosporins, carbapenems and fluoroquinolones (Hirsch and Tam 2010; Chatterjee et al. 2016).
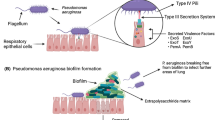
The chronic infections of P. aeruginosa found to cause serious clinical complications due to its inherent ability to form a biofilm. Biofilm is an integrated community of sessile microorganisms protected within self-producing polymeric substance, provides homeostasis and stability during the progress of infection in host. This condition supports and promotes the resistance development by structurally limiting the permeability of the antimicrobial agents and facilitates the adaptive resistance mechanism in P. aeruginosa. It is extremely difficult to eradicate the biofilm state of P. aeruginosa by routine antimicrobial agents as biofilms are highly tolerant to most of the available antimicrobial agents as compared to its planktonic counterparts (Furiga et al. 2016). For example, the notorious biofilm-forming Gram-negative pathogen, P. aeruginosa has the inherent ability to develop both intrinsic and acquired drug resistance, as discussed earlier. It was evaluated that the biofilm and stationary phase of the planktonic cultures were exposed to the sublethal concentration of known antibiotics, ciprofloxacin and observed for the development of mutational resistance. It was reported that the bacteria in the biofilm are less probable to develop the mutational resistance as compared to the planktonic cultures, since biofilm residing bacteria are poorly exposed the antimicrobial agent (Ahmed et al. 2018).
In matured biofilm, bacteria accounts only for 10% while the rest 90% is composed of self-producing extracellular polymeric substances (EPS) matrix containing polysaccharides, proteins, extracellular DNA and lipids. The development of biofilm and stability of the biofilm is generally governed by different intrinsic and extrinsic factors such as genetic makeup of the isolate, hydrodynamic conditions, motility behaviours of pathogenic bacteria, nutrient availability, intracellular communication and the health of host immune system (Maurice et al. 2018). In healthcare sectors, biofilm infections remain highly problematic, since it could occur in host tissues or the surface of the host and medical instrumentals including joint and organ replacement, catheters and ventilators (Taylor et al. 2014). Indeed biofilm of P. aeruginosa being a sensation among the scientific community and potential efforts have been initiated to eradicate them. Three major strategies have been proposed as primary methods to eradicate biofilms including (1) controlling the microbial attachment to the surface, (2) interrupting the biofilm formation and architecture which enhance the internalization of antimicrobial agents and (3) Disruption of mature biofilm (Rasamiravaka et al. 2015; Rossi Gonçalves et al. 2017). There is an urgent call for the development of desired antimicrobial strategies in worldwide scientific community. This chapter presents key features of multidrug-resistant P. aeruginosa, biofilm formation, associated infection and current and future options which could help in eradicating the infection by multidrug-resistant P. aeruginosa strains.
13.2 Pseudomonas aeruginosa and Associated Infections
As a matter of fact, P. aeruginosa is a very common bacterium, which is frequently isolated from the human with a disease. Although it acts as an opportunistic pathogen, it has the potential enough to cause infection to any part of human organs or tissues. However, the incidence of disease progression is very common among the individual’s underlying disease, age or impaired immune system. In recent years, the incidence of healthcare-associated infection by P. aeruginosa was increased worldwide which subsequently evolved as a major threat to the patients as well as the economy (Wu et al. 2019). The hospitalized patients are frequently encountered by the pathogenic microorganisms, since the environmental setup of the healthcare unit potentially contaminated by widespread pathogens. Although standard procedures are followed by the healthcare sectors to maintain the sterility, most of the pathogens have managed to persist. Inpatients are repeatedly infected with some pathogens through direct or indirect contact with the environmental surfaces of healthcare units which were contaminated by infected roommate or prior room occupant (Kohlenberg et al. 2010). It is interesting that P. aeruginosa resides in the environment of healthcare units are capable of thriving in the adverse condition with wide range of the temperature, relatively nutrient-limited conditions and continuous exposure to disinfectants. Especially, water system in the healthcare unit has a high tendency to be contaminated with P. aeruginosa infection and facilitates the successful establishment of biofilm that subsequently remains as a major reason for the transmission of the pathogen (Garvey et al. 2016). Inpatients are at high risk of acquiring different kinds of infections such as pneumonia, burn wound infection, acute and chronic pulmonary infection (Kerr and Snelling 2009).
13.2.1 Burn Wound Infection
Burn wounds could be specified as skin abrasion mediated by heat exposure, electrical shock, exposure to chemicals and radiations. Burn wounds could be categorized from moderate to severe based on their degree of damage on the skin. The incidence of burn wound infections (moderate to severe burns) have been increased every year worldwide and are very common in developing countries (Mofazzal Jahromi et al. 2018). As compared to earlier strategies, the success and the survival rate of the burn individual in the present scenario have been increased due to the advanced development in burn wound care units. As a result, significant percentage of the burn-related deaths were reduced, still complete eradication of burn-related death remains a dream due to the infection caused by the pathogenic microorganisms.
As per the recent trends in burn wound infections, more than 75% of the mortality is caused by microbial infections followed by burn shock and hypovolemia (Wang et al. 2018). P. aeruginosa and S. aureus are the most prevalent pathogens causing serious infections to burn wound patients and potentially reduced the success rate of burn wound treatment process (Gonzalez et al. 2016). The incidence between the burn wound individual and the pathogen could happen either by acquiring from the normal flora of the host or hospital environment. The drug-resistant strains make the condition even more worse and subsequently altering the therapeutic process. For instance, the recovering or less critical burn wounded individuals are also at high risk of acquiring chronic infections which makes the situation more complicated. This indicates the complication and the threat integrated with the prevention of burn wound infections inside the burn unit (Simões et al. 2018). Moreover, the antibiotic susceptibility pattern of normal microflora colonized in the burn wound is possibly changing their pattern as antibiotic resistance during the period of treatment process. Recently, serious investigations are executed in emergency basis for the development of potential drug candidates and therapeutic medication to address the problems associated with the burn wound infections. The following burn wound car unit protocol could possibly control the severity of the burn wound infection. These include surveillance on microbiota on the wound in needed basis, strict enforcement of patient and staff hygiene, isolation of infected individual and continuous monitoring of administration of antibiotics as well as gaining complete susceptibility profile against wound microbiota (Rafla and Tredget 2011).
13.2.2 Hospital- and Ventilator-Associated Pneumonia
Pneumonia is a respiratory disease characterized by inflammation of lungs with congestion caused by the most common nosocomial pathogen, P. aeruginosa. Pneumonia gains considerable attention in healthcare sectors where this disease is diagnosing repeatedly with hospitalized individuals whom have underlying medical condition or highly susceptible for P. aeruginosa infections (Fujii et al. 2014). Nosocomial pneumonia could be further classified based on the location and conditions of infection onset. If the development of disease observed and clinical manifestation results with inflammation in lung parenchymal after at least 48 h of hospitalization, it is termed as hospital-associated pneumonia (HAP) (Niederman 2010). Similarly, in ventilator-acquired pneumonia (VAP) where the pathogens are sourced from the ventilators. Ventilators are the mechanical device that was designed to breathe for patients where ventilator tube was inserted into the human through windpipe and facilitate the patient to breathe.
As discussed earlier, P. aeruginosa is the most common pathogen forming microcolonies on the medical device and infect the airway of an individual acquiring mechanical breath using ventilators. Ventilators acquiring pneumonia creates sever effect which leads to the death of the patient (Aarts et al. 2003). Despite the considerable development in antibiotics and its associated therapeutic practises, HAP and VAP remain a potential threat to the global healthcare sectors (Wilke et al. 2011; Bouglé et al. 2017). Diagnosing these HPA and VAP is possible by observing the clinical manifestation including fever, gradual increase in white blood cell count and microbiological procedures for pulmonary infections. The microbiological procedure confirms disease condition by bacterial loads in the samples including brush specimens, bronchoalveolar lavage fluid and endotracheal aspirate specimen. The recently multidrug-resistant strains of P. aeruginosa are prevalence in causing both HAP and VAP and remain reason for mortality associated with these diseases (Tumbarello et al. 2013; Micek et al. 2015).
13.2.3 Pseudomonas aeruginosa Infections in Cystic Fibrosis
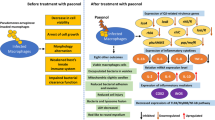
Cystic fibrosis is caused due to the occurrence of mutation of cystic fibrosis transmembrane conductance regulator (CFTR) gene residing on chromosome VII and a cAMP-dependent chloride channel. These mutations resulted in the dehydrated and thickened airway surface liquid that hinders mucociliary clearance from the airway (Gellatly and Hancock 2013). The initial incidence of the pathogen in the modified airway surface liquid allows the establishment of an acute infection resulting in significant enhancement of inflammatory responses. This condition potentially impairs the immune system which consequently impairs the host to control the inflammation and results in chronic lung inflammation (Alhazmi 2015). Although P. aeruginosa not a causative agent for the CF since it being an autosomal receive genetic disorder but Pseudomonas plays an major role in increases of mortality rate in persons with CF. P. aeruginosa is capable to overcome from the treatment for acute infection and can able to adapt to the lung environment and potential establishing the biofilm resulting in chronic infection (Markou and Apidianakis 2014). Interestingly, P. aeruginosa strains isolated from the acute phase of lung infection are not similar to that of the strain observed in the chronic lung infection (Fig. 13.1). Herein, most of the virulent phenotypes observed in the isolates of acute lung infections are absent in their counterpart isolate of chronic infections (Sousa and Pereira 2014). For example, the inflammatory response stimulating virulence factors of P. aeruginosa including flagella and pili are not expressed in the isolates from chronic infection. Similarly, several other virulence factors are downregulated including type III secretion system. On the other hand, P. aeruginosa from chronic lung infections prevalently overexpresses the genes that take part in the successful establishment of biofilm, consequently production of the exopolysaccharide and alginate were drastically increased and the colonies become mucoid (Gellatly and Hancock 2013). The individual with CF disorder has a high tendency to be infected with chronic pseudomonal lung infections. Hyperactive inflammatory response remains a clinical manifestation of CF was potentially stimulated to worse by the toxins from the pathogens and result in gradual deterioration of lung function and subsequently leads to death of an individual (Furiga et al. 2016).
13.3 Pseudomonas aeruginosa Pathogenesis and Major Virulence Factors
Though the intensive advancement in the antimicrobial therapies addressed the answers to numerous deadly infections and diseased conditions, bacterial infections remain an uphill challenge for the scientific community which endorsed the ever-increasing burden of multidrug-resistant strains deteriorate the efficacy of the routine antibiotics. This brings a situation where the severely infected individuals continuously failed to respond to the appropriate antibiotic even when infected with susceptible microbial pathogens (Hauser 2011). To address upon the present problem, efforts are being made to gain appropriate knowledge about the bacterial pathogenesis and their adaptive mechanism toward antimicrobial agents. What follows is a maximum knowledge about known virulence determines suspected of contributing to P. aeruginosa pathogenesis (Fig. 13.2).
13.3.1 Lipopolysaccharide
Lipopolysaccharide (LPS) is widely accepted as a crucial virulence trait observed in P. aeruginosa. It is a complex glycolipid which is the major constituent of outer leaflet in the outer membrane of Gram-negative bacteria. The LPS plays a significant role in bacterial pathogenesis process by protecting the pathogen from the host defense mechanisms. Furthermore, it displays several other roles in bacterial pathogenicity including antigenicity, the inflammatory response, exclusion of external molecules and obstructing the interaction with antimicrobial agents (Gellatly and Hancock 2013). Since LPS gains biological importance in P. aeruginosa pathogenesis, intense study has been conducted to obtain vast knowledge about its biosynthesis pathways and other virulence impact of LPS. Enhancing the knowledge about the structure, function and participant genes of P. aeruginosa in the synthesis process of LPS could potentially increase the global understanding of the pathogenic profile of P. aeruginosa (Goldberg and Pier 1996). The LPS of P. aeruginosa constitutes three domains including lipid A which holds the disaccharide backbone that support the attachment of large amount of fatty acid chain subsequently facilitate a ground to the LPS to stable at the outer membrane. The structural difference of the lipid A in their number, position and properties of connected acyl groups is based on isolated and environmental conditions (Pier 2007). This lipid A glycolipid participates in the activation of host signalling pathways including TLR4 to NFκB by binding to coreceptors such as MD2 and CD14. Activation these signalling pathways leads to the synthesis of pro-inflammatory cytokines, chemokines, inflammation and subsequently induce endotoxic shock.
The variation that occurs in the lipid A could potentially alter the susceptibility nature of P. aeruginosa to different antimicrobial agents and also modify the inflammatory behaviours. For instance, the isolates from the chronically infected CF individual showed significantly enhanced inflammatory response and the extent of these alterations appears to gain high disease severity (King et al. 2009). Nevertheless, these modifications support P. aeruginosa to develop resistance against routinely administered antimicrobial agents. The second domain refers the polysaccharide core region, which is constructed by attaching nine or ten sugar molecules with lipid A and branched with oligosaccharide. The above-mentioned two domains are most common in the LPS molecules on the surface of the bacterial cells, which are also referred as lipid A-core. The third domain refers as O-specific polysaccharide, O antigen or O chain, which shows high variability due to its chemical constituent where it made up of continuous carbohydrate polymer, covalently attached to the core. Interestingly, P. aeruginosa is potential enough to synthesize two different O antigens which distinctly shows a variation in their structure, serology and their biosynthesis pathway in the same cells. Previously, these have been known to be called as A- and B-band O antigens, but recently according to the structural nomenclature and the O-serotyping pattern of P. aeruginosa the B-band O antigen is called as O-Specific Antigen, whereas the A-band O antigen is called as Common Polysaccharide Antigen (PIER 2007). The Common Polysaccharide Antigen production is very common, but not in all the strain of the P. aeruginosa. This antigens are the complex of homopolymer of d-rhamnose, which promote the weak antibody response. On the other hand, O-Specific Antigens are heteropolymer with continuous O unit with various sugar moieties. Unlike Common Polysaccharide Antigen, the O-Specific Antigens are potentially immunogenic and readily enhance the antibody response (Maldonado et al. 2016). The chemical structure, physiological behaviour and the number of sugar moieties in the O-Specific Antigen’s structure could be varied from isolates to isolates and also based on their growth conditions (Werneburg et al. 2012).
13.3.2 Flagella and Type IV Pili
The attachment of bacterial pathogen to the host surface is an initial and critical step involved in the establishment of P. aeruginosa infection. Flagella and type IV pili are two major P. aeruginosa adhesins, which play a significant role in the initial attachment process. Furthermore, these two virulence phenotypes remain the major attributes for the spreading of disease from one organ to the other and also support the successful establishment of P. aeruginosa biofilm (Campodonico et al. 2010; Bucior et al. 2012). Flagella are one among organs of bacteria which is found to be highly complex and conserved even among diverse bacterial species. The synthesis process of flagella was encoded by more than 50 genes. The flagella in pathogen plays several roles in development of infection like maintaining the chemotaxis and mediating the motility. In some pathogen motility remains an important phenotype for survival, because the flagella provide an opportunity for the pathogen to acquire essential nutrients from the environment (Feldman et al. 1998).
Type IV pili are nanoscale protein filament with hair-like appendages decorated on the surface of P. aeruginosa. These type IV pili initiate and support the bacterial attachment process, facilitate the cell–cell aggregation, initiation and maturation of biofilm and providing twitching motility (Beaussart et al. 2014). P. aeruginosa has the inherent potential to exhibit different types of motility including swimming, twitching and swarming. The swimming motility frequently observed in aqueous conditions or in the medium plate with less agar concentration. This swimming motility is mediated by the flagella and hence called as flagellum-mediated motility. Similarly, twitching motility is mediated by type IV pili and mostly observed on the solid or interface medium. The third motility is termed as swarming motility that promotes coordinated and rapid movement of the P. aeruginosa on the semisolid media. Recently, it was reported that swarming motility could be mediated by both flagella and type IV pili and in some cases even the amount of rhamnolipids present in the medium also affects the swarming motility of P. aeruginosa (Overhage et al. 2007). The role of the motility in the pathogenicity of the P. aeruginosa was investigated by several in vivo studies where the mutant strain with defective of both flagella and pili infected to burn wound mouse (Feldman et al. 1998).
13.3.3 Type III Secretion System
In bacteria, a secretion is a biological event in which the macromolecules of bacteria are transferred across the cellular envelope into the surrounding environment. This biological event believed to be more difficult in Gram-negative bacteria where macromolecules tasked to cross two membrane barriers. The evolution in Gram-negative bacteria addressed a path to overcome these complications by facilitating the development of specialized secretion systems. Till date, six secretion systems were reported in Gram-negative bacteria, P. aeruginosa. Type III secretion systems (T3SS) are widespread membrane-embedded nanomachinery that are found in different Gram-negative bacteria and facilitate transfer of toxin molecules also termed as effectors from bacterial cytosol to cytoplasm of the targeted eukaryotic cells (Puhar and Sansonetti 2014). This makes the difference among the secretion system that most of them secrete the effectors into the extracellular surroundings, where they bind to the distant target cell surface receptors. The effect of the toxin distinctly varies where in some cases it supports the symbiotic relationship between the host and the bacteria and in most cases it mediates the pathogenic features including membrane disruption, activation of apoptosis process and structural rearrangements in cytoskeleton (Burkinshaw and Strynadka 2014). However, T3SS apparatus are highly conserved at both structural and functional level among the bacterial species but potentially inject the effectors into the distinct hosts such as humans, plants and animals (Izoré et al. 2011). The apparatus of T3SS consists of three major parts including multiring basal body (made up of more than 25 different proteins and bridges all the layers of the bacteria including inner bacterial membrane), the peptidoglycan layer and the outer bacterial membrane; a hollow needle-like structure in which the bacteria mobilizes effector toxin from basal body in semi-unfolded form, it believed to have both inner and outer membrane component. Herein, the inner membrane established of the lipoprotein PscJ and outer membrane component made of oligomerized secretin PscC; and translocon, pore or the needle that inserted into the host cells and the release the toxin effectors and made up of two T3SS hydrophobic protein that play significant role the host immune system alteration (Anantharajah et al. 2016; Cascales 2017).
Most of the pathogenic strains of P. aeruginosa have the inherent ability to overcome the phagocytic clearance which is dependent upon the behaviour of T3SS (Burstein et al. 2015). Several clinical investigations also supported the statement that T3SS of P. aeruginosa has a crucial role in inducing pathogenicity and in most cases it leads to the death of the patient. Furthermore, in in vitro condition, T3SS mutant strain failed to establish the infection in most of the host unlike its counterpart wild strain (Galle et al. 2012). The T3SS of P. aeruginosa participates in the production and injection four effector toxins such as ExoS, ExoT, ExoY and ExoU. Recently, nucleoside diphosphate kinase was also reported to be transferred from pathogen to host cells via T3SS (Zhu et al. 2016). Although it seems T3SS involved in the production of limited numbers of effector toxins, but these limited effector toxins furnished P. aeruginosa to cause infection among wide range of hosts (Yamazaki et al. 2012). Interestingly, the strain nullified with all the four effector toxin genes is still capable to cause infections indicating the possibility of other effectors beyond these four effectors toxin which is yet be discovered (Galle et al. 2012). The activation of T3SS in P. aeruginosa occurred as responsive mechanism to the environmental signals including the attachment of pathogen on to the host and comparatively low concentration level of calcium in the growth medium or in the growing environment. However, the exact signalling mechanism is yet to be known (Anantharajah et al. 2016).
13.3.4 Type IV Secretion System (T4SS)
Similar to the T3SS, the T4SS also possesses core complex bridging both inner and outer bacterial membranes and pili that extends out into the extracellular surroundings (Depluverez et al. 2016). Based on the function, the T4SS could be classified into three distinct categories. The first category system involves the mobilization of single-stranded DNA into the host cells by conjugation process which potentially facilitates the pathogen to adapt to the host environmental condition, without surprising mediates the development of resistance mechanisms (Juhas et al. 2008). The first category T4SS system is activated only when the pathogen is in contact with the host. Similarly, second category system functions as transport to several protein molecules directly into the host cells, thus system involves successful establishment of infection by introducing pathogenic secretion into the host tissues from the pathogen (Christie et al. 2014). The third category of T4SS influences the adaptation of DNA from the external environment. It also involves in releasing both DNA and protein molecules to the exterior surface of bacteria. The ability to transfer both DNA and protein to the host cells make the T4SS system unique among the other secretion system (Trokter et al. 2014).
P. aeruginosa is one among the Gram-negative bacteria which employs T4SS for horizontal gene transfer that help to create a pathogenic island which subsequently plays a major role in its pathogenicity (Ma et al. 2003). In P. aeruginosa, pKLC and PAPI well-known pathogenicity islands that use T4SS for the spreading of pathogenic gene in Pseudomonas. The pathogenicity island, pKLC is constituted by combining phage and plasmid origin, which potentially transfer genetic information with increased excision rate from the chromosome. It being 103,532 bp long island with broad-spectrum open reading frames and genes encoding for type IV sex pili. Furthermore, it holds the gene encoded for chvB, which is found to be a major virulence trait in P. aeruginosa that commonly named as glucan synthetase (Juhas 2015). The other pathogenic island PAPI displayed as a larger island with 108 kb that encodes for several pathogenic traits. For the transfer genetic information from PAPI, it forms extrachromosomal circular intermediate before it is administrated into the recipient cells (Carter et al. 2010).
13.3.5 Proteases
Proteases are the enzymes synthesized by a microorganism which helps the microorganism, especially microbial pathogen in several ways including providing peptide nutrients and shows its effective contribution in the pathogenesis of infectious disease by different mechanism (Lantz 1997). These proteases are potentially capable of causing direct or indirect damage to host by lysing the cell surface and tissue protein or actively participates in inactivating the important proteins for host defence mechanism, respectively (Herwald and Egesten 2009; Musicki et al. 2009). Although microbial proteases are widely accepted as virulence factors due to their significant role in the pathogenesis of infectious diseases, limited reports are still available to confirm that microbial protease specifically participates in the microbial pathogenesis (Ingmer and Brøndsted 2009). Among the pathogenic microorganisms, P. aeruginosa was widely studied for the secretion of proteases and their participation in pathogenicity. In most cases, the ocular infection and sepsis condition by P. aeruginosa was mediated by proteases which potentially involved in the denaturation of immunoglobulins, fibrins and subsequently disrupt the epithelial cells (Alionte et al. 2001). P. aeruginosa is well known for its ability in producing alkaline protease via type I secretion system which readily participates in the degradation process of host complement protein and fibronectin. Nevertheless, they also produced two different elastolytic enzymes such as LasA and LasB. The production of these enzymes is mediated by the quorum sensing mechanisms of the P. aeruginosa and produced via type II secretion system. LasA proteases are serine protein and act on the cell wall of Staphylococci, hence often called as “Staphylolysin”. LasB protease, often called as “elastase” which potentially acts on the lung surfactant proteins A and D (Matsumoto 2004; Gellatly and Hancock 2013). In recent years, different protease inhibitors including α2-macroglobulin and specific elastase inhibitors were reported which control the P. aeruginosa infections and fatal septic shock which are mediated by bacterial proteases enzymes (Hobden 2002).
13.3.6 Exotoxin A
As discussed earlier, P. aeruginosa is capable of producing several extracellular virulence factors. Exotoxin A is one among them which attributes a significant role in the pathogenesis of P. aeruginosa infections (Al-Dahmoshi et al. 2018). P. aeruginosa utilizes a type II secretion system for the production of exotoxin A and the functional characters of this toxin are identical to Diphtheria toxin. The host receives this toxin through a surface receptor called α2-macroglobulin receptor (Morlon-Guyot et al. 2009). Exotoxin A manages to internalize into a cytoplasm via previously mentioned transport mechanism and potentially affects the elongation factor, which subsequently inhibits protein synthesis in the host and suppresses the host immune response. Furthermore, the exotoxin A is also capable of inducing apoptosis process in host which finally leads to the death of the host cells (Pillar and Hobden 2002). This behaviour of exotoxin A allows the research community to conduct investigations to use these exotoxin A as apoptosis-inducing factor on cancer cells. The production of exotoxin A in P. aeruginosa mainly dependent on the availability of iron and the gene regA is directly involves in the transcriptional regulation in the production of exotoxin A. Similarly the genes vfr and lasR are indirectly participating in the transcriptional regulation process coupled with the synthesis of exotoxin in P. aeruginosa (Michalska and Wolf 2015).
13.3.7 Alginate
In P. aeruginosa infection, neutrophils remain an initial line of defence mechanism via phagocytosis. On the other hand, the alginate of P. aeruginosa protects the bacteria from several host defence mechanism including phagocytosis, oxygen radicals and other host immune defence mechanisms as well as from the treatment of antimicrobial agents (Colbert et al. 2018). The importance of clinical complications because of the presence of the alginate was reported that it is capable to create a condition of inefficient pulmonary clearance (Franklin et al. 2011). Moreover, they actively involved in spreading the infection from one organ to another, for instance alginate could able to mediate the spreading of lung infection to spleen (McCaslin et al. 2015). Interestingly, the pathogenic isolates of P. aeruginosa, PAO1 found to be non-mucoid. At the same time, isolates colonize on the host confronts several host immune feedback and exposed to different antimicrobial agents. These adverse conditions make the isolates to alter its transcriptional regulators and convert to a mucoid phenotype by synthesizing alginate. However, in some cases the mucoid phenotype resides in the lung infected area are failed to hold similar phenotype when they culture in laboratory conditions (Limoli et al. 2017).
13.3.8 Quorum Sensing and Biofilm Formations
Quorum sensing (QS) is a communication mechanism lies among the microbial community which facilitates a coordinated adaptation of microbial community to certain environmental condition, including adapting towards infection site within the host. This communication is mediated by a small membrane-diffusible signalling molecule also termed as autoinducers. These signals are coordinately synthesized by the microbial cells which act as cofactors for most of the transcriptional regulators coupled with virulent traits, when the concentration of this signalling molecules reaches its threshold level in the environment (Fila et al. 2018). However, the level of signalling molecules in the surrounding environment directly depends upon the number of microbial cells in the same environment. These signalling molecules coordinated the entire community to deliver a similar response. Furthermore, these communication networks, widely determined as a major cause for the evolution of multidrug resistance (Kalaiarasan et al. 2017).
P. aeruginosa utilizes this microbial communication to coordinate among their community. However, P. aeruginosa is well known for its ability to produce three signalling molecules including 3-oxo-dodecanoyl homoserine lactone (3-oxo-C12 HSL), butyryl homoserine lactone (C4 HSL) and 2-heptyl-3-hydroxy-4-quinolone (PQS). The first signalling molecule 3-oxo-C12 HSL, is synthesized due the activation of LasI AHL synthase and binds with LasR transcriptional receptor protein. Similarly, the second signalling molecule, C4 HSL, is produced by initiation of RhlI AHL synthase and bind with specific transcriptional receptor, RhlR. The synthesis process of the third signalling molecule is mediated by complex multistep biosynthesis pathway (Harmsen et al. 2010). This microbial communication network facilitates the P. aeruginosa several ways including survival of the bacteria, allows development of biofilm and controlling the production of different virulence factors. Several reports are supporting this context where the strain in absence of these signalling mechanisms was failed to produce different virulence determines and establishment of infections (Kariminik et al. 2017; Defoirdt 2018).
Biofilm could be simply defined as a biological gathering where microbial community attached with one another and develop a well-organized complex structure that is possible to happen under governs of quorum sensing system of bacteria (Palanisamy et al. 2014). In biofilms, all the bacterial cells are compactly packed inside the polymeric matrix also termed as extracellular polymeric substance (EPS). These polymeric matrixes contain several other biological substances including polysaccharides, nucleic acid, lipids and proteins (Koh et al. 2013). Among the total weight of biofilm more than 50–90% weight is shared by the polymeric matrix rest of the weights are shared by the bacterial cell in the biofilm. These thick complex matrixes facilitate the biofilm in several ways including protecting from both physical and chemical factors that subsequently affect the biofilm (Bai and Rai 2011). Due to the complex nature of the biofilm, potentially resist the penetration of any chemical from external environment to biofilm which makes most of the routine antibiotics inefficient to eradicate the biofilm. Interestingly, the bacterial cells show very slow growth than planktonic cells due to the establishment of the nutrient-limited environment inside the biofilm (Kalia et al. 2019). However, biofilm residing shows high resistance than its counterpart planktonic cells. These conditions made the P. aeruginosa infections remain as an unsolved problem in human healthcare sectors.
13.4 Antibiotic Resistance Mechanism of Pseudomonas aeruginosa
In recent years, treating the disease caused by P. aeruginosa becomes a potential issue worldwide due to the development of a resistance mechanism against available antibiotics. Based on recent WHO report, P. aeruginosa strain with carbapenem-resistant gene was found to cause most curial infection. Continuous instances of antimicrobial-resistant strains were observed due to the excessive use of antimicrobial agents in the infection treatment process (Chatterjee et al. 2016). P. aeruginosa demonstrates resistance to major classes of antibiotics such as aminoglycosides (e.g. gentamicin), quinolones (e.g. ciprofloxacin), β-lactams (e.g. cephalosporin) and polymyxins (e.g. colistin). The mechanisms underline with the antimicrobial resistance in P. aeruginosa lies with multifactorial processes like synthesis of antibiotic altering enzymes such as β-lactamases, metallo-β-lactamases and other enzymes which effectively alter the aminoglycoside; expression of the efflux pumps which potentially protect the bacterium from the internalization of antimicrobial agent into cytoplasm; capable to gain antibiotic resistance genes (Taylor et al. 2014). Furthermore, self-mutation also contribute to the successful establishment of the resistance mechanism in P. aeruginosa. Nevertheless, development of biofilm by the bacterium creates a micronutrient environment which potentially limits the penetration of the antimicrobial agent into the biofilm structure and direct multiresistance mechanism to cells reside in the biofilm matrix (Pang et al. 2019). The resistance mechanism could be majorly categorized into three different resistance mechanisms including intrinsic, acquiring and adaptive.
13.4.1 Intrinsic Resistance
The intrinsic resistance mechanism toward antimicrobial agents in P. aeruginosa could be defined as innate capability of a bacterium to suppress the activity of the antimicrobial agent by altering its structural and functional motifs. The intrinsic antibiotic resistance is found often in P. aeruginosa against different classes of antibiotics which could be achieved by the bacterium by controlling the permeability nature of an outer membrane, activation of efflux pump system which readily recognize the antibiotics and pumps out from the cell and expressing the genes encodes for the production of antibiotic inactivating enzymes (Fig. 13.3) (Valentini et al. 2018). The condition of lower permeability lies on the outer membrane of the bacterial cells making it difficult for antimicrobial agents to penetrate into the bacterial cytoplasm. Some of structurally small antimicrobial agents possessing the hydrophilic potential and readily dissolve in water molecules and pass thorough the water-filled porin channels (Breidenstein et al. 2011). The process involved in this mode of penetration of the antimicrobial agents into bacterial cells found relatively slow which subsequently facilitate the option to develop more intrinsic resistance to different antimicrobial agents. Although the outer membrane plays a significant role in controlling the antibiotic entry, this mechanism alone is not sufficient to control the entry of the antibiotics. The other intrinsic resistance mechanism, efflux system provides a promising contribution that resists the internalization of the antimicrobial agents into the cells (Oliver et al. 2015). The synergizes mode of both discussed intrinsic mechanisms proceeding to the development of high level of the resistance to currently available antimicrobial agents relevant for P. aeruginosa.
13.4.2 Acquired Resistance
The major difference between the intrinsic and acquired resistance is the prior exposure to the antimicrobial agent. Wherein the intrinsic resistance it is not necessary that bacterium should previously exposed to antimicrobial agents, but in case of acquired resistance, it could be initiated once the bacterium experienced the antimicrobial agents (Friedman et al. 2016). In acquired resistance mechanism, once the bacteria experience the antimicrobial agent it undergoes several chromosomal gene modifications and develops resistance against same antimicrobial agent. In other way bacteria also undertake the acquired resistance via acquiring the genetic resistance information from different molecular elements including plasmids, transposons, interposons and integrons (Satpathy et al. 2016). The major mechanism of horizontal gene transfer necessitates transformation, transduction and conjugation. These genetic alterations on the bacterial system could potentially control the uptake of antimicrobial agents and changing the target of the antibiotics (Macia et al. 2014). Nevertheless, the gene modification also facilitates the functional behaviours of intrinsic resistance such as expression of efflux pump and production of antibiotic inactivating enzymes.
13.4.3 Adaptive Resistance
Adaptive resistance mechanism supports P. aeruginosa to overcome and continue to survive even after antibiotic treatment by altering the genetic information of the protein synthesis according to the environmental signals. Adaptive mechanism of the P. aeruginosa gains significant consideration in the clinical sectors due to its notable role in evading from the antimicrobial treatment process (Yelin and Kishony 2018). P. aeruginosa adaptive resistance was widely studied in which biofilm plays a significant role in creating persistent cells that subsequently case the persistent infection and result in poor prediction of the course of a disease. Unlike the other resistance mechanisms, the adaptive resistance mechanism is completely dependent upon the environmental circumstances which could regulate the different transcriptional factors of P. aeruginosa and surprisingly, the susceptibility nature of the bacterium most often reverted when the adverse environmental circumstances are normalized or removed (Bjarnsholt et al. 2005). Different environmental factors like temperature, pH, oxygen demand condition, DNA stress and nutrient deficiency as well as the motility of the organisms also remain as potential causes for the development of adaptive resistance.
Formation of biofilm remains an important factor for the initiation of an adaptive resistance mechanism. As discussed earlier, biofilm is an aggregation of microbial cells that attached one another and encapsulated in self-producing polymeric matrix and capable to attach to both biotic and abiotic surfaces (de la Fuente-Núñez et al. 2013). The P. aeruginosa cells reside in the biofilm are comparatively less sensitive to antibiotics and immunes defence mechanisms of host than its counterpart planktonic cells. Interestingly, the sensitivity of the P. aeruginosa against the antimicrobial agents would restore when the biofilm residing cells happen to grow outside of the biofilm (Jamal et al. 2018). This behavioural change is elucidating that biofilm-mediated resistance is independent of genomic modification and adaptive resistance mechanisms. The biofilm condition makes several physiological and phenotypic alterations in bacterium. For instance, the non-mucoid strain of P. aeruginosa happens to enter into the biofilm matrix would transcriptionally altered as from non-mucoid- to mucoid-producing strain (Macia et al. 2014).
13.5 Novel Strategies for Treating Drug-Resistant P. aeruginosa Infection
Although there are continuous instances of the antibiotic resistance strain, still antibiotics are used to control the infection caused by P. aeruginosa. Almost all the currently available antimicrobial agents are involved in either slowing down the organism's growth also termed as bacteriostatic agents or causing death which is also called as bactericidal agents (Christensen et al. 2012). Each antibiotic specifically interfered with some of the mechanisms underlined with the metabolic pathway notice in bacteria. For instance, certain class of antibiotic acts on the metabolic pathway coupled with bacterial cell wall synthesis, some of other class participates as an inhibitor in bacterial protein and nucleic acid synthesis metabolic pathways. Furthermore, certain group of also plays significant role in hindering nature of bacterial metabolism and cell membrane (Van Acker et al. 2014). As discussed earlier, due to indiscriminate use of antibiotics potentially causing emergence of resistance behaviours among the pathogen, which subsequently brings to the situation where the currently available antimicrobial agent no longer effective to control the bacterial infection (Zaidi et al. 2017). This makes for an urgent call to discover a novel antimicrobial agent to substitute the current antibiotics and potentially address the negative impact that lies with current antibiotics.
In recent years, several different approaches were initiated to improve the efficacy of the currently available antibiotics by means of different formulation procedures (Wagner et al. 2016; Smith et al. 2017). For example, antibiotics are prepared as an inhalation solution and used for long-term treatment processes which are most common in chronic infection of P. aeruginosa. However, this formulation process showed better recalcitrant efficacy, but still created a concern about increased antibiotic resistance. Similarly, some of the antibiotics are also formulated as a dry powder which is administered by inhalation. This produces significantly enhancing success rate of treatment procedure due to its ability to penetrate the sputum and potentially reach the infection site (Hurley et al. 2012). Furthermore, decoration of the antimicrobial agents on to the nanomaterial gains significant attention in recent years. As a result, the antibiotics like amikacin and ciprofloxacin are encapsulated in liposome and investigated for its outdo in controlling the bacterial infection as compared to routine administration procedure that lies with the utilization of antibiotics (Jeevanandam et al. 2016).
13.5.1 Anti-Quorum Sensing
Target the signalling system of the P. aeruginosa found to be an effective therapeutic option and the drug which involves in the interference of QS is also called as quorum sensing inhibitors (QSIs) (Bacha et al. 2016). The QSIs are novel class of antimicrobial agents because these drug molecules show less possibility to experience and development of bacterial resistance which remains a major drawback among antibiotics. The novel QSI should have some desired properties like should be small molecules which potentially inhibit the expression of QS-related genes; the drug should be more specific toward QS system of the bacteria; these QSI should not ant toxic symptom to the host cells during treatment process; the drug should not interfere with any other metabolic pathway like protein and DNA synthesis than QS circuit of the bacteria; the drug should be chemically stable enough to retain the host cell for sufficient time to enable the effective therapeutic actions (Chatterjee et al. 2016; Asfour 2018). Recently, several biological metabolites including phytochemicals and microbial metabolites are employed as potential anti-QS agents (Bhardwaj et al. 2013). For example, the phytochemical, mosloflavone was investigated for their ability in the hindering the QS of P. aeruginosa and suppression of virulence traits. The study release that the phytochemical, mosloflavone effectively blocks the QS system of P. aeruginosa and significantly reduces the production of different virulence factors. The study also reveals the toxicity profile of mosloflavone using in vivo model, Caenorhabditis elegans (Hnamte et al. 2019).
13.5.2 Bacteriophage Therapy
Bacteriophages are generally termed as bacterial viruses which are ubiquitous and found in diverse organisms and grow vigorously in the bacterial host (Rohde et al. 2018). The life cycle of bacteriophage is widely classified into virulent phage (lytic phage) which potentially causes death to the host and temperate phages (Pires et al. 2015). The lytic phages are initially attaching to the surface of the host cell and it extends its infection by injecting DNA into the host allows it DNA to replicate along with the host. After the complete establishment of infection, it induces the host to undergo for apoptosis process and subsequently kill the host. The releasing phages from the lysed host cell would potentially initiate the infection cycle in other host cells (Waters et al. 2017). On the other hand temperate phages also initiate the infection by attaching to the host cell and gradually integrate its genetic material into the host chromosome. In cases, they integrate its genetic material in bacterial plasmid and allows the transmission during the cell division without causing much adverse effect on the growth of host cell (Chan et al. 2018).
The idea of utilizing the phages as antimicrobial agent was reported in previous century but due to certain limitations and the phenomenal growth in antibiotics significantly reduced the popularity of the phage-mediated antimicrobial therapy. As a consequence of resistance mechanisms, alternative therapeutic options gained considerable interest in phages-mediated antimicrobial therapy (Hill et al. 2018; Law et al. 2019). The bacteriophage-mediated antimicrobial therapy found to be an potential alternative currently available antimicrobial agents in several ways: bacteriophages never initiate self-amplification process in absence of it susceptible bacteria; they are capable to penetrate into the biofilm matrix and infect the bacteria; their potential in killing even the persistent cells were found highly difficult using conventional antibiotics (Chan et al. 2018). A recent study showed that phage therapy could be an effective therapeutic aid against P. aeruginosa infection and study proved the result inference using zebrafish model system (Cafora et al. 2019).
13.5.3 Antibiofilm Peptides
As discussed earlier, the biofilm matrix of P. aeruginosa limits the efficacy of most of the antibiotics. The infections associated with biofilm bring the situation where the treatment such disease found utterly difficult to treat (Teerapo et al. 2019). There is an urgent call for the development of novel antimicrobial agent while the conventional antibiotic are increasingly inefficient in the controlling the bacterial infection. With this aspect, antimicrobial peptides are placed in the centre point of the attraction as an alternative approach for the treatment of bacterial infections (Pletzer and Hancock 2016). In general, antimicrobial peptides are the highly conserved molecules observed in diverse organisms and having greater concern with innate immunity of all species (Dostert et al. 2019). The antimicrobial peptides gained significant consideration due their specialized characteristic features including rapid action and wide spectrum of antimicrobial activity against different microorganisms such as bacteria, viruses, fungi and protozoa. Furthermore, antimicrobial peptides are less potential in the development resistance within the microbial cells, hence this has been displayed as promising antimicrobial agent to combat the multidrug-resistant strain-mediated infections (Grassi et al. 2019).
13.6 Conclusion
Prevention of individuals from P. aeruginosa infection remains significant burden for humans. Different resistances mechanisms such as intrinsic, acquired and adaptive possibly strengthen the multidrug-resistant behaviours of P. aeruginosa, which subsequently displays a potential tolerant to almost all the available antimicrobial agents. Moreover, the biofilm-forming nature of P. aeruginosa delivers several persistent cells which make the condition highly unfavourable for regular antimicrobial treatment process. Over several decades, several remedies were discovered to enhance the delivery and the efficacy of the antimicrobial agents to host that eventually control the microbial infection. However, P. aeruginosa is highly capable to adapt to most of the adverse conditions which lead to the repeated incidence of resistance development. In recent years, non-antibiotic therapies including anti-virulence, phage and antimicrobial peptide therapies are widely investigated for their antimicrobial potential which showed less potential in developing resistance. Still considerable knowledge about the host–microbial interactions is required to design innovative and effective antimicrobial agents to control infection caused by multidrug-resistant microorganisms.
References
Aarts M-A, Hancock JN, Heyland D, Marshall JC (2003) Antibiotics for ventilator-associated pneumonia. In: Aarts M-A (ed) The Cochrane database of systematic reviews. Wiley, Chichester
Ahmed MN, Porse A, Sommer MOA et al (2018) Evolution of antibiotic resistance in biofilm and planktonic Pseudomonas aeruginosa populations exposed to subinhibitory levels of ciprofloxacin. Antimicrob Agents Chemother 62:1–12. https://doi.org/10.1128/AAC.00320-18
Al-Dahmoshi HOM, Al-Khafaji NS, Abdulzahra Jeyad A et al (2018) Molecular detection of some virulence traits among Pseudomonas aeruginosa isolates, Hilla-Iraq. Biomed Pharmacol J 11:835–842. https://doi.org/10.13005/bpj/1439
Alhazmi A (2015) Pseudomonas aeruginosa – pathogenesis and pathogenic mechanisms. Int J Biol 7:44–67. https://doi.org/10.5539/ijb.v7n2p44
Alionte LG, Cannon BM, White CD et al (2001) Pseudomonas aeruginosa LasA protease and corneal infections. Curr Eye Res 22:266–271. https://doi.org/10.1076/ceyr.22.4.266.5509
Anantharajah A, Mingeot-Leclercq M-P, Van Bambeke F (2016) Targeting the type three secretion system in Pseudomonas aeruginosa. Trends Pharmacol Sci 37:734–749. https://doi.org/10.1016/j.tips.2016.05.011
Asfour H (2018) Anti-quorum sensing natural compounds. J Microsc Ultrastruct 6(1):1. https://doi.org/10.4103/JMAU.JMAU_10_18
Azam MW, Khan AU (2019) Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov Today 24:350–359. https://doi.org/10.1016/j.drudis.2018.07.003
Bacha K, Tariku Y, Gebreyesus F et al (2016) Antimicrobial and anti-quorum sensing activities of selected medicinal plants of Ethiopia: implication for development of potent antimicrobial agents. BMC Microbiol 16:139. https://doi.org/10.1186/s12866-016-0765-9
Bai AJ, Rai VR (2011) Bacterial quorum sensing and food industry. Compr Rev Food Sci Food Saf 10:183–193. https://doi.org/10.1111/j.1541-4337.2011.00150.x
Bassetti M, Vena A, Croxatto A et al (2018) How to manage Pseudomonas aeruginosa infections. Drugs Context 7:1–18. https://doi.org/10.7573/dic.212527
Battle SE, Rello J, Hauser AR (2009) Genomic islands of Pseudomonas aeruginosa. FEMS Microbiol Lett 290:70–78. https://doi.org/10.1111/j.1574-6968.2008.01406.x
Beaussart A, Baker AE, Kuchma SL et al (2014) Nanoscale adhesion forces of Pseudomonas aeruginosa type IV pili. ACS Nano 8:10723–10733. https://doi.org/10.1021/nn5044383
Bhagirath AY, Li Y, Somayajula D et al (2016) Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm Med 16:174. https://doi.org/10.1186/s12890-016-0339-5
Bhardwaj AK, Vinothkumar K, Rajpara N (2013) Bacterial quorum sensing inhibitors: attractive alternatives for control of infectious pathogens showing multiple drug resistance. Recent Pat Antiinfect Drug Discov 8:68–83. https://doi.org/10.2174/1574891X11308010012
Bjarnsholt T, Jensen PØ, Burmølle M et al (2005) Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373–383. https://doi.org/10.1099/mic.0.27463-0
Bouglé A, Foucrier A, Dupont H et al (2017) Impact of the duration of antibiotics on clinical events in patients with Pseudomonas aeruginosa ventilator-associated pneumonia: study protocol for a randomized controlled study. Trials 18:37. https://doi.org/10.1186/s13063-017-1780-3
Breidenstein EBM, de la Fuente-Núñez C, Hancock REW (2011) Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. https://doi.org/10.1016/j.tim.2011.04.005
Bucior I, Pielage JF, Engel JN (2012) Pseudomonas aeruginosa pili and flagella mediate distinct binding and signaling events at the apical and basolateral surface of airway epithelium. PLoS Pathog 8:e1002616. https://doi.org/10.1371/journal.ppat.1002616
Burkinshaw BJ, Strynadka NCJ (2014) Assembly and structure of the T3SS. Biochim Biophys Acta, Mol Cell Res 1843:1649–1663. https://doi.org/10.1016/j.bbamcr.2014.01.035
Burstein D, Satanower S, Simovitch M et al (2015) Novel type III effectors in Pseudomonas aeruginosa. MBio 6:2–7. https://doi.org/10.1128/mBio.00161-15
Cafora M, Deflorian G, Forti F et al (2019) Phage therapy against Pseudomonas aeruginosa infections in a cystic fibrosis zebrafish model. Sci Rep 9:1527. https://doi.org/10.1038/s41598-018-37636-x
Campodonico VL, Llosa NJ, Grout M et al (2010) Evaluation of flagella and flagellin of Pseudomonas aeruginosa as vaccines. Infect Immun 78:746–755. https://doi.org/10.1128/IAI.00806-09
Carter MQ, Chen J, Lory S (2010) The Pseudomonas aeruginosa pathogenicity island PAPI-1 is transferred via a novel type IV pilus. J Bacteriol 192:3249–3258. https://doi.org/10.1128/JB.00041-10
Cascales E (2017) Inside the chamber of secrets of the type III secretion system. Cell 168:949–951. https://doi.org/10.1016/j.cell.2017.02.028
Chan BK, Turner PE, Kim S et al (2018) Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health 2018:60–66. https://doi.org/10.1093/emph/eoy005
Chatterjee M, Anju CP, Biswas L et al (2016) Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. Int J Med Microbiol 306:48–58. https://doi.org/10.1016/j.ijmm.2015.11.004
Christensen LD, van Gennip M, Jakobsen TH et al (2012) Synergistic antibacterial efficacy of early combination treatment with tobramycin and quorum-sensing inhibitors against Pseudomonas aeruginosa in an intraperitoneal foreign-body infection mouse model. J Antimicrob Chemother 67:1198–1206. https://doi.org/10.1093/jac/dks002
Christie PJ, Whitaker N, González-Rivera C (2014) Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta, Mol Cell Res 1843:1578–1591. https://doi.org/10.1016/j.bbamcr.2013.12.019
Colbert B, Kumari H, Piñon A et al (2018) Alginate-regulating genes are identified in the clinical cystic fibrosis isolate of Pseudomonas aeruginosa PA2192. bioRxiv:1–33. https://doi.org/10.1101/319004
da Silva Filho LVRF, Ferreira FDA, Reis FJC et al (2013) Pseudomonas aeruginosa infection in patients with cystic fibrosis: scientific evidence regarding clinical impact, diagnosis, and treatment. J Bras Pneumol 39:495–512. https://doi.org/10.1590/S1806-37132013000400015
de la Fuente-Núñez C, Reffuveille F, Fernández L, Hancock RE (2013) Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol 16:580–589. https://doi.org/10.1016/j.mib.2013.06.013
Defoirdt T (2018) Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol 26:313–328. https://doi.org/10.1016/j.tim.2017.10.005
Depluverez S, Devos S, Devreese B (2016) The role of bacterial secretion systems in the virulence of gram-negative airway pathogens associated with cystic fibrosis. Front Microbiol 7:1–8. https://doi.org/10.3389/fmicb.2016.01336
Dostert M, Belanger CR, Hancock REW (2019) Design and assessment of anti-biofilm peptides: steps toward clinical application. J Innate Immun 11:193–204. https://doi.org/10.1159/000491497
Feldman M, Bryan R, Rajan S et al (1998) Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun 66:43–51
Fila G, Krychowiak M, Rychlowski M et al (2018) Antimicrobial blue light photoinactivation of Pseudomonas aeruginosa: quorum sensing signaling molecules, biofilm formation and pathogenicity. J Biophotonics 11:e201800079. https://doi.org/10.1002/jbio.201800079
Franklin MJ, Nivens DE, Weadge JT, Lynne Howell P (2011) Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol 2:1–16. https://doi.org/10.3389/fmicb.2011.00167
Friedman ND, Temkin E, Carmeli Y (2016) The negative impact of antibiotic resistance. Clin Microbiol Infect 22:416–422. https://doi.org/10.1016/j.cmi.2015.12.002
Fujii A, Seki M, Higashiguchi M et al (2014) Community-acquired, hospital-acquired, and healthcare-associated pneumonia caused by Pseudomonas aeruginosa. Respir Med Case Rep 12:30–33. https://doi.org/10.1016/j.rmcr.2014.03.002
Furiga A, Lajoie B, El Hage S et al (2016) Impairment of Pseudomonas aeruginosa biofilm resistance to antibiotics by combining the drugs with a new quorum-sensing inhibitor. Antimicrob Agents Chemother 60:1676–1686. https://doi.org/10.1128/AAC.02533-15
Galle M, Carpentier I, Beyaert R (2012) Structure and function of the type III secretion system of Pseudomonas aeruginosa. Curr Protein Pept Sci 13:831–842. https://doi.org/10.2174/138920312804871210
Garvey MI, Bradley CW, Tracey J, Oppenheim B (2016) Continued transmission of Pseudomonas aeruginosa from a wash hand basin tap in a critical care unit. J Hosp Infect 94:8–12. https://doi.org/10.1016/j.jhin.2016.05.004
Gellatly SL, Hancock REW (2013) Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173. https://doi.org/10.1111/2049-632X.12033
Goldberg JB, Pier GB (1996) Pseudomonas aeruginosa lipopolysaccharides and pathogenesis. Trends Microbiol 4:490–494. https://doi.org/10.1016/S0966-842X(97)82911-3
Gonzalez MR, Fleuchot B, Lauciello L et al (2016) Effect of human burn wound exudate on Pseudomonas aeruginosa virulence. mSphere 1:1–14. https://doi.org/10.1128/mSphere.00111-15
Grassi L, Batoni G, Ostyn L et al (2019) The antimicrobial peptide lin-SB056-1 and its dendrimeric derivative prevent Pseudomonas aeruginosa biofilm formation in physiologically relevant models of chronic infections. Front Microbiol 10:1–14. https://doi.org/10.3389/fmicb.2019.00198
Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T (2010) An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol Med Microbiol 59:253–268. https://doi.org/10.1111/j.1574-695X.2010.00690.x
Hauser AR (2011) Pseudomonas aeruginosa: so many virulence factors, so little time∗. Crit Care Med 39:2193–2194. https://doi.org/10.1097/CCM.0b013e318221742d
Herwald H, Egesten A (2009) Bacterial proteases disarming host Defense. J Innate Immun 1:69–69. https://doi.org/10.1159/000181143
Hill C, Mills S, Ross RP (2018) Phages & antibiotic resistance: are the most abundant entities on earth ready for a comeback? Future Microbiol 13:711–726. https://doi.org/10.2217/fmb-2017-0261
Hirsch EB, Tam VH (2010) Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res 10:441–451. https://doi.org/10.1586/erp.10.49
Hnamte S, Parasuraman P, Ranganathan S et al (2019) Mosloflavone attenuates the quorum sensing controlled virulence phenotypes and biofilm formation in Pseudomonas aeruginosa PAO1: in vitro, in vivo and in silico approach. Microb Pathog 131:128–134. https://doi.org/10.1016/j.micpath.2019.04.005
Hobden JA (2002) Pseudomonas aeruginosa proteases and corneal virulence. DNA Cell Biol 21:391–396. https://doi.org/10.1089/10445490260099674
Huang Z, Jiang Y, Liang J (2011) Pathogenesis could be one of the anti-cheating mechanisms for Pseudomonas aeruginosa society. Med Hypotheses 76:166–168. https://doi.org/10.1016/j.mehy.2010.09.007
Hurley MN, Cámara M, Smyth AR (2012) Novel approaches to the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. Eur Respir J 40:1014–1023. https://doi.org/10.1183/09031936.00042012
Ingmer H, Brøndsted L (2009) Proteases in bacterial pathogenesis. Res Microbiol 160:704–710. https://doi.org/10.1016/j.resmic.2009.08.017
Izoré T, Job V, Dessen A (2011) Biogenesis, regulation, and targeting of the type III secretion system. Structure 19:603–612. https://doi.org/10.1016/j.str.2011.03.015
Jamal M, Ahmad W, Andleeb S et al (2018) Bacterial biofilm and associated infections. J Chin Med Assoc 81:7–11. https://doi.org/10.1016/j.jcma.2017.07.012
Jeevanandam J, Chan YS, Danquah MK (2016) Nano-formulations of drugs: recent developments, impact and challenges. Biochimie 128–129:99–112. https://doi.org/10.1016/j.biochi.2016.07.008
Juhas M (2015) Type IV secretion systems and genomic islands-mediated horizontal gene transfer in Pseudomonas and Haemophilus. Microbiol Res 170:10–17. https://doi.org/10.1016/j.micres.2014.06.007
Juhas M, Crook DW, Hood DW (2008) Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol 10:2377–2386. https://doi.org/10.1111/j.1462-5822.2008.01187.x
Kalaiarasan E, Thirumalaswamy K, Harish BN et al (2017) Inhibition of quorum sensing-controlled biofilm formation in Pseudomonas aeruginosa by quorum-sensing inhibitors. Microb Pathog 111:99–107. https://doi.org/10.1016/j.micpath.2017.08.017
Kalia VC, Patel SKS, Kang YC, Lee J-K (2019) Quorum sensing inhibitors as antipathogens: biotechnological applications. Biotechnol Adv 37:68–90. https://doi.org/10.1016/j.biotechadv.2018.11.006
Kariminik A, Baseri-Salehi M, Kheirkhah B (2017) Pseudomonas aeruginosa quorum sensing modulates immune responses: an updated review article. Immunol Lett 190:1–6. https://doi.org/10.1016/j.imlet.2017.07.002
Kerr KG, Snelling AM (2009) Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect 73:338–344. https://doi.org/10.1016/j.jhin.2009.04.020
Kibret KT, Moges Y, Memiah P, Biadgilign S (2017) Treatment outcomes for multidrug-resistant tuberculosis under DOTS-Plus: a systematic review and meta-analysis of published studies. Infect Dis Poverty 6:7. https://doi.org/10.1186/s40249-016-0214-x
King JD, Kocíncová D, Westman EL, Lam JS (2009) Review: Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun 15:261–312. https://doi.org/10.1177/1753425909106436
Klockgether J, Tümmler B (2017) Recent advances in understanding Pseudomonas aeruginosa as a pathogen. F1000Res 6:1261. https://doi.org/10.12688/f1000research.10506.1
Koh CL, Sam CK, Yin WF et al (2013) Plant-derived natural products as sources of anti-quorum sensing compounds. Sensors (Basel) 13:6217–6228. https://doi.org/10.3390/s130506217
Kohlenberg A, Weitzel-Kage D, van der Linden P et al (2010) Outbreak of carbapenem-resistant Pseudomonas aeruginosa infection in a surgical intensive care unit. J Hosp Infect 74:350–357. https://doi.org/10.1016/j.jhin.2009.10.024
Kung VL, Ozer EA, Hauser AR (2010) The accessory genome of Pseudomonas aeruginosa. Microbiol Mol Biol Rev 74:621–641. https://doi.org/10.1128/MMBR.00027-10
Lantz MS (1997) Are bacterial proteases important virulence factors? J Periodontal Res 32:126–132. https://doi.org/10.1111/j.1600-0765.1997.tb01393.x
Law N, Logan C, Yung G et al (2019) Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 47:665–668. https://doi.org/10.1007/s15010-019-01319-0
Lee DG, Urbach JM, Wu G et al (2006) Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol 7:R90. https://doi.org/10.1186/gb-2006-7-10-r90
Limoli DH, Whitfield GB, Kitao T et al (2017) Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. MBio 8:1–18. https://doi.org/10.1128/mBio.00186-17
Lorè NI, Cigana C, De Fino I et al (2012) Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PLoS One 7:e35648. https://doi.org/10.1371/journal.pone.0035648
Ma Q, Zhai Y, Schneider JC et al (2003) Protein secretion systems of Pseudomonas aeruginosa and P. fluorescens. Biochim Biophys Acta Biomembr 1611:223–233. https://doi.org/10.1016/S0005-2736(03)00059-2
Macia MD, Rojo-Molinero E, Oliver A (2014) Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin Microbiol Infect 20:981–990. https://doi.org/10.1111/1469-0691.12651
Maldonado RF, Sá-Correia I, Valvano MA (2016) Lipopolysaccharide modification in gram-negative bacteria during chronic infection. FEMS Microbiol Rev 40:480–493. https://doi.org/10.1093/femsre/fuw007
Markou P, Apidianakis Y (2014) Pathogenesis of intestinal Pseudomonas aeruginosa infection in patients with cancer. Front Cell Infect Microbiol 3:1–5. https://doi.org/10.3389/fcimb.2013.00115
Matsumoto K (2004) Role of bacterial proteases in pseudomonal and serratial keratitis. Biol Chem 385:1007–1016. https://doi.org/10.1515/BC.2004.131
Maurice NM, Bedi B, Sadikot RT (2018) Pseudomonas aeruginosa biofilms: host response and clinical implications in lung infections. Am J Respir Cell Mol Biol 58:428–439. https://doi.org/10.1165/rcmb.2017-0321TR
McCaslin CA, Petrusca DN, Poirier C et al (2015) Impact of alginate-producing Pseudomonas aeruginosa on alveolar macrophage apoptotic cell clearance. J Cyst Fibros 14:70–77. https://doi.org/10.1016/j.jcf.2014.06.009
Micek ST, Kollef MH, Torres A et al (2015) Pseudomonas aeruginosa nosocomial pneumonia: impact of pneumonia classification. Infect Control Hosp Epidemiol 36:1190–1197. https://doi.org/10.1017/ice.2015.167
Michalska M, Wolf P (2015) Pseudomonas exotoxin A: optimized by evolution for effective killing. Front Microbiol 6:1–7. https://doi.org/10.3389/fmicb.2015.00963
Mofazzal Jahromi MA, Sahandi Zangabad P, Moosavi Basri SM et al (2018) Nanomedicine and advanced technologies for burns: preventing infection and facilitating wound healing. Adv Drug Deliv Rev 123:33–64. https://doi.org/10.1016/j.addr.2017.08.001
Morlon-Guyot J, Mere J, Bonhoure A, Beaumelle B (2009) Processing of Pseudomonas aeruginosa exotoxin A is dispensable for cell intoxication. Infect Immun 77:3090–3099. https://doi.org/10.1128/IAI.01390-08
Musicki B, Liu T, Lagoda GA et al (2009) Endothelial nitric oxide synthase regulation in female genital tract structures. J Sex Med 6:247–253. https://doi.org/10.1111/j.1743-6109.2008.01122.x
Nguyen L, Garcia J, Gruenberg K, MacDougall C (2018) Multidrug-resistant pseudomonas infections: hard to treat, but hope on the horizon? Curr Infect Dis Rep 20:23. https://doi.org/10.1007/s11908-018-0629-6
Niederman MS (2010) Hospital-acquired pneumonia, health care–associated pneumonia, ventilator-associated pneumonia, and ventilator-associated tracheobronchitis: definitions and challenges in trial design. Clin Infect Dis 51:S12–S17. https://doi.org/10.1086/653035
Oliver A, Mulet X, López-Causapé C, Juan C (2015) The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21–22:41–59. https://doi.org/10.1016/j.drup.2015.08.002
Overhage J, Lewenza S, Marr AK, Hancock REW (2007) Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J Bacteriol 189:2164–2169. https://doi.org/10.1128/JB.01623-06
Palanisamy N, Ferina N, Amirulhusni A et al (2014) Antibiofilm properties of chemically synthesized silver nanoparticles found against Pseudomonas aeruginosa. J Nanobiotechnology 12:2. https://doi.org/10.1186/1477-3155-12-2
Pang Z, Raudonis R, Glick BR et al (2019) Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv 37:177–192. https://doi.org/10.1016/j.biotechadv.2018.11.013
Pena C, Cabot G, Gomez-Zorrilla S et al (2015) Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis 60:539–548. https://doi.org/10.1093/cid/ciu866
Pier G (2007) Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int J Med Microbiol 297:277–295. https://doi.org/10.1016/j.ijmm.2007.03.012
Pillar CM, Hobden JA (2002) Pseudomonas aeruginosa exotoxin A and keratitis in mice. Investig Ophthalmol Vis Sci 43:1437–1444
Pires DP, Vilas Boas D, Sillankorva S, Azeredo J (2015) Phage therapy: a step forward in the treatment of Pseudomonas aeruginosa infections. J Virol 89:7449–7456. https://doi.org/10.1128/jvi.00385-15
Pletzer D, Hancock REW (2016) Antibiofilm peptides: potential as broad-spectrum agents. J Bacteriol 198:2572–2578. https://doi.org/10.1128/JB.00017-16
Puhar A, Sansonetti PJ (2014) Type III secretion system. Curr Biol 24:R784–R791. https://doi.org/10.1016/j.cub.2014.07.016
Rafla K, Tredget EE (2011) Infection control in the burn unit. Burns 37:5–15. https://doi.org/10.1016/j.burns.2009.06.198
Rasamiravaka T, Labtani Q, Duez P, El Jaziri M (2015) The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int 2015:1–17. https://doi.org/10.1155/2015/759348
Rohde C, Wittmann J, Kutter E (2018) Bacteriophages: a therapy concept against multi-drug–resistant bacteria. Surg Infect 19:737–744. https://doi.org/10.1089/sur.2018.184
Rossi Gonçalves I, Dantas RCC, Ferreira ML et al (2017) Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz J Microbiol 48:211–217. https://doi.org/10.1016/j.bjm.2016.11.004
Salter SJ (2015) Keeping an eye on P. aeruginosa. Nat Rev Microbiol 13:69–69. https://doi.org/10.1038/nrmicro3422
Satpathy S, Sen SK, Pattanaik S, Raut S (2016) Review on bacterial biofilm: an universal cause of contamination. Biocatal Agric Biotechnol 7:56–66. https://doi.org/10.1016/j.bcab.2016.05.002
Silby MW, Winstanley C, Godfrey SAC et al (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680. https://doi.org/10.1111/j.1574-6976.2011.00269.x
Simões D, Miguel SP, Ribeiro MP et al (2018) Recent advances on antimicrobial wound dressing: a review. Eur J Pharm Biopharm 127:130–141. https://doi.org/10.1016/j.ejpb.2018.02.022
Smith WD, Bardin E, Cameron L et al (2017) Current and future therapies for Pseudomonas aeruginosa infection in patients with cystic fibrosis. FEMS Microbiol Lett 364:1–9. https://doi.org/10.1093/femsle/fnx121
Sousa A, Pereira M (2014) Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs—a review. Pathogens 3:680–703. https://doi.org/10.3390/pathogens3030680
Stefani S, Campana S, Cariani L et al (2017) Relevance of multidrug-resistant Pseudomonas aeruginosa infections in cystic fibrosis. Int J Med Microbiol 307:353–362. https://doi.org/10.1016/j.ijmm.2017.07.004
Streeter K, Katouli M (2016) Pseudomonas aeruginosa: a review of their pathogenesis and prevalence in clinical settings and the environment. Infect Epidemiol Med 2:25–32. https://doi.org/10.18869/modares.iem.2.1.25
Taccetti G, Campana S, Neri AS et al (2008) Antibiotic therapy against Pseudomonas aeruginosa in cystic fibrosis. J Chemother 20:166–169. https://doi.org/10.1179/joc.2008.20.2.166
Taylor PK, Yeung ATY, Hancock REW (2014) Antibiotic resistance in Pseudomonas aeruginosa biofilms: towards the development of novel anti-biofilm therapies. J Biotechnol 191:121–130. https://doi.org/10.1016/j.jbiotec.2014.09.003
Teerapo K, Roytrakul S, Sistayanarain A, Kunthalert D (2019) A scorpion venom peptide derivative BmKn–22 with potent antibiofilm activity against Pseudomonas aeruginosa. PLoS One 14:e0218479. https://doi.org/10.1371/journal.pone.0218479
Trokter M, Felisberto-Rodrigues C, Christie PJ, Waksman G (2014) Recent advances in the structural and molecular biology of type IV secretion systems. Curr Opin Struct Biol 27:16–23. https://doi.org/10.1016/j.sbi.2014.02.006
Tumbarello M, De Pascale G, Trecarichi EM et al (2013) Clinical outcomes of Pseudomonas aeruginosa pneumonia in intensive care unit patients. Intensive Care Med 39:682–692. https://doi.org/10.1007/s00134-013-2828-9
Valentini M, Gonzalez D, Mavridou DA, Filloux A (2018) Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr Opin Microbiol 41:15–20. https://doi.org/10.1016/j.mib.2017.11.006
Van Acker H, Van Dijck P, Coenye T (2014) Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol 22:326–333. https://doi.org/10.1016/j.tim.2014.02.001
Wagner S, Sommer R, Hinsberger S et al (2016) Novel strategies for the treatment of Pseudomonas aeruginosa infections. J Med Chem 59:5929–5969. https://doi.org/10.1021/acs.jmedchem.5b01698
Wang Y, Beekman J, Hew J et al (2018) Burn injury: challenges and advances in burn wound healing, infection, pain and scarring. Adv Drug Deliv Rev 123:3–17. https://doi.org/10.1016/j.addr.2017.09.018
Waters EM, Neill DR, Kaman B et al (2017) Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax 72:666–667. https://doi.org/10.1136/thoraxjnl-2016-209265
Werneburg M, Zerbe K, Juhas M et al (2012) Inhibition of lipopolysaccharide transport to the outer membrane in Pseudomonas aeruginosa by peptidomimetic antibiotics. Chembiochem 13:1767–1775. https://doi.org/10.1002/cbic.201200276
Wiehlmann L, Wagner G, Cramer N et al (2007) Population structure of Pseudomonas aeruginosa. Proc Natl Acad Sci 104:8101–8106. https://doi.org/10.1073/pnas.0609213104
Wilke M, Grube R, Bodmann K (2011) Guideline-adherent initial intravenous antibiotic therapy for hospital-acquired/ventilator-associated pneumonia is clinically superior, saves lives and is cheaper than non guideline adherent therapy. Eur J Med Res 16:315. https://doi.org/10.1186/2047-783X-16-7-315
Wolfgang MC, Kulasekara BR, Liang X et al (2003) Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc Natl Acad Sci 100:8484–8489. https://doi.org/10.1073/pnas.0832438100
Wu Y-L, Yang X-Y, Ding X-X et al (2019) Exposure to infected/colonized roommates and prior room occupants increases the risks of healthcare-associated infections with the same organism. J Hosp Infect 101:231–239. https://doi.org/10.1016/j.jhin.2018.10.014
Yamazaki A, Li J, Zeng Q et al (2012) Derivatives of plant phenolic compound affect the type III secretion system of Pseudomonas aeruginosa via a GacS-GacA two-component signal transduction system. Antimicrob Agents Chemother 56:36–43. https://doi.org/10.1128/AAC.00732-11
Yelin I, Kishony R (2018) Antibiotic resistance. Cell 172:1136–1136.e1. https://doi.org/10.1016/j.cell.2018.02.018
Zaidi S, Misba L, Khan AU (2017) Nano-therapeutics: a revolution in infection control in post antibiotic era. Nanomedicine 13:2281–2301. https://doi.org/10.1016/j.nano.2017.06.015
Zhu M, Zhao J, Kang H et al (2016) Modulation of type III secretion system in Pseudomonas aeruginosa: involvement of the PA4857 gene product. Front Microbiol 7:1–13. https://doi.org/10.3389/fmicb.2016.00007
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Parasuraman, P., Syed, A., Siddhardha, B. (2020). Pathogenesis and Drug Resistance of Pseudomonas aeruginosa. In: Siddhardha, B., Dyavaiah, M., Syed, A. (eds) Model Organisms for Microbial Pathogenesis, Biofilm Formation and Antimicrobial Drug Discovery. Springer, Singapore. https://doi.org/10.1007/978-981-15-1695-5_13
Download citation
DOI: https://doi.org/10.1007/978-981-15-1695-5_13
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-1694-8
Online ISBN: 978-981-15-1695-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)