Abstract
On April 6, 2016, the SJ-10 recoverable microgravity experimental satellite (SJ-10 satellite) was launched from Jiuquan in China, which conducted a mission of space microgravity experiments. As a recoverable satellite, the SJ-10 satellite provided an effective, open, and comprehensive platform to study space life and microgravity science. The SJ-10 satellite program consisted of 27 experiments including both fields of microgravity and space life sciences. Among the experiments, “three-dimensional (3D) cell culture and tissue restoration of NSCs under microgravity” proposed by Dr. Jianwu Dai and his staff was selected from more than 200 applications. This project was characterized by two aspects: neural stem cells and 3D culture. It was the first time that in vitro-cultured NSCs experienced a microgravity environment in space. 3D culture provided a specialized environment to benefit the in vitro tissue constructs. NSC-based therapy has attracted attention in recent years, which may be a promising treatment for many neurological diseases such as spinal cord injury, Alzheimer’s disease, stroke, and Parkinson’s disease. The 3D culture of NSCs under microgravity may provide valuable data for tissue reconstruction of the nervous system. To communicate the background and progress of this research, this review focuses on the key points of NSCs, 3D culture, and microgravity.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Neural Stem Cells
Neural stem cells are a type of progenitor cell of nervous systems, which self-renew and generate both neurons and glia. They are the source of neurons and glia in the CNS, although the regenerative ability of the CNS is limited. The CNS in mammals including humans consists of neurons and glia.
1.1 Main Cell Types of Nervous Systems
Neurons, a nervous system cell type, function in transmitting information through electronic and chemical signals. Neurons connect to each other through synapses between axons or axons and dendrites to form neural networks. Neurons are responsible for transmitting sensory signals from the whole body to the brain or spinal cord, which are the central processors of nervous systems, and precisely convey the ordered signals of the brain or spinal cord to the whole body. The processes from neuron cells are defined as axons and dendrites by their different shapes, lengths, and functions. Dendrites always taper off and are shorter, whereas axons tend to maintain a constant radius and are relatively long. In terms of functions, axons transmit electrochemical signals and dendrites receive them. The structure of the connection of the axon and dendrite is called a synapse. As the central functional unit of nervous systems, damage to neurons would greatly affect the functions of nervous systems. Furthermore, neurons of the CNS cannot regenerate by themselves, which is the main reason that CNS diseases rarely receive a favorable prognosis.
Astroglia, also called astrocytes, are a subtype of glial cells in the CNS. An important function of astrocytes is supporting the physical structure of the CNS. Neurotrophins secreted by astrocytes facilitate neuron functions. The capability of glycogenesis in astroglia maintains the dynamic balance of glucose to fuel neurons. Upon CNS injury, astrocytes fill the space with a glial scar. On one hand, the glial scar protects neural cells from further cell death, and on the other hand, the glial scar forms a barrier that blocks regenerating axons from crossing the injury site.
Oligodendrocytes are another subtype of glial cell in the CNS. Their main functions are to provide support and insulation to axons in the CNS, which are equivalent to the functions performed by Schwann cells in the PNS. Oligodendrocytes ensheath axons by creating myelin that is 80% lipids and 20% proteins. When separated from Schwann cells, a single oligodendrocyte can extend its processes to up to 50 axons. Schwann cells only wrap around one axon. Because of the close interaction of axons and myelin, several myelin-associated proteins and axon guidance molecules are expressed by oligodendrocytes to regulate the development of axons. However, once injury occurs in the CNS, oligodendrocytes are destroyed and a large amount of myelin is released in the injury site. Researchers have shown that myelin-associated proteins, such as Nogo, MAG, and OMGP, and axon guidance molecules, such as ephrin B3 and semaphorin 4D, act as strong inhibitors of neurite outgrowth from postnatal neurons in vitro.
1.2 Regulation of Neural Stem Cells
During development, NPCs generate different types of neural cells, such as neurons, astrocytes, and oligodendrocytes, which comprise the nervous system (McConnell 1995; Okano and Temple 2009). NSCs give rise to all neurons of the mammalian CNS (Gotz and Huttner 2005). Neurogenesis, defined as the process of generating functional neurons from precursors, is very important for nervous systems and traditionally seen as occurring only during the embryonic and perinatal stages in mammals (Ming and Song 2005). However, current studies suggest that neurogenesis is not limited to the early stages of embryonic development. Neurogenesis in adults can also occur and is generally believed to be very limited under normal physiological conditions, but can be induced after injury (Gould 2007). Some quiescent neural progenitor cells, which exit the cell cycle, activate and undergo a new cell cycle. These new activated neural progenitor cells proliferate to maintain the neural progenitor cell pool, while some neural progenitor cells carry out differentiation processes to produce new functional neurons. The number of neurons in the mature nervous system is determined by the balance among cell proliferation, differentiation, and death (Fogarty et al. 2016). Before neurogenesis, the neural plate and tube consist of a single layer of neuroepithelial cells, which forms the neuroepithelium (Gotz and Huttner 2005). Mammalian neurogenesis begins on day 10 of embryonic development. Neural epithelial cells begin to undergo asymmetric division starting from embryonic day (E) 10. From E10, asymmetrical divisions begin to induce neurogenesis. Most of these cells are called NPCs. At the same time, increasingly more NPCs finish their self-renewal process and enter into asymmetric divisions, which result in neuron differentiation. Early efforts revealed that cells undergoing mitosis are generally found apically in the VZ, lining the ventricles, whereas differentiating neurons are located basally near the brain surface (Fishell and Kriegstein 2003). Some radial glial cells occupy most of the nerve epithelium in the apical direction, which can undergo interkinetic nuclear migration that is dependent on the cell cycle. Neural precursor cells in the ventricular surface divide to maintain their population and give rise to neurons that migrate to remote areas. After neurogenesis, cells that maintain self-renewal stay in the ventricular zone. However, differentiated cells that have finished the cell cycle prefer to move along the pial fiber to construct the cortical plate. During adult neurogenesis, several types of intermediate cells are located between the cortex and ventricles. These intermediate cells are produced through asymmetrical divisions and then undergo symmetrical divisions to produce neurons (Noctor et al. 2004).
1.3 Neurogenesis in the Brain
Previous studies have demonstrated that NSCs produce functional neurons. In the adult brain, kinetic studies of neurogenesis are still insufficient. A conservative estimate in rats and mice suggests that one neuron is produced among 2000 existing neurons every day (Kempermann et al. 1997). With aging, the neurogenesis rate decreases, while neurogenesis persists in the dentate gyrus at the same time in elderly rodents and humans (Gage 2000). From embryonic day 10 onwards in mice and humans, NPCs divide asymmetrically to renew themselves and generate other neural cell types. After birth, most neurons in the nervous system have been generated. Only a few types of NSCs are maintained in some specialized niches (Fishell and Kriegstein 2003). In the brain, NSCs are mainly located in two regions. The SGZ in the dentate gyrus of the hippocampus, where new dentate granule cells appear (Gage 2000; Kempermann and Gage 2000). During adulthood, neurons can be generated from NSCs in the SGZ (Goncalves et al. 2016). In the adult SGZ, there are two cell types. Type 1 cells can be detected by Nestin, GFAP and Sox2 expression, which is similar to the way that NSCs are detected. Type 2 cells may be derived from type 1 cells. GFAP protein is not expressed in type 2 cells; however, some type 2 cells that only express Sox2 can undergo a self-renewal cycle and then differentiate into neurons and astrocytes. Intermediate progenitors generated from some radial and non-radial precursors give rise to neuroblasts, and intermediate neurons migrate to the granule cell layer and differentiate into DG cells. The DG neurons extend processes into molecular layer and project axons towards CA3. Another region is the SVZ of the lateral ventricles, where new neurons are generated and migrate to the olfactory bulb to become interneurons through the RMS (Gage 2000). In the adult SVZ, there is a population of ependymal cells in the ependyma, which are regarded as unique progenitors during adult neurogenesis. Proliferating radial glial-like cells give rise to transient amplifying cells that in turn generate neuroblasts. In the RMS, neuroblasts form a chain and migrate toward the olfactory bulb through a tube formed under the assistance of astrocytes. These progenitor cells in the SVZ also contribute to constant neurogenesis in the olfactory bulb. Once these progenitors reach the core of the olfactory bulb, immature neurons detach from the RMS and migrate radially toward glomeruli where they differentiate into different subtypes of interneurons (Lledo et al. 2006). In fact, in the early central nervous system, their fates have been determined. There are still three cell types: type A cells are considered as migrating neuroblasts, type B cells are GFAP-positive progenitor cells, and type C cells are a class of transit amplifying cells. All of these cells constitute the basis of adult brain neurogenesis.
Adult brain neurogenesis regions are the SGZ in the dentate gyrus of the hippocampus and SVZ of the lateral ventricles. Neurons generated in the SVZ migrate through the rostral migratory stream and reach the olfactory bulb. In the SGZ, intermediate progenitors are generated from radial and non-radial glia, and intermediate neurons migrate into the DG zone. During adult neurogenesis, many factors are involved in regulating the niche. In the SGZ region, mature neurons, newborn neurons, astrocytes, and oligodendroglial cells construct a complex microenvironment that is vital for CNS development, especially adult neurogenesis. Astrocytes in the hippocampus promote the occurrence of hippocampal neurons and the integration of newborn neurons, which may be achieved through the Wnt pathway. In addition, astrocytes from the hippocampus, which are not derived from the spinal cord, promote neural progenitor proliferation and neuronal fate determination of multipotent adult NSCs in culture (Lim and Alvarez-Buylla 1999; Song et al. 2002). Neurogenesis regulation by astrocytes results from some membrane-attached factors being expressed by astrocytes. These factors play a key role in regulating neural precursors, such as migration and synapse formation (Barkho et al. 2006). Microglia play a positive role by enriching the neurogenesis environment in the SGZ region. They recruit T cells to function to the most extent (Ziv et al. 2006). In the SVZ region, there is a class of ventricular cells with high Noggin expression, which promotes neurogenesis through bone morphogenetic protein signaling pathways. Ventricular cells facilitate neural stem cell self-renewal together with pigment epithelium-derived factors. Some dopamine fibers in SVZ progenitors promote NSC proliferation by dopamine receptors (Lim et al. 2000; Platel et al. 2010). In the brain, factors regulating neurogenesis are complex and diverse. Calcium channel agonists increase adult hippocampal neuron differentiation. Neurogenesis regulation also requires the Sonic hedgehog pathway. Better regulation of the microenvironment is essential for the occurrence of nerves in the brain.
1.4 Neurogenesis in the Spinal Cord
In the last decade, NSCs have been found in specialized zones of the nervous system during adulthood. These endogenous NSCs are capable of constantly differentiating into neurons (Shihabuddin 2008). However, studies have generally concentrated on the activation and recruitment of brain endogenous neurogenesis, whereas few reports concern spinal cord endogenous neurogenesis. Thus far, studies have not only identified NSCs in the subgranular zone of the dentate gyrus and the subventricular zone of the lateral ventricles in brain, but also among ependymal cells of the central canal in the spinal cord (Coskun et al. 2008; Gage 2000; Gross 2000). In the spinal cord, the ependymal cell is the only cell type with a potential differentiation ability. The ependymal cell originates from embryonic day 14–16 radial glial cells. At 1 week postnatally, these ependymal cells begin to differentiate and appear to form two distinct subpopulations. The first subpopulation is derived from radial glial progenitor cells during embryonic development. The second subpopulation forms postnatally, and the first ventricular tube occurs in the first 8–15 days after birth (Delgehyr et al. 2015). However, it is unclear whether these astrocytes and oligodendroglia originate from the same cell clone or different ependymal subpopulations. Ependymal cells are ciliated cells lining the ventricular system of the central spinal canal. They are responsible for moving cerebrospinal fluid and forming a barrier in the brain and spinal cord parenchyma. In the intact spinal cord, few ependymal cells divide. Once cultured in vitro, they vigorously divide and produce astrocytes, oligodendrocytes, and neurons (Burda and Sofroniew 2014). There are three dividing cell subpopulations in the adult mammalian spinal cord. Eighty percent of proliferating cells are oligodendrocyte progenitors. These cells maintain a strong ability to proliferate. Their major function is to generate myelinated oligodendrocytes in the adult spinal cord, especially after spinal cord injury (Barnabe-Heider et al. 2010). Less than 5% of cells are astrocytes. These populations divide infrequently to maintain their population. Upon spinal cord injury, astrocytes participate in glial scar formation (Barnabe-Heider et al. 2010; Lee-Liu et al. 2013). In addition, ependymal cells occupy less than 5%, but they are a class of cells with cilia. They are the only cell type with multipotency in the spinal cord, which allows differentiation and the three types of cells to appear. All of these factors contribute to spinal cord neurogenesis and functional recovery.
1.5 Neurogenesis After Injury
Neurogenesis in other adult CNS regions is generally believed to be very limited under normal physiological conditions, but can be induced after injury (Gould 2007). After spinal cord injury, ependymal cells begin fast divisions by self-renewal and generate a large number of astrocytes that participate in scar formation. In addition, they generate a small number of oligodendrocytes capable of myelinating axons. Therefore, ependymal cells in the adult spinal cord represent a potential NSC population (Burda and Sofroniew 2014). After injuries including peripheral injury, some specialized genes are highly expressed. The appearance of their gene products around the central process periphery promotes regeneration of the central nervous system. ATF3 is expressed in injured neurons and combines with promoters together with C-Jun to promote neuron regeneration. Sox11b also regulates neural progenitor cell proliferation and promotes neural stem cell differentiation to neurons.
1.6 Regulation of Neurogenesis
During adult neurogenesis, a superior neurogenesis microenvironment is regarded as the center where neurogenesis occurs easily. Increasingly more scientists have begun to focus on research of neurogenesis microenvironments (Schofield 1978). In the adult brain, reactive NSCs are restricted by the microenvironment components. Some cellular factors have been identified (Inghilleri and Iacovelli 2011; Riquelme et al. 2008). Mature neurons, such as interneurons, release GABA that regulates the proliferation of sporadic neural stem cells. The process of neuron maturation, dendritic development, and newborn neuron integration is also regulated (Ge et al. 2006). Newborn neuron survival is affected by glutamate, and the NMDAR is involved in such regulation (Tashiro et al. 2006). To investigate mechanisms of adult neurogenesis, many scientists have investigated transcription factors, cytoplasmic factors, epigenetic regulators, and niche receptors (Ma et al. 2010). In terms of the NSC differentiation mechanism, previous studies have reported two pathways. First, there is self-regulation including negative and positive regulation. The Notch signaling pathway directs NSCs to divide symmetrically and increase NSC numbers, which allows neural stem cells to maintain stem cell traits. Therefore, notch plays a negative role during neural differentiation. However, the bHLH transcription factor family plays an important role in positive regulation. The bHLH transcription factor family includes Mash1, NeuroD, Ngn1/Ngn2, and Math. These factors induce NSCs to undergo asymmetric division and produce new neurons. In addition, transcription inhibitor N-CoR prevents NSCs from differentiating into glial cells (Imayoshi and Kageyama 2014). Wnt also plays a key role during neural stem cell proliferation and differentiation. Wnt3 promotes neuronal fate commitment and proliferation of neural precursors in the adult SGZ (Kleber and Sommer 2004; Lie et al. 2005). Shh is activated in RG cells (Ahn and Joyner 2005) and required for their maintenance and establishment in the SVZ and SGZ of the adult brain (Bonaguidi et al. 2005). Second, exogenous signal regulation is also necessary, which includes cytokines and the niche. EGF and bFGF are involved in maintenance of NSC self-renewal, whereas PDGF, BDNF, IL-1, and LIF function during NSC differentiation. Regarding the niche, many classes of cells exist around neural stem cells. Some neural cells, glial cells, and matrix, including all kinds of glycoproteins and mucoproteins, construct a complex regulation network through crosslinked connections. Interestingly, the Notch signaling pathway also prefers to regulate niche components to prevent ependymal cells from differentiating into niche astrocytes through EphB2 in the adult SVZ region (Nomura et al. 2010). Further research is still necessary. In contrast, BMPs tend to promote NSC differentiation into glial cells rather than neurons in the adult brain (Bonaguidi et al. 2005; Lim et al. 2000). The BMP negative function can be antagonized by noggin and ngn-1 that are expressed by ependymal cells in the SVZ, and astrocytes and granule cells in the SGZ, respectively (Lim et al. 2000; Ueki et al. 2003). Inhibition of BMP signaling in adult SGZ neural precursors may result in their activation and an increase in neurogenesis. However, it can also lead to a decrease in precursors and loss of neurogenesis. Therefore, regulation of the BMP equilibrium is very important. Methamphetamine reduces dentate gyrus stem cell self-renewal by delaying the cell cycle, inducing more neural differentiation. During this process, it may rely on the NMDA signaling pathway. Sox2-positive cells are also decreased (Baptista et al. 2014). Hepatocyte growth factor promotes fibroblast proliferation and survival of cortical neurons, and regulates the maturation of neurons. Factors involved in the regulation and control of neuron occurrence are diverse, complex, and mutually connected, and more research should be performed for further exploration.
1.7 Prospects
Over the past several years, the field of adult neurogenesis has turned its focus from depicting the neurogenesis phenomenon and its regulation to explaining molecular mechanisms of neuronal development, stem cell regulation, and functional contributions. Previous studies have shown tremendous progress in understanding adult neurogenesis. The discovery of continuous neurogenesis in the adult nervous system has overturned a century old dogma and provided us with a new opinion on the plasticity of adult neurogenesis. Currently, we know the neural progenitor cell locations and their occurrence processes. However, it is unclear whether we know all progenitor cell locations. If a new progenitor cell pool was found, it may guide us to perform deeper research for more progress. Although many factors have been demonstrated to regulate adult neurogenesis, the relationship between factor regulation and functional recovery is still being determined. Proving the theory that the regulation mechanism guides regeneration after injury is our ultimate aim. Cognitive testing has provided a good method to understand human DG functions, which may indicate a possible relationship between neurogenesis and behavioral pattern separation. However, new methods are needed to evaluate neurogenesis in vivo (Tamura et al. 2016). More information about neurogenesis after injury will facilitate solving the problem from a new perspective. Gene expression advances have already provided us with a new insight into the molecular mechanisms and signaling cascades involved in neural differentiation and newborn neuron functional integration. Our goal is to search for more effective treatment strategies against neurological disease and further resolve our healthcare difficulties.
2 3D Culture
Cell culture plays important roles in biological research, drug discovery, and industrial applications. As the first cell line, Hela cells are an established cell culture and tool of biology. Traditional cell cultures are 2D, in which cells are grown on flat dishes as a monolayer. However, failures of 2D-cultured cells are due to the lack of natural microenvironments. Recently, increasing evidence has indicated that 3D cell culture systems more accurately represent the actual microenvironment compared with 2D culture systems. Thus, the 3D-cultured cell behavior is more reflective of in vivo cellular responses. In fact, studies have shown that the morphology and physiology of 3D-cultured cells are different from those of cells in 2D culture environments.
With the increase in the number of cell lines, parallel advancement in cell culture techniques, imaging, data acquisition, and analysis methods is being applied to 3D cell culture. Using such systems, the cells are co-cultured in different material structures with a variety of different types of cells in vitro. Cells in a vector with a 3D spatial structure can migrate, grow, and perform many functions. 3D culture methods mimic in vivo conditions and have become increasingly important in biological research and tissue regeneration.
2.1 3D Culture Methods
Traditional 2D culture usually grows cells as a monolayer on glass plates or, more commonly, polystyrene tissue culture flasks. However, 3D cell cultures grow cells as 3D aggregates or spheroids using a scaffold/matrix or in a scaffold-free manner as shown in Fig. 1 (Edmondson et al. 2014). 3D culture provides a biological microenvironment for cell proliferation, differentiation, and specific extracellular matrix secretion, which can be potentially used in many applications. 3D scaffolds are generated using various natural materials (e.g. collagen, gelatin, elastin, chitosan, chitin, fibrin, and fibrinogen) and synthetic substances (e.g. polystyrene, PCL, and 2-hydroxyethyl methacrylate). The most commonly used scaffolds are collagen, gelatin, agarose, fibronectin, and laminin, especially collagen. In 3D culture systems, a type I collagen matrix is commonly used because of many advantages such as easy processing, flexibility for live cell manipulation, and low cost. By changing the collagen concentration or introducing chemical cross-linking compounds, we can vary the pore sizes of collagen scaffolds, as well as the ligand density and stiffness, making it easy to change the gel structural properties (Baker et al. 2009).
2.2 3D Cultures Models and Scaffolds
A wide variety of techniques are currently applied to culture cells in 3D structures. These techniques can be classified into two main categories: scaffold based and non-scaffold based. Scaffold-based methods include biological scaffolds, polymeric hard scaffolds, and micropatterned microplate surfaces. Non-scaffold-based 3D culture methods include the hanging drop method, microfluidic 3D cell culture, and spheroids (Ravi et al. 2015).
2.2.1 Scaffold-Based 3D Cell Culture
2.2.1.1 Biological Scaffolds
Scaffolds can be made of natural or biological components such as proteins commonly found in in vivo microenvironments. They resemble in vivo microenvironments such as exposure to soluble growth factors, hormones, and other molecules that cells interact with in vivo, which can alter gene and protein expression. Biological scaffolds are advantageous compared with polymeric scaffolds, because the latter lack endogenous factors that act mainly to permit cell functions, but not promote appropriate cell behaviors. For example, the viability and proliferation of numerous cell types grown in hydrogels are increased because they are derived from natural sources, which promote many cellular functions (Caliari and Burdick 2016).
2.2.1.2 Polymeric Hard Scaffolds
3D tumor and tissue models designed to mimic in vivo environments can be created by seeding cells on pre-fabricated scaffolds or matrices. 3D cultures form because cells attach, migrate, and fill the interstices within the polymeric hard scaffold (Erickson et al. 2009). These scaffolds provide a physical support system for cell culture in vitro and have shown utility in in vivo tissue regeneration, because they have the potential to recreate the natural physical and structural environment of living tissue. Polymeric hard scaffolds are used for regenerative medicine and preclinical testing.
2.2.1.3 Micropatterned Microplate Surfaces
Micropatterned plate surfaces contain micrometer compartments regularly arrayed on the bottom of each well. Wells are square or round. The plate wells allow reduced consumption of culture medium in conventional cell culture. The well materials are mostly rigid and impermeable, and facilitate long term cell culture. They allow precise control of the size, shape, and location of the cultured cell in the micropattern combined with microtechnology as microengineered templates. Micropatterned cell culture provides numerous cell samples for subsequent analysis, which are cultured under the same condition.
2.2.2 Non-scaffold-Based 3D Cell Culture
2.2.2.1 Hanging Drop Method
HDP takes advantage of natural cell attachment by lacking surfaces to which cells can attach. The cells assemble spontaneously into a 3D structure. The method can be used to co-culture two or more different cell populations, elucidating the role of cell-cell or cell-matrix interactions in specifying spatial relationships between cells. It facilitates studies of embryonic development and tumor-stromal cell interactions in malignant invasion, and applications in tissue engineering. This simple method provides a tool to generate tissue-like cellular aggregates for molecular and biochemical analysis in a physiologically relevant model.
2.2.2.2 Spheroids
Microplates used for spheroids can be optically clear round bottomed with a black opaque body and have a covalent ultralow attachment surface to reduce cellular adhesion to the well surface. It is easy to exchange medium or apply drugs. The 3D spheroid cellular model can better simulate natural cellular interactions and mimic in vivo microarchitecture for more biologically relevant information. This method promotes formation of single spheroids by robust circular formation.
2.2.2.3 Microfluidic 3D Cell Culture
Microfluidic techniques allow spatial control of fluids in micrometer-sized channels, which have been explored to extend the physiological relevance of 3D culture models. Microfluidic 3D cell culture using microfluidic perfusion plates enables high quality cell culture in a 3D matrix. Cells in microfluidic platforms can be overlaid or embedded in the gel for long term perfusion culture and be used to create similar heterogeneous models. Multiple 3D ECM experiments can be performed at a fraction of the time. It also provides an additional level of complexity to the cellular environments in which cells have continuous nutrition and oxygen as well as waste removal through culture medium.
2.3 3D Culture Advantages
In 2D cell culture, cells are grown on flat polystyrene dishes. The unnatural material is very stiff. This stiffness may affect cell functions including cell-to-cell and cell-to-matrix attachments, as well as cell proliferation and differentiation (Ravi et al. 2015). Some important areas for which 3D cell culture systems are excellent models include studies involving drug discovery, cytotoxicity, genotoxicity, cell growth, apoptosis, survival, gene and protein expression, differentiation, and developmental changes. Similarly, co-cultures in 3D systems provide a better understanding of the cell interactions.
2.3.1 Cell Attachments
Cells adhere and spread on a surface and form unnatural cell attachments on a synthetic surface in conventional 2D culture. In contrast, cells attach to another cell and form natural cell-to-cell attachments in 3D cell culture. The cells synthesize and secrete extracellular matrix in three dimensions, which is the natural material to which cells attach. It is flexible and soft like natural tissues. It consists of native complex proteins and provides important biological cues to the cells. In 3D cell culture environments, cells exert forces on one another, move, and migrate. These cell-to-cell interactions in 3D cell culture include gap junctions that directly couple one cell to another. Gap junctions are much more widespread in 3D cell culture. Cells in a 3D system are closely proximate that enables surface adhesion molecules and surface receptors to bind to surface adhesion molecules. In addition, 3D culture maximizes cell-to-cell communication and signaling critical for cell functions. The phenotype or functions of 3D-cultured cells are more complex and similar to the functions of native tissues than those of 2D-cultured cells. For example, liver cells perform more active functions in 3D versus 2D. Muscle cells perform more muscle cell functions in 3D cell culture than 2D cell culture. Moreover, cartilage cells form more differentiated cartilage tissue in 3D compared with 2D.
2.3.2 Cell Proliferation
The proliferation of 3D cultured cells is rapid. The characteristics of 3D-cultured stem cells can be regulated by growth factors to direct differentiation to the desired lineage in a specific 3D environment. The physical, chemical and biological characteristics of a scaffold can be modulated by the features of the materials. In addition, cells cultured in a 3D environment show a different status from those in traditional 2D culture. Currently, 3D culture is used for stem cell research, cancer research, and drug screening. 3D culture has a huge potential for applications in scientific research as well as clinical applications.
2.4 Applications of 3D Cell Cultures
3D culture approaches facilitate better understanding of in vivo conditions. Cells in 3D environments are good models as “near-to-in vivo” systems and provide useful insights in various manners (Cukierman et al. 2001). The major advantage of 3D over 2D systems is the decrease in the gap between the cell culture system and cellular physiology. In conventional 2D conditions, some interactions are lost, including extracellular matrix components, cell-to-cell attachments, and cell-to-matrix interactions, which are important for cell proliferation, differentiation, and cellular functions in vivo. In addition, the integration of signaling pathways is mutual when cells are grown in basement membrane-like gels (Lee et al. 2007). 3D cell culture systems are applied to differentiation studies. For example, 3D systems are useful to understand the mechanisms of human osteoblast differentiation into osteocytes (Mc Garrigle et al. 2016) and study the role of osteocytes in bone metastasis and tissue engineering, as well as cancer research, gene and protein expression studies, drug discovery, and pharmacological applications.
2.4.1 Cell Function Applications
2.4.1.1 Cell Morphology Studies
Distinct differences in the morphology of cells have been found when grown as monolayers or in 3D cultures. A panel of breast cancer cell lines exhibited similar morphology as monolayers. However, in 3D culture, their morphologies could be classified into four types. For example, cell lines that formed grape-like morphology (e.g. MDAMB-361, AU565, and CAMA-1) become loosely associated with reduced cell-cell adhesion, which might explain why they have a highly metastatic potential as the tumor progresses.
2.4.1.2 Cytoskeleton Studies
The compositions of the cytoskeleton and extracellular matrix are very different in cells grown as monolayers or in 3D scaffolds. For example, smooth muscle cells on a 3D matrix have fewer focal adhesions, actin stress fibers, and a reduced cell surface area because of restriction by the surrounding matrix.
2.4.1.3 Cell Proliferation Studies
Some studies indicate that physiological stiffness is a well conserved inhibitor of mitogenesis at the elastic moduli range of 600–4,300 Pa in tissues isolated from mammary glands, thoracic aortae, and femoral arteries of mouse (Klein et al. 2009).
2.4.1.4 Cell Adhesion and Signaling Studies
In multicellular organisms, cells are surrounded by various other cells and a well-structured matrix. When grown in a complex environment, communication and adhesion to diverse elements appear to control cell behaviors. For example, morphology of the A549 cell line significantly differs in conventional monolayers and laminin-enriched 3D cultures. For the first 4 days, the cell doubling times are similar. During the next 4 days, resistance to X-rays increases. Global gene expression of cells in monolayers and 3D cultures is different. The genes may be involved in biological adhesion, cell adhesion, immune responses, or organ development. Cancer biologists and clinicians are increasingly interested in signaling pathways because of their roles in tumor development and influence on therapeutic modalities. For example, mTOR and tIGF-1R signaling cascades are activated in EWS patients because of their roles in chemotherapy resistance. When 2D EWS monolayers and 3D scaffolds are compared, 3D cultures cells show a high amount of phosphorylated IGF-1R, indicating abnormal IGF-1R/mTOR gene expression (Fong et al. 2013).
2.4.1.5 Cell Apoptosis and Motility Studies
Interactions of the cytoskeleton and the extra materials in 3D have important effects on various cell activities, including inhibition or acceleration of apoptosis.
The movement of cells into biomaterial scaffolds is a prerequisite for tissue repair and regeneration. There is a requirement for engineered biomaterials to optimize 3D-cultured cell migration.
2.4.1.6 Cell Physiology and Microenvironment Studies
3D cell cultures facilitate expounding the various cell functions, such as proliferation, adhesion, viability, morphology, and microenvironment, and responses to drugs. The environments surrounding cells play a key role in deciding cell differentiation fates and biological functions. Tumor microenvironments have been extensively studied, which, along with abnormal conditions, emphasizes the importance of a healthy milieu for cell behaviors. When oral squamous cell carcinoma cells were cultured on synthetic PLG scaffolds, major changes occurred. A study showed increases in secretion of bFGF, VEGF, and IL-8 by 23-, 2-, and 98-fold, respectively. Certain factors secreted in 3D cultures important for IL-8 expression are lost in 2D models.
2.4.1.7 Tumor Models and Cancer Biology Studies
3D cultures have gained attention in the field of regenerative medicine for their usefulness as in vitro models of solid tissues. Many cell types grown as 3D tumor spheroids have three layers, inner quiescent, central necrotic, and outer proliferating layers, which mimic the microenvironment of human solid tumors. In terms of tumorigenesis mechanisms, several studies show that 3D cell organizations are more novel, unanticipated visions, and represent an integral missing component in in vitro cancer studies.
2.4.2 Gene and Protein Expression
It will be interesting when we compare gene and protein expression in 3D cultures. Studies show significant differences in gene and protein expression of cells in 3D cultures compared with that of in vivo cancers.
2.4.3 Drug Discovery and Drug Response Studies
Apart from studies on direct drug effects, 3D systems have proven to be efficient models to study the synergistic effects of biologically important substances on cells. A 3D approach is simple but effective. Researchers treated oral squamous cell carcinomas in 3D PLG and 2D monolayers with the cytotoxic PI3-kinase inhibitor LY294002. The monolayers were sensitive to the drug, whereas 3D-cultured tumor cells with their microenvironment had significant resistance (Fischbach et al. 2007). Numerous drug candidates fail in clinic trials every year because of low efficacy, adverse events, and other reasons. Such failures indicate that the results obtained from 2D cultures may not predict in vivo results. For example, studies have failed to reach the primary endpoint of Zalutumumab, a fully human EGFR monoclonal antibody that has been demonstrated clinical benefit in refractory head and neck cancer (Luca et al. 2013). The differences in cellular responses of 2D and 3D cultures are possibly due to five aspects. First, there are some differences in the physical and physiological properties of 3D and 2D cultures. 2D-cultured cells are stretched out on an unnatural flat surface, but cells maintain normal morphology when cultured in scaffold regardless of biological or synthetic materials. Furthermore, therapeutic agents are often designed to target specific cell surface receptors. However, there are differences in the structure, localization, and arrangement of cell surface receptors on 2D- and 3D-cultured cells, which will affect the binding efficiency and their expression. The quantity and position of surface receptors in the same cell type are also different in the two culture environments. Third, gene and protein expression profiles are distinct between 2D- and 3D-cultured cells. 2D-cultured cells grow as a monolayer that is completely different from the in vivo environment, and their reactions to drugs are not accurate. Fourth, 2D-cultured cells are all in the same proliferation stage, whereas 3D-cultured cells are in different stages. Studies show that large spheroids are usually heterogeneous: the outer region is proliferating cells and the inner region is quiescent cells because of the lack of nutrients and gas exchange. Activation of cell proliferation may be needed for reactions to some drugs. Fifth, differences appear in the drug distance to cells and local pH. In a 2D monolayer, drugs diffuse to cells equally, but in a spheroid, the drug diffusion to cells may lead to variable concentrations. This effect depends on how deep the cells are from the surface. The depth of a cell is also related to the local pH.
2.5 Prospects
With the high demand for scientific research, cell culture systems have advanced rapidly over the past decades. As 3D culture systems emerge, they are exhibiting many features that better mimic those of in vivo environments. 3D culture systems are closer to animal models in many aspects (Yamada and Cukierman 2007). 3D culture systems have also been successfully introduced into translational medicine. To date, 3D culture systems have been reported to be appropriate for more than 380 cells lines. The 3D structure provides the possibility to analyze complex cell interactions, which was unable to be fulfilled by traditional 2D culture systems. 3D culture systems have good prospects for both basic and applied research. They have attracted broad interest to develop new support materials. It is evident that 3D cell culture models are better models than traditional 2D monolayer cultures, because 3D cultures improve cell-cell and cell-extracellular matrix interactions, and cell populations and structures resemble in vivo architectures. This approach provides an environment that mimics the in vivo environment. In the past several years, a number of 3D cell culture systems have been developed as experimental tools for diverse research purposes. Certainly, 3D culture systems have very high potential for application in many fields of cell research, disease modeling, and drug discovery.
In summary, 3D systems mimic in vivo conditions compared with 2D systems, making 3D culture more useful for studies of real cell functions and drug applications. 3D cell culture technologies generate more healthy cells and promote cell proliferation, which provide convenient approaches for further studies.
3 3D Neural Stem Cell Culture
Since Reynolds et al. first successfully isolated NSCs from the adult mammalian CNS and induced them to proliferate and differentiate into neurons and astrocytes, NSCs have been defined as self-renewing cells that generate neurons and glia of the nervous systems of all animals during embryonic development (Brent and Reynolds 1992). NSCs differentiate into neurons, astrocytes, and oligodendrocytes (Gage 2000). The classical method for isolating and culturing NSCs is to isolate cells from a particular region of neural tissue in the brain or spinal cord, or from embryonic and neonatal neural tissues. Conventional NSC isolation and culture methods can be divided into two types according to the NSC growth mode: the neurosphere culture method (three-dimensional culture) and monolayer culture method (two-dimensional culture) (Reynolds and Weiss 1992).
3.1 Overview of 3D NSC Culture
The earliest 3D NSC culture appeared in 1992. Reynolds et al. first used a neurosphere assay to promote NSC proliferation in vitro. The striata of adult mice were enzymatically dissociated, and cells were seeded in a culture dish in the absence of a supplementary substrate or adhesion factors (Fig. 2).
After 2 days in vitro, most cells had died, a few cells were undergoing cell division, and proliferating clusters of cells detached and formed a sphere of nestin-positive cells. This neurosphere culture method is simple to perform and the most common method for isolating, propagating, and studying embryonic and adult NSCs. However, in the neurosphere assay, both NSCs and NPCs proliferate and form clonal spheres. Therefore, to discriminate NSCs from NPCs in the suspension neurosphere assay, Louis et al. established the neural colony-forming cell assay in 2008 (Louis et al. 2008).
3D NSC culture is an artificially created environment in which NSCs are allowed to grow and interact with their surroundings in all three dimensions. Unlike a 2D environment, 3D cell culture permits cells to branch out in all directions in vitro, which is similar to how they would behave in vivo. In addition to the abovementioned neurosphere assay, there are commercially available culturing tools that claim to provide advantages for 3D NSC culture in vitro (Fig. 3).
In general, 3D NSC-culturing platforms can be classified as two types: scaffold-free and scaffold techniques (Cheng et al. 2013). Scaffold-free techniques apply another approach independent from using scaffolds, including the use of low adhesion plates, rotating bioreactors, magnetic levitation, and magnetic 3D bioprinting (Cheng et al. 2013; Souza et al. 2010). Scaffold techniques include the use of solid scaffolds, hydrogels, collagen, and other natural or artificial materials (Yang et al. 2012). Pivotal design requirements for polymer scaffolds used to 3D culture NSCs can be categorized as physical (e.g. rigidity and degradation rate), biochemical (e.g. biological activity), and practical (e.g. cost and reproducibility). Soft matrices are needed because they facilitate cell attachment and axon regrowth. They also demonstrate mimetic mechanical properties observed in vivo, which are required for neural tissue engineering. Scaffolds can be artificially modified for better binding of neurotrophic factors or NSCs. For example, in our previous study, a porous collagen scaffold was modified with chemical moieties for covalent crosslinking with cetuximab, an anti-EGFR antibody. When the functionalized collagen scaffold was implanted into a rat complete transection model, it promoted grafted NSCs to differentiate into neuronal lineages rather than astrocytes (Li et al. 2013). In addition, the anti-EGFR antibody in the collagen scaffold bound to EGFR expressed on the surface of NSCs. As a result, the functionalized collagen scaffold retained grafted NSCs at an injury site, preventing them from diffusing into cerebrospinal fluid.
In vitro 2D and 3D culture models of NSCs show differences in cell morphology, gene expression, cell-cell interactions, cell-matrix interactions, proliferation, migration, and differentiation (Knowlton et al. 2016; Prewitz et al. 2012). For example, outgrowth of axons from NSCs demonstrates a more aligned profile in a biomaterial scaffold, because it provides an aligned ECM structure that controls neurite morphology (Han et al. 2010). A significant difference is noted in gene expression between 2D and 3D NSC cultures, specifically for genes encoding cytoskeleton, ECM, and neuronal function proteins. Cells connected to surrounding cells through intercellular signaling and cell-cell interactions are regulated by the extracellular matrix (ECM). A great advantage of changing from traditional 2D monolayer culture to 3D culture is that the 3D condition stimulates these complicated cell-cell and cell-ECM interactions more than the 2D condition. Cell-cell and cell-ECM signaling define the tissue specificity and drives homeostatic maintenance. According to the organizing principle proposed by Bissell and colleagues, cell-cell and cell-ECM interactions regulate progression of a cell’s life cycle, including proliferation, migration, and apoptosis. These interactions are also vital for precise tissue formation during developmental stages (Gibbons et al. 2013). In general, compared with 2D culture, 3D NSC cultures as in vitro models aim to fill the gap between 2D NSC studies and the in vivo environment, especially for neural tissue regeneration therapies where there is little regenerative possibility. These 3D culture models are indispensable to support NSCs, allowing a natural flow of oxygen, nutrients, and growth factors, and possibly favoring neural cell regrowth (Cunha et al. 2011). Applications of these 3D cultures may be very useful for basic research of neural tissue structures and functions, designing disease models, engineering tissue for drug development, and generating replacement tissues with a patient’s genetic makeup.
3.2 Applications
The mammalian CNS has little capability for self-repair, and mature neurons are not able to proliferate after severe injury. Recent progress in the field of neural biology and tissue engineering offers a deeper understanding of CNS diseases. A primary example of the intersection between biology and tissue engineering is neural tissue engineering, a domain that gains a great deal from implementing the recent progress in 3D NSC culture in vitro. However, the demand for biomimetic neural tissue models and effective therapies remains unmet. The recent promotion of expanding 2D neural tissue engineering to the third dimension shows great potential to advance the field. Here, we present several of the most representative achievements in 3D culture of NSCs, which is classified into scaffold-free and scaffold techniques (Fig. 4).
3.2.1 Applications of 3D NSC Aggregates
NSCs tend to proliferate and form neurospheres when cultured under scaffold-free conditions. The applications of neurosphere culture in vitro include transplantation for the treatment of CNS diseases and constructing CNS disease models for diagnosis and screening potential drugs by acting as a predictor of clinical outcomes.
3.2.1.1 Transplanting Neurospheres for the Treatment of Neural Diseases
Studies have reported that functional recovery can be induced after transplantation of neurospheres into the injured brain or spinal cord of rodents. After transplantation in vivo, grafted neurospheres survive, migrate, and differentiate into three neural lineages at the injured site. Furthermore, they enhance angiogenesis to ameliorate hypoxic and ischemic conditions in brain disease or promote axonal regeneration and synapse formation with host tissue neurons in spinal cord injury. A previous study introduced the method of supplying neurospheres to the injured spinal cord through injection into the cerebrospinal fluid from the fourth ventricle or cisterna magna. The results showed that a large quantity of injected cells migrated to the lesion site and integrated with the host spinal cord tissue for repair. In another study, the authors transplanted hiPSC-NSs into a mouse Th10 lesion epicenter at 9 days after inducing the impact injury. The grafted hiPSC-NSs survived and migrated into the host spinal cord tissue. Moreover, they differentiated into neurons, astrocytes, and oligodendrocytes at 2 weeks post-surgery. About 50% of the grafted hiPSC-NSs had differentiated into neurons of which half were mature. The authors also observed synapse formation between hiPSC-derived neurons and host neurons as well as enhanced axonal regrowth and angiogenesis after injury. The transplanted hiPSC-NSs promoted motor functional and electrophysiological recovery after spinal cord injury (Nori et al. 2011).
3.2.1.2 Constructing CNS Disease Models for Diagnosis and Drug Screening
Recent progress in modeling CNS diseases from patient-derived cells, which are robust enough to produce large quantities of relevant cells, has greatly advanced clinical molecular and functional analyses. For example, Matigian et al. designed schizophrenia and Parkinson’s disease models based on patient-derived cells from the olfactory mucosa, which are regenerated from NSCs throughout life. Mucosa biopsies from patients and control subjects grew as neurospheres in vitro, and cell lines derived from neurospheres were obtained to analyze gene expression, protein expression, and cell functions including neurodevelopmental pathways in schizophrenia, and xenobiotic metabolism, oxidative stress and mitochondrial functions in Parkinson’s disease. The authors claimed to have identified new candidate genes and pathways for future investigation, and the disease model created from patient-derived neurospheres may provide better understanding of the disease etiology, diagnostics, and drug discovery (Matigian et al. 2010).
3.2.1.3 Neurospheres as a Predictor of Clinical Outcomes
Considering that formation of renewable neurospheres cultured in vitro could be a defining characteristic of certain CNS diseases, such as brain glioma, studies have been designed to evaluate the relationships among neurosphere formation, tumorigenic capacity of patient-derived glioma, and clinical outcomes. Tumor samples were cultured under neurosphere conditions and transplanted into mouse brains to examine whether they were tumorigenic. The authors observed that renewable neurosphere formation and tumorigenic capacity both predicted an increased risk of rapid tumor progression and patient death. In general, this study suggested that the ability of neural stem-like cells from patient-derived brain glioma to form neurospheres in vitro showed potential as a predictor of clinical outcomes (Laks et al. 2009).
3.2.2 Application of NSC-Loaded Scaffolds to Neural Tissue Engineering
Although the development of tissue engineering approaches for nervous systems is in its infancy, a growing number of important applications have emerged. NSCs encapsulated or adhered to natural or artificial scaffolds can be used in the field of modeling and treating nervous system diseases. They can also be used to fabricate biomimetic neural tissue to replace damaged tissue in patients.
3.2.2.1 Injured Neural Tissue Interacts Reciprocally with NSC-Seeded Scaffolds for Repair
There are several advantages of NSC-seeded scaffold transplantation over NSC aggregate grafts for treating neurological disorders. For example, scaffolds can be designed to incorporate the extracellular matrix composition of neural tissue, which provide mechanical properties for cell attachment. When the scaffold is transplanted in vivo, it will have good biocompatibility with host neural tissue and guide cells to grow in an ordered direction. In addition, scaffolds can be functionalized with different kinds of bioactive molecules to regulate adverse niches caused by injury, facilitating neuronal differentiation of NSCs at injury sites. Park et al. seeded NSCs onto a polyglycolic acid polymer scaffold and implanted the scaffold into infarction cavities of the mouse brain injured by hypoxia-ischemia. The authors observed that an intricate meshwork of many highly branched neurites of endogenous and grafted NSC-derived neurons emerged, and some anatomical connections appeared to be reconstituted. The results showed that transplantation of NSC-seeded scaffolds may gradually augment the constitutive reparative response by improving a series of interactions between grafted and host NSCs, including promotion of neurogenesis, connectivity, and reformation of injured cortical tissue.
3.2.2.2 Generation of Functional Artificial Neural Tissue to Replace Damaged or Diseased Tissues
Among the various neural regenerative procedures, one of the most promising is filling the injury site with artificial neural tissue. Lin et al. designed a multicomponent and micropatterned conduit seeded with NSCs and transplanted the nerve guidance conduit into a 10 mm-long defect of the sciatic nerve in male SD rats. The authors found that the biodegradable nerve guidance conduit provided both physical and biological guidance for axon regeneration at the injured site and offered an alternative for repairing sciatic nerve injury. Another study combined HUCB-NSCs and biodegradable scaffolds to form artificial neural tissue and transplanted the HUCB-NSC-seeded scaffold for hypoxic/ischemic brain injury repair. The results indicated that the 3D environment facilitated HUCB-derived NSC maturation and could be considered as a promising regenerative medicine application for nervous system repair (Jurga et al. 2009).
3.3 Challenges
Although it has been reported that NSC aggregates and NSC-loaded scaffolds can be successfully applied to neural tissue engineering, some challenges related to the transition from 2D to 3D culture systems still exist. For example, when NSCs are grown as spheroids in suspension culture, cells in the innermost layer have little access to nutrients and oxygen, and it is inconvenient to expel waste and CO2. Therefore, the growth of NSCs is suppressed in the center, and large neurospheres are unable to grow past a certain size in standard culture environments, which makes it difficult to research the later stages of development in vitro. The lack of sufficient vascularization is a crucial factor of 3D neural tissue engineering to be considered in future work (Novosel et al. 2011). An abundant blood supply will transport nutrients for NSC metabolism and provide enough oxygen to expel waste and CO2. Furthermore, formation of new blood vessels in and around grafted artificial neural tissue may contribute to integration of the transplant with host tissue.
Future work in this field will aim at improving long-term survival in 3D-cultured tissue by introducing vasculature. Current techniques include promoting the formation of new blood vessels from existing vessels (Chen et al. 2012). Other novel strategies for enhancing vasculature are electrospinning fibrous meshes, encapsulating blood vessels into synthetic scaffolds, or making use of existing blood vessels in natural scaffolds. Efforts should be directed to allowing mass delivery of oxygen and nutrients to achieve long term viability in neural tissue engineering.
3.4 Prospects
A long-term aim of using 3D culture models of NSCs is the application of personalized neural regenerative tissue engineering. Using a patient’s own NSCs to replace an impaired or diseased tissue or organ will decrease issues associated with limited donor supply. Moreover, implementation of 3D fabrication techniques will aid in the formation of biomimetic structures in neural tissue. Therefore, biomimetic tissue could be transplanted into the injured site and replace destroyed neural tissue, thereby ultimately ameliorating the quality of life for patients with developmental neurodegenerative disorders.
4 Microgravity of 3D-Cultured Neural Stem Cells
Recently, increasingly more attention has been focused on the influence of microgravity in tissue engineering. One of the most important characteristics of space is microgravity, i.e. the absence of gravity. There is universal gravity on Earth, which greatly affects cellular metabolism, morphology, signaling pathways, and secretion. It is of great importance to explore how microgravity affects physical, chemical, and biological processes during the exploration and interpretation of space.
Under microgravity, a large variety of fundamental physical phenomena are significantly changed or even eliminated completely. Spaceflight provides a real microgravity environment, but spaceflight opportunities are very rare and the costs are very high. Technical limitations of spaceflight studies seriously restrict tissue engineering research for space biomedicine. Spaceflight studies require perfect technical simulation systems to mimic the microgravity in space. To satisfy research needs, many instruments have been designed to simulate microgravity by the NASA Johnson Space Center for cell culture, such as the slow turning lateral vessel, high aspect ratio vessel, rotating wall bioreactor, and rotary cell culture system. The more widely used bioreactor machines imitate microgravity (Nickerson et al. 2004).
The principle of these microgravity-simulating rotation bioreactors is dependent on the balance between gravitation and the centrifugal force. These devices are commercially available to generate simulated microgravity and conveniently provide a good platform for microgravity studies. A great deal of microgravity-related experiments in cell biology have been performed using these instruments. Simulated microgravity provides a better approach for space studies concerned with biomedical research on Earth. Using these instruments, we may better understand the effect of microgravity on the functions of stem cells.
4.1 Effect of Microgravity on the Structure and Functions of Stem Cells
Stem cells are widely used in tissue engineering and cell therapies to treat or prevent many diseases. They might play an important role in guiding and shaping the future of clinical medicine. Stem cell technologies are used to induce directed differentiation and maintain stem cell self-renewal. Proper exploitation of these methods may lead to the generation of 3D human tissue models that have great potential for future medical therapies. It has been reported that stem cells tend to gradually lose their polarity and differentiation ability in long term 2D culture. In vitro 3D cultures are emerging as novel systems to study tissue development, organogenesis, and stem cell behavior in vitro. Understanding the effect of microgravity on stem cell functions will provide important information for treatment and preventive strategies to deal with medical problems during space exploration.
In comparison with traditional static adherent culture, microgravity provides a very specialized environment for cell culture. Many researchers have pointed out that microgravity switches 2D cell culture to 3D. In microgravity, cells will not settle at the bottom of the culture dish. They float freely in the culture medium, which contributes to interacting each other and assembly together to form multicellular 3D spherical structures without a scaffold in microgravity. Dissociated cells tend to assemble together for tissue-like self-organization. Microgravity provides an opportunity for dissociated floating cells to self-aggregate and interact with each other freely. Cells are inclined to float in the culture medium and not sediment in a low shear-modeled microgravity environment. In particular, 3D cell aggregates formed under microgravity are larger than those engineered in conventional bioreactors or 2D cultures.
Recent studies have shed light on some molecular mechanisms concerning how microgravity affects the morphology and function of cells. A thorough understanding of gene expression is important to reveal the molecular mechanisms driving microgravity-induced alterations. Various cell types alter their gene expression profiles when exposed to microgravity, but the underlying mechanism has not been clearly elucidated yet. Revealing the molecular mechanisms of stem cells in microgravity will contribute to understanding the intricate regulation of cell growth and functions. Understanding the molecular and cellular mechanisms of alterations in microgravity is urgently needed to meliorate space medicine. It will cause a profound biotechnological effect on regenerative medicine. Many studies have shown that microgravity alters the global expression profile of genes. When murine bone marrow stromal cells were cultured in osteo-inducing medium during spaceflight, microarray results revealed changes in the expression of 1,599 genes. The expression of most cell cycle-related genes was decreased, suggesting that cell proliferation was inhibited in microgravity. Moreover, the cells tended to express neural associated genes (Monticone et al. 2010). The same conclusion was reached by Professor Ma and his team. Their results indicated that rat mesenchymal stem cells tend to differentiate into neurons in simulated microgravity (Chen et al. 2011). Further studies revealed that the gene expression of EBs during differentiation was changed in microgravity compared with normal conditions. The results of quantitative real-time PCR indicated that expression of most differentiation-related genes was changed (Blaber et al. 2015).
Studies of stem cells in microgravity have made some progress. It is well known that microgravity affects the cell proliferation rate, metabolism, and physiology. Previous studies have shown that microgravity promotes the proliferation of stem cells by sustaining their differentiation ability (Yuge et al. 2006). Cells will not settle at the bottom of a culture dish and float freely in the culture medium, which contributes to interactions each other and assembly together to form multicellular 3D spherical structures without a scaffold in microgravity (Grimm et al. 2014). mESCs were the first ESCs applied to study biological effects of microgravity using a multidirectional G force generator device that simulates microgravity. The major changes of mESCs in microgravity were a decrease of adhesion, increased apoptosis, and delay in the DNA repair process (Wang et al. 2011). The differentiation of ESCs is also significantly affected by microgravity. The formation speed of embryoid bodies was faster in simulated microgravity than in conventional culture, which was accompanied by multidirectional differentiation into hepatocyte-like cells, heart muscle cells, endothelial cells, blood vessel cells, and various other cells types. Furthermore, rat embryonic stem cells form spheres that grow in cultivation medium without LIF, serum, or a feeder layer in microgravity. LIF and a feeder layer of MEFs play pivotal roles in maintaining the pluripotency of ESCs. The results indicated that simulated microgravity may provide a better environment for ESC culture (Kawahara et al. 2009). These findings provide valuable information on ESC-related regenerative medicine. 3D aggregates may be grown to form tissue-like structures in the future. When grown in a 3D environment, ESCs self-organize and differentiate into various cell types of the three germ layers, such as endoderm- and ectoderm-derived cells, mimicking their in vivo counterparts.
As another extensively studied type of adult stem cell, MSCs are widely used in tissue engineering. MSCs can be induced to differentiate into various cell types such as myocardial cells, osteocytes, adipocytes, and hepatocytes. When dog MSCs were cultured under microgravity conditions, both the morphology and growth rate of the cells were better than those of cells cultured under normal conditions. Microgravity decreased the proliferation potential of murine BMSCs in the KUBIK aboard space mission ISS 12S (Soyuz TMA-8 + Increment 13) from March 30 to April 8, 2006 (Monticone et al. 2010). Several studies have demonstrated that the differentiation of MSCs is strongly affected by gravity. Osteogenic differentiation is suppressed by microgravity, whereas differentiation of adipocyte, cartilage, nerve, and endothelial lineages was improved in microgravity. Directed differentiation of MSCs into the cartilage phenotype is significantly enhanced by microgravity compared with normal gravity (Wang et al. 2007). When MSCs were cultured in microgravity, the osteogenic differentiation-related gene RUNX2 was suppressed, whereas adipocyte differentiation-related genes, such as glucose transporter 4, adipsin, leptin, and PPARγ2, were upregulated. Neural differentiation-related genes, such as those involved in neural development, neuron morphogenesis, transmission of nerve impulses, and synapses, were also activated during spaceflight (Monticone et al. 2010). The changes in the differentiation potential of MSCs under microgravity indicated that microgravity is expected to facilitate the clinical application of MSCs.
Neural stem cells play pivotal roles during CNS development by differentiating into multiple cell types such as neurons, astrocytes, and oligodendrocytes. The proliferation of human neural stem cells is also affected by microgravity. Expression of β-adrenoreceptor is induced by microgravity. Furthermore, the formation of cAMP upregulates CREB pathways, and PKA is activated by microgravity. The amount of mitochondrial mass and the ATP level are both elevated by microgravity, demonstrating that microgravity promotes proliferation of neural stem cells by improving the function of mitochondria (Chiang et al. 2012).
The microenvironment of stem cells is formed by the interaction among cells and the relationship between cells and the extracellular matrix. Under microgravity conditions, stem cell niches are apparently different from those under traditional 2D conditions. Stem cell niches play pivotal roles in maintaining the balance of self-renewal and differentiation of stem cells, which determines the fate of stem cells. Currently, scientists have begun focus on the effect of simulated or real microgravity on the structure and function of stem cells.
4.2 Effect of Microgravity on Tissue Engineering
Tissue engineering has provided a broad clinical prospect to treat tissue or organ defects. The study of tissue engineering in simulated or real microgravity environments is a hot research area in space medicine. Space provides low shearing stress microgravity, and microgravity induces many changes in organisms during spaceflight. Recent spaceflight studies have verified the role of microgravity in modulating stem cell-related tissue regeneration. When EBs were exposed to microgravity conditions for 15 days, their differentiation ability was inhibited, whereas self-renewal was maintained. When EB cells cultured in microgravity were returned to normal gravity, they showed greater stemness and more readily differentiated into cardiomyocytes (Blaber et al. 2015). This alteration of the mESC differentiation capacity could have significant implications in the field of human tissue engineering and the use of stem cells to regenerate adult tissues.
In vitro 3D culture has gained a lot of research interest in tissue engineering. As an emerging novel system, 3D culture has been used to study tissue development, organogenesis, and stem cell behavior in vitro. 3D cell culture is widely used in tissue engineering and regenerative medicine. When stem cells are cultured in a 3D environment, they form organoids and can be propagated for a long period in vitro. To better understand the mechanism of the formation of multicellular 3D structures, we performed analyses from various perspectives, including signal transduction, cell adhesion, apoptosis, and the extracellular matrix induced by altered gravity conditions. Many studies have suggested that the formation of 3D aggregates under microgravity can be divided into three phases of regulation from 2D to 3D growth: (1) cells undergo changes in interactions with the ECM; (2) under microgravity conditions, the expression of proapoptotic factors increases, which increases the apoptosis rate; (3) many signaling and kinase-dependent pathways are altered during the transition from 2D to 3D (Grimm et al. 2014). Wnt signaling can be altered under microgravity conditions. The majority of Wnt pathway-related genes are downregulated by microgravity (Lin et al. 2009). Many kinds of signaling pathways form a complicated network to govern cell actions. Numerous signaling pathways are influenced by microgravity in the interior of the cell (Puca et al. 2012).
Tissues such as 3D constructs can be maintained for long periods in simulated microgravity created by RWV bioreactors. Potential advantages of using a simulated microgravity environment for tissue engineering have been demonstrated by several studies. Devices that generate simulated microgravity are widely used in tissue engineering, which provide better in vitro model systems for spaceflight studies. Various cell types exposed to real space or simulated microgravity conditions can be grown in the form of 3D tissues. Primary porcine hepatocytes gathered spontaneously to form high density 3D aggregates and maintained important metabolic functions when cultured in the NASA rotary bioreactor for 21 days (Nelson et al. 2010). The random positioning machine system produces simulated microgravity, which can be used to engineer cartilage grafts with less cells (Stamenkovic et al. 2010).
Space medicine research can facilitate development of new strategies for tissue engineering. The increasing scientific and medical relevance of such research is evidenced by the growing number of reports in which advanced bioreactors are used for in vitro studies in physiologically relevant cell and tissue models. Tissue engineering in microgravity has had a tremendous effect on space research, biomedical sciences, and its applications on Earth.
5 Systematic Analysis of Molecular Mechanisms Involved in the Proliferation and Differentiation of NSCs During Spaceflight
The SJ-10 satellite provided a platform to elucidate the molecular mechanisms of cell alterations in biological, physical and chemical processes in space. The recovery cabin maintained orbit for 12 days in space. The aim of the SJ-10 satellite was to understand the effect of spaceflight on the growth and differentiation of NSCs. 3D cell culture systems have advantages by providing more physiologically relevant information and predictive data, which closely mimic in vivo conditions compared with traditional 2D cell culture systems. A biomaterial-based 3D culture system was adopted for differentiation analysis during spaceflight.
5.1 Cell Culture and Harvesting
Rat NSCs were isolated from telencephalon tissue and cultured in proliferation or differentiation media for 12 days during spaceflight of the SJ-10 satellite. Simultaneously, cells were cultured on the ground under similar culture conditions as the control group (Fig. 5). During spaceflight, the cells were cultured in fully automated bioreactors designed by the Shanghai Institute of Technical Physics of the Chinese Academy of Sciences. The automatic cell culture device changed the culture medium, collected images, and fixed cell automatically. When the recovery cabin of the satellite returned to Earth, some cells were fixed by RNA ladder stabilization reagent or paraformaldehyde, while live cells were used to detect cell viability.
5.2 Preparation of 3D Cell Cultures
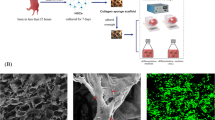
A collagen sponge scaffold was chosen for 3D NSC culture, which was prepared from bovine collagen. The diameter of the round collagen sponge sheet was approximately 5 mm and its thickness was about 1 mm. Our previous studies had shown that cells maintain a good condition when attached on collagen sponge scaffolds by SEM. Therefore, the collagen sponge scaffold had good biocompatibility with NSCs. Before departure of the satellite, a cell suspension (3 × 105 cells per collagen scaffold) was seeded on the scaffold for cell attachment and then cultured in differentiation medium.
5.3 Remote Sensing Data of Cell Proliferation
For proliferation analysis, the single cell suspension grew as spheroids in the proliferation medium without outside interference. The diameter and size of the spheroids can be used as key indicators of cell proliferation. During the spaceflight, real-time image data, which reflected the dynamics of cell proliferation, were acquired by the automated imaging system at regular times, and then the images were instantly transmitted back to the telemetry data center on the ground. Based on the images, the size of neurospheres in the space group was smaller than those in the ground group cultured in the same system on Earth. The smaller volume of the neurospheres indicated that the growth rate of NSCs was inhibited in space.
5.4 Cell Viability Analysis
The viability of cells after spaceflight was evaluated by FDA/PI staining, a fluorescence-based universal method. Cells were incubated with the staining solution (100 µg/ml FDA and 60 µg/ml PI) for about 5 min and then transferred to PBS. In the staining process, living cells are stained with FDA, while dead cells are stained with PI. The results of FDA/PI staining indicated that both the proliferative cells of neurospheres and differentiated cells cultured on the scaffold exhibited high cell viability after returning from space.
5.5 Analysis of Immunofluorescence Staining
Immunofluorescence staining was performed by a slightly modified version of published procedures (Cui et al. 2016). Three pivotal differentiation-related markers, Tuj1 (neuron-specific tubulin III, early neuron marker), GFAP (astrocyte marker), and Map2 (mature neuron marker), were chosen as the evaluation indicators of differentiation. First, the cells were incubated with primary antibodies overnight at 4 °C and then incubated with the secondary antibodies for 1 h at room temperature. Fluorescence images were obtained by a scanning laser confocal fluorescence microscope. In the space group, the expression of Map2 was significantly increased compared with the ground group. In contrast, the expression of GFAP was decreased during spaceflight. Nevertheless, the spaceflight had no effect on the expression of Tuj1. The expression of Tuj1 was steady in both space and ground groups. These results led us to speculate that NSCs tended to differentiate into neurons, while the differentiation of NSC to astrocytes was inhibited in space.
5.6 RNA-Seq Preparation and Sequencing
To understand the molecular mechanism driving the differentiation and proliferation of NSCs in space, we performed RNA-seq to understand the alteration of the genome-wide transcription profile caused by spaceflight. Total RNA was extracted using Trizol reagent (Sigma Chemical, St. Louis, MO) from all groups of NSCs. Each group included three biological replicates. An RNeasy Plant Mini Kit (Qiagen Sciences, Valencia, CA) was used to purify the RNA extracts. The quality of RNA was assessed by agarose gel electrophoresis and the ND-1000 NanoDrop spectrophotometer 2000 (NanoDrop Technologies, Wilmington, DE). The concentration and integrity of RNA indicated that the RNA quality of all samples was high. The Illumina X Ten platform was chosen to construct RNA libraries and perform RNA-seq of cell samples cultured in space and ground groups. Transcriptome libraries were constructed from 1 µg total RNA with the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA).
Numerous studies have shown that the Illumina platform offers significantly increased sequencing depth, and it may generate millions of 150 bp paired-end high quality sequence reads. In the proliferation group of NSCs, we obtained 46,081,266 and 46,759,526 paired-end reads from space and ground group libraries, respectively. We also obtained 48,036,974 and 48,395,940 paired-end reads for space and ground group libraries in the differentiation group of NSCs, respectively. Low quality reads were filtered and removed. Filtered data were logarithm-transformed and normalized by the quantile method. Clean reads resulting from a filter step were pooled together and then mapped to the reference genome. The matching rates of the read pairs, which were mapped to the rat reference genome in the Ensembl database, release 82, in the four groups were 89%, 85.46%, 89.46%, and 87.69%, respectively. The reference genome sequence of 22,268 Rattus norvegicus genes and annotation data were downloaded from the UCSC website (http://genome.uscs.edu). The relative expression of differentially expressed genes between space and ground groups was measured in fragments per kilobase of transcript per million mapped reads, and represented as the FPKM unit. Based on expected number of fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) with a false discovery rate (FDR)-adjusted p-value of ≤0.05 and |log2 ratio|, ≥1, 3279 annotated differentially expressed genes were identified in the differentiation group of NSCs. Moreover, 1589 differentially expressed genes were screened in the proliferation group of NSCs between space and ground groups.
The original sequencing reads were processed by several steps. First, the quality of sequencing data was checked by the NGS QC Toolkit, and then low quality reads, which contained ploy-N regions, were removed. Tophat was used for mapping these clean reads to a reference rat genome to identify known and novel splice junctions and generate read alignments for each sample (Kim et al. 2013). Next, cufflinks was used to calculate and normalize the FPKM value of each gene (Trapnell et al. 2012). The read counts of genes were processed and analyzed by htseq-count (Anders et al. 2015). The gene fusions from the RNA-Seq data were identified by the deFuse program (McPherson et al. 2011). The identification of DEGs was analyzed by functional estimator SizeFactors and nbinomTest (Anders et al. 2012). The thresholds used to screen the significantly differentially expressed genes were set as p-value < 0.05 and absolute fold change >2.
5.7 Hierarchical Clustering Analysis
Hierarchical clustering analysis of major DEGs was performed using the FPKM values of proliferating or differentiated NSCs in space and ground groups. In the proliferation group, the heatmap of hierarchical clustering showed that the differential gene expression patterns of selected DEGs were related to stemness, the cell cycle, or proliferation (fold-change >2 or P < 0.5) using the FPKM values. The expression of cell proliferation marker Ki67, cell cycle regulators (CDKN2a, CDKN2b. and CDK1), and the transcription factor Pax6 was significantly changed during spaceflight. As important cell cycle regulators, the expression of cyclin-dependent kinase CDK1 was downregulated, while the expression of negative cell cycle regulators CDKN2a and CDKN2b was upregulated in the space group. The alteration of cell cycle regulators may induce cell cycle arrest and influence cell proliferation. The expression of cell proliferation marker Ki67 was downregulated in the space group. In addition to these genes, many self-renewal- and neurogenesis-related genes, such as Pax6, Rest, and Klf4, were upregulated in the space group (Fig. 6a). In the differentiation group, the heatmap indicated that the expression of differentiation-related genes and many epigenetic regulator genes in the space group was also significantly altered. The expression of two neuron markers, Map2 and Tuj1, was elevated, while expression of astrocyte marker GFAP was downregulated. In line with the expression of neuron markers, the expression of PTEN, HDAC2, Wnt5A, Neurog2, and Ezh2 genes was also upregulated in the space group (Fig. 6b). Previous studies have demonstrated that these genes may facilitate neural differentiation of NSCs (Barton and Fendrik 2013; Bengoa-Vergniory and Kypta 2015; Chen et al. 2015; Lange et al. 2006; Sher et al. 2012; Sun et al. 2011). Considering that Id3 partly determines neural stem cell differentiation into astrocytes, the downregulation of Id3 shown in the heatmap of the space group may explain why the astrocyte marker GFAP was downregulated.
5.8 Gene Ontology and KEGG Enrichment Analysis
Considering that functional and pathway analyses are powerful tools to gain insights into the underlying biology of differentially expressed genes, GO enrichment and pathway analyses of DEGs were performed using the GO website and KEGG. The Entrez IDs of differentially expressed genes were entered into the R package cluster Profiler to perform GO enrichment analysis (Yu et al. 2012). Functional annotation analysis of DEGs was performed using the DAVID tool (http://david.abcc.ncifcrf.gov/) for biological interpretation (Huang et al. 2009). GO analysis was carried out using the DAVID functional annotation tool, according to the enrichment scores. KEGG enrichment analysis was also used to analyze the pathways of DEGs. Cluster Profiler was used to perform KEGG enrichment analysis after establishing the parameters. Optional interaction genes were extracted among the screened DEGs using BioGrid and String databases (Chatr-Aryamontri et al. 2017; Szklarczyk et al. 2015).
In the proliferation group of NSCs, GO enrichment analysis showed that the DEGs were enriched in the terms of cell proliferation, calcium ion binding, apoptotic process, cell junction, cytoskeleton, and microtubule. Pathway analysis based on the KEGG database showed that these genes were significantly enriched in MAPK, Rap1, and Ras signaling pathways. Additionally, GO enrichment and KEGG pathway analyses showed that DEGs in the proliferation group of NSCs were remarkably enriched in the cell cycle, apoptosis, cytoskeleton, and cell adhesion (Fig. 7).
In the differentiation group of NSCs, GO enrichment analysis showed that the DEGs were most enriched in oxidative stress, apoptosis, cell adhesion, and some signaling pathways. Moreover, KEGG enrichment analysis revealed that the enriched pathways were Wnt signaling, cell adhesion, and TNF and NF-κB signaling pathways (Fig. 8).
The analyses indicated that many biological processes and signaling pathways were affected by space. The GO terms and KEGG classifications served as indications of significantly different biological processes in NSCs between space and ground environments, which could guild further studies to determine their functions in space. Among the variable cell signaling pathways, Wnt and mTOR signaling pathways have attracted much more attention, because the expression of many important genes involved in the Wnt signaling pathway varied in the space group of NSCs (Fig. 9).
5.9 Real-Time PCR Analysis of DEGs
To confirm the accuracy of RNA-Seq data, we performed qRT-PCR on 16 randomly selected DEGs to validate the RNA-Seq results. All 16 genes were related to self-renewal and differentiation. First, HiScript II Q RT SuperMix for qPCR System (Vazyme, R223-01) was used to reverse transcribe RNA into cDNA. Then, amplification was performed in 96-well optical reaction plates (Applied Biosystems, Darmstadt, Germany) using a StepOnePlus amplification and detection system (Applied Biosystems Inc., Foster City, CA). The standard reaction mixture contained 100 ng cDNA, a pair of primers, and the SYBR Green I PCR master mix (Applied Biosystems) in a total volume of 25 µl. The standard conditions used for real-time PCR were 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C for denaturation and 30 s at 55 °C for annealing and elongation. Data were analyzed using StepOne v2.0 software (Applied Biosystems). Data of each sample were normalized to ACTB as an internal control. In accordance with RNA-seq data, the results of qRT-PCR showed that the expression of six genes (Sox2, Notch1, Pax6, Cdkn2b, Cdkn2a, and Cdk1) in the proliferation group was upregulated in the space group. In addition, the expression of proliferation marker ki67 in the proliferation group was downregulated in the space group. Seven genes (Pten, Hdac2, Wnt5a, Neurog2, Ezh2, Ncam1, and Id3) involved in neural differentiation were selected to perform Q-PCR. The expression of these seven genes was elevated in the space group, which has been reported to facilitate neural differentiation of NSCs (Barton and Fendrik 2013; Bengoa-Vergniory and Kypta 2015; Chen et al. 2015; Lange et al. 2006; Sher et al. 2012; Sun et al. 2011). The Q-PCR results validated the accuracy of RNA-seq data.
5.10 Differential Expression and Variation Analysis of miRNAs
As a critical post-transcriptional regulator of gene expression, miRNA plays pivotal roles in regulating differentiation of NSCs. Because numerous gene targets can be regulated by a single miRNA, miRNAs are now considered as important regulators in the proliferation and differentiation processes of NSCs. We also performed small RNA sequencing to gain a large amount of information about the microRNA transcriptome. Differentially expressed miRNAs were screened using DESeq 2 with the thresholds of a multiple hypothesis-corrected p-value of <0.05 and absolute fold change of >2. Quantitative analysis of the differentially expressed miRNAs and systematically organizing the miRNA-target interactions are the basis for thoroughly understanding miRNA-mediated regulatory networks and the underlying molecular mechanisms. In the proliferation group of NSCs, 93 miRNAs were downregulated, which was accompanied by 90 upregulated miRNAs in the space group compared with the ground group. A total of 51 miRNAs were upregulated and 91 miRNAs were downregulated in the differentiation group of NSCs in the space group compared with the ground group. A large number of differentially expressed miRNAs mainly regulated the proliferation, migration, and differentiation of cells. Furthermore, the target genes of these differentially expressed miRNAs were involved in the cell cycle, cell adhesion, and many cell signaling pathways such as Notch and Wnt, indicating that miRNAs serve as important regulators by regulating downstream target genes related to the cell cycle, cell growth, metabolic processes, and cell signaling in the differentiation and proliferation of NSCs.
Among these differentially expressed miRNAs, the expression of let-7, miR-128, miR-378, miR-138, miR-338, and miR-330 was increased in the space group of NSCs which has been reported to have the an inhibitory effect on the proliferation of NSCs (Godlewski et al. 2008; Zhao et al. 2010; Barca-Mayo and Lu 2012; Choi et al. 2016; Cimadamore et al. 2013; Huang et al. 2015). In addition, the expression of differentiation-related miRNAs miR-125, miR-124, miR-134, miR-17, and miR-21 was elevated in the space group of NSCs (Cui et al. 2012; Xu et al. 2012; Gioia et al. 2014; Jiang et al. 2016; Mao et al. 2016).
5.11 Functional Enrichment Analysis of DEG miRNAs and Construction of the miRNA-mRNA Regulatory Network
To predict the potential functions and target genes of differentially expressed miRNAs identified in the space group of NSCs, functional enrichment of predicted target genes was performed. The systematic analysis results of miRNA-mRNA regulatory functions contributes to exploring the possible molecular mechanisms involved in spaceflight. The expression of differentially expressed miRNAs and their target genes was diverse. miRNA target prediction software miRanda and TargetScan were used to obtain information about target genes of these miRNAs. The principle of screening was to satisfy two conditions: matching score >150 and binding energy <−30 kcal/mol. The data were further filtered using expression correlation. Following preliminary analysis of differentially expressed genes, integrated analysis of miRNA and mRNA expression profiles involved in the proliferation or differentiation of NSCs was performed. A core miRNA-mRNA regulatory network that participated in the proliferation or differentiation of NSCs was constructed. The miRNA-mRNA network-based analysis could be useful to reveal the regulating relationships of DE miRNAs and their target genes. The systematic analysis of the expression and functional interaction involving miRNA and mRNA enables identification of the predicted miRNA-target gene network pattern and functional candidates of miRNA-mRNA pairs associated with spaceflight. The miRNA-target gene regulatory network was constructed with the miRNA-target gene pairs by Cytoscape software with the prefuse force directed layout algorithm (Fig. 10). This network may contribute to better understanding of the roles of spaceflight in regulating the proliferation and differentiation processes of NSCs.
Differentially expressed miRNA genes between space and Earth groups were analyzed by databases such as GO and KEGG for functional enrichment analysis. The gene expression data of these genes were extracted and imported into Cytoscape, and then the ExpressionCorrelation plugin computed the coexpression network based on chosen genes (Shannon et al. 2003). By integrating large scale analysis of the transcriptome, miRNAome data and bioinformatics analysis, the GO and KEGG analyses indicated that the proliferation, cell cycle, differentiation, adhesion, apoptosis, and migration of NSCs are affected by spaceflight. Furthermore, the GO analysis of DEGs combined with mRNA-miRNA regulatory network analysis indicated that many pivotal genes, miRNAs, and signaling pathways were involved in the proliferation or differentiation of NSCs during spaceflight. In the proliferation group of NSCs, many target genes involved in regulating the proliferation of NSCs were regulated by these differentially expressed miRNAs between space and ground groups, such as cyclin D1, Ccnd2, Lin28b, E2F2, Hmga1, and Hmga2. Moreover, several neural differentiation-related genes, such as Nestin, Smad4, Stat3, and sp1, were regulated by some differentially expressed miRNAs in the differentiation group of NSCs between space and ground groups. The results also showed that Pax6 and RunX2 genes involved in self-renewal of NSCs were elevated in the space group. In contrast, the expression of oligodendrocyte markers Gal and Olig2 was decreased.
Apart from direct regulation of target genes involved in the differentiation or proliferation of NSCs, miRNAs may also regulate differentiation or proliferation processes via regulating cell signaling pathways. For example, the neural differentiation-related genes Plxna1, Myo6, and Lif are downstream targets of miR-125. Moreover, the Wnt signaling pathway can be regulated by miR-125 in the differentiation group of NSCs. In addition, many target genes of the differentially expressed miRNAs are involved in this pathway. Using bioinformatics analysis, we constructed a network among stem cell- and Wnt signaling-related genes to further elucidate the role of Wnt signaling during spaceflight. The correlation analysis indicated that certain Wnt signaling-related genes, which have been demonstrated to facilitate neuron differentiation, were significantly upregulated in the space group of NSCs (Lange et al. 2006). Taken together, the mRNA-miRNA networks supported the presumption that exposure to space caused alterations of proliferation and differentiation processes via different regulatory mechanisms in NSCs. Neural differentiation of NSCs was facilitated in space, whereas cell proliferation was suppressed during spaceflight.
6 Summary
As a recoverable satellite, the recovery cabin of the SJ-10 satellite maintained orbit for 12 days in space. The satellite provided an effective, open, and comprehensive platform to study space life and microgravity science, which allows elucidation of the molecular mechanisms of cell alterations in biological, physical and chemical processes in space. When NSCs are exposed to space, the cells maintained self-renewal in the proliferation medium despite a decrease in their growth rate. In addition, the cells were inclined to differentiate into neurons when cultured in differentiation medium in space. Detailed genomic analyses of NSCs after spaceflight may help us to reveal the molecular mechanisms of their differentiation and proliferation states in space environments.
Abbreviations
- 3D:
-
Three-dimensional
- 2D:
-
Two-dimensional
- BDNF:
-
Brain-derived neurotrophic factor
- bFGF:
-
Basic fibroblast growth factor
- bHLH:
-
Basic helix-loop-helix
- BMP:
-
Bone morphogenetic protein
- BMSCs:
-
Bone marrow stromal cells
- CNS:
-
Center nervous systems
- CREB:
-
CAMP response element-binding protein
- DE:
-
Differentially expressed
- DEGs:
-
Differentially expressed genes
- DG:
-
Dentate gyrus
- ECM:
-
Extracellular matrix
- EGF:
-
Epidermal growth factor
- EGFR:
-
Epidermal growth factor receptor
- ESCs:
-
Embryonic stem cells
- EWS:
-
Ewing sarcoma
- FDA:
-
Fluorescein diacetate
- FPKM:
-
Fragments per kilobase of transcript per million mapped reads
- GFAP:
-
Glial fibrillary acidic protein
- GO:
-
Gene Ontology
- HDP:
-
Hanging drop plates
- HGF:
-
Hepatocyte growth factor
- hiPSC-NSs:
-
HiPSC-derived neurospheres
- HUCB-NSCs:
-
Human umbilical cord blood-derived NSCs
- IGF-1R:
-
Insulin-like growth factor-1 receptor
- IL:
-
Interleukin
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- MAG:
-
Myelin-associated glycoprotein
- Map2:
-
Microtubule-associated protein-2
- MEFs:
-
Mouse embryonic fibroblasts
- mESCs:
-
Mouse embryonic stem cells
- MSCs:
-
Mesenchymal stem cells
- mTOR:
-
Mammalian target of rapamycin
- NSCs:
-
Neural stem cells
- NPCs:
-
Neural progenitor cells
- OMGP:
-
Oligodendrocyte myelin glucoprotein
- PBS:
-
Phosphate buffered saline
- PDGF:
-
Platelet growth factor
- PI:
-
Propidium iodide
- PLG:
-
Poly(lactide-co-glycolide)
- PNS:
-
Peripheral nervous system
- RG:
-
Radial glial-like
- RMS:
-
Rostral migratory stream
- SJ-10 satellite:
-
SJ-10 recoverable microgravity experimental satellite
- SGZ:
-
Subgranular zone
- SEM:
-
Scanning electron microscope
- SVZ:
-
Subventricular zone
- Tuj1:
-
Neuron-specific tubulin III
- VEGF:
-
Vascular endothelial growth factor
- VZ:
-
Ventricular zone
References
Ahn S, Joyner AL (2005) In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature 437:894–897
Anders S, Reyes A, Huber W (2012) Detecting differential usage of exons from RNA-seq data. Genome Res 22:2008–2017
Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169
Baker EL, Bonnecaze RT, Zaman MH (2009) Extracellular matrix stiffness and architecture govern intracellular rheology in cancer. Biophys J 97:1013–1021
Baptista S, Lasgi C, Benstaali C et al (2014) Methamphetamine decreases dentate gyrus stem cell self-renewal and shifts the differentiation towards neuronal fate. Stem Cell Res 13:329–341
Barca-Mayo O, Lu QR (2012) Fine-tuning oligodendrocyte development by microRNAs. Front Neurosci 6:13
Barkho BZ, Song H, Aimone JB et al (2006) Identification of astrocyte-expressed factors that modulate neural stem/progenitor cell differentiation. Stem Cells Dev 15:407–421
Barnabe-Heider F, Goritz C, Sabelstrom H et al (2010) Origin of new glial cells in intact and injured adult spinal cord. Cell Stem Cell 7:470–482
Barton A, Fendrik AJ (2013) Sustained vs. oscillating expressions of Ngn2, Dll1 and Hes1: a model of neural differentiation of embryonic telencephalon. J Theor Biol 328:1–8
Bengoa-Vergniory N, Kypta RM (2015) Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cell Mol Life Sci 72:4157–4172
Blaber EA, Finkelstein H, Dvorochkin N et al (2015) Microgravity reduces the differentiation and regenerative potential of embryonic stem cells. Stem Cells Dev 24:2605–2621
Bonaguidi MA, Mcguire T, Hu M et al (2005) LIF and BMP signaling generate separate and discrete types of GFAP-expressing cells. Development 132:5503–5514
Brent A, Reynolds SW (1992) Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255:1707–1710
Burda JE, Sofroniew MV (2014) Reactive gliosis and the multicellular response to CNS damage and disease. Neuron 81:229–248
Caliari SR, Burdick A (2016) A practical guide to hydrogels for cell culture. Nat Methods 13:405–414
Chatr-Aryamontri A, Oughtred R, Boucher L et al (2017) The BioGRID interaction database: 2017 update. Nucleic Acids Res 45:D369–D379
Chen J, Liu R, Yang Y et al (2011) The simulated microgravity enhances the differentiation of mesenchymal stem cells into neurons. Neurosci Lett 505:171–175
Chen YC, Lin RZ, QiH et al (2012) Functional human vascular network generated in photocrosslinkable gelatin methacrylate hydrogels. Adv Funct Mater 22:2027–2039
Chen X, Wang W, Zhang J et al (2015) Involvement of caspase-3/PTEN signaling pathway in isoflurane-induced decrease of self-renewal capacity of hippocampal neural precursor cells. Brain Res 1625:275–286
Cheng TY, Chen MH, Chang WH et al (2013) Neural stem cells encapsulated in a functionalized self-assembling peptide hydrogel for brain tissue engineering. Biomaterials 34:2005–2016
Chiang MC, Lin H, Cheng YC et al (2012) Beta-adrenoceptor pathway enhances mitochondrial function in human neural stem cells via rotary cell culture system. J Neurosci Methods 207:130–136
Choi I, Woo JH, Jou I et al (2016) PINK1 deficiency decreases expression levels of mir-326, mir-330, and mir-3099 during brain development and neural stem cell differentiation. Exp Neurobiol 25:14–23
Cimadamore F, Amador-Arjona A, Chen C et al (2013) SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc Natl Acad Sci USA 110:E3017–3026
Coskun V, Wu H, Blanchi B et al (2008) CD133+ neural stem cells in the ependyma of mammalian postnatal forebrain. Proc Natl Acad Sci USA 105:1026–1031
Cui Y, Xiao Z, Han J et al (2012) MiR-125b orchestrates cell proliferation, differentiation and migration in neural stem/progenitor cells by targeting Nestin. BMC Neurosci 13:116
Cui Y, Han J, Xiao Z et al (2016) The miR-20-Rest-Wnt signaling axis regulates neural progenitor cell differentiation. Sci Rep 6:23300
Cukierman E, Pankov R, Stevens DR et al (2001) Taking cell-matrix adhesions to the third dimension. Science 294:1708–1712
Cunha C, Panseri S, Villa O et al (2011) 3D culture of adult mouse neural stem cells within functionalized self-assembling peptide scaffolds. Int J Nanomed 6:943–955
Delgehyr N, Meunier A, Faucourt M et al (2015) Ependymal cell differentiation, from monociliated to multiciliated cells. Methods Cell Biol 127:19–35
Edmondson R, Broglie JJ, Adcock AF et al (2014) Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol 12:207–218
Erickson IE, Huang AH, Chung C et al (2009) Differential maturation and structure-function relationships in mesenchymal stem cell- and chondrocyte-seeded hydrogels. Tissue Eng Part A 15:1041–1052
Fischbach C, Chen R, Matsumoto T et al (2007) Engineering tumors with 3D scaffolds. Nat Methods 4:855–860
Fishell G, Kriegstein AR (2003) Neurons from radial glia: the consequences of asymmetric inheritance. Curr Opin Neurobiol 13:34–41
Fogarty LC, Song B, Suppiah Y et al (2016) Bcl-xL dependency coincides with the onset of neurogenesis in the developing mammalian spinal cord. Mol Cell Neurosci 77:34–46
Fong EL, Lamhamedi-Cherradi SE, Burdett E et al (2013) Modeling Ewing sarcoma tumors in vitro with 3D scaffolds. Proc Natl Acad Sci USA 110:6500–6505
Gage FH (2000) Mammalian neural stem cells. Science 287:1433–1438
Ge S, Goh EL, Sailor KA et al (2006) GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature 439:589–593
Gibbons MC, Foley MA, Cardinal KO (2013) Thinking inside the box: keeping tissue-engineered constructs in vitro for use as preclinical models. Tissue Eng Part B Rev 19:14–30
Gioia U, Di Carlo V, Caramanica P et al (2014) Mir-23a and mir-125b regulate neural stem/progenitor cell proliferation by targeting Musashi1. RNA Biol 11:1105–1112
Godlewski J, Nowicki MO, Bronisz A et al (2008) Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res 68:9125–9130
Goncalves JT, Schafer ST, Gage FH (2016) Adult neurogenesis in the hippocampus: from stem cells to behavior. Cell 167:897–914
Gotz M, Huttner WB (2005) The cell biology of neurogenesis. Nat Rev Mol Cell Biol 6:777–788
Gould E (2007) How widespread is adult neurogenesis in mammals? Nat Rev Neurosci 8:481–488
Grimm D, Wehland M, Pietsch J et al (2014) Growing tissues in real and simulated microgravity: new methods for tissue engineering. Tissue Eng Part B Rev 20:555–566
Gross CG (2000) Neurogenesis in the adult brain: death of a dogma. Nat Rev Neurosci 1:67–73
Han Q, Jin W, Xiao Z et al (2010) The promotion of neural regeneration in an extreme rat spinal cord injury model using a collagen scaffold containing a collagen binding neuroprotective protein and an EGFR neutralizing antibody. Biomaterials 31:9212–9220
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Huang Y, Liu X, Wang Y (2015) MicroRNA-378 regulates neural stem cell proliferation and differentiation in vitro by modulating Tailless expression. Biochem Biophys Res Commun 466:214–220
Imayoshi I, Kageyama R (2014) bHLH factors in self-renewal, multipotency, and fate choice of neural progenitor cells. Neuron 82:9–23
Inghilleri M, Iacovelli E (2011) Clinical neurophysiology in ALS. Arch Ital Biol 149:57–63
Jiang D, Du J, Zhang X et al (2016) miR-124 promotes the neuronal differentiation of mouse inner ear neural stem cells. Int J Mol Med 38:1367–1376
Jurga M, Lipkowski AW, Lukomska B et al (2009) Generation of functional neural artificial tissue from human umbilical cord blood stem cells. Tissue Eng Part C Methods 15:365–372
Kawahara Y, Manabe T, Matsumoto M et al (2009) LIF-free embryonic stem cell culture in simulated microgravity. PLoS ONE 4:e6343
Kempermann G, Gage FH (2000) Neurogenesis in the adult hippocampus. Novartis Found Symp 231:220–235; discussion 235–241, 302–226
Kempermann G, Kuhn HG, Gage FH (1997) Genetic influence on neurogenesis in the dentate gyrus of adult mice. Proc Natl Acad Sci USA 94:10409–10414
Kim D, Pertea G, Trapnell C et al (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Kleber M, Sommer L (2004) Wnt signaling and the regulation of stem cell function. Curr Opin Cell Biol 16:681–687
Klein EA, Yin L, Kothapalli D et al (2009) Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr Biol 19:1511–1518
Knowlton S, Cho Y, Li XJ et al (2016) Utilizing stem cells for three-dimensional neural tissue engineering. Biomater Sci 4:768–784
Laks DR, Masterman-Smith M, Visnyei K et al (2009) Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells 27:980–987
Lange C, Mix E, Rateitschak K et al (2006) Wnt signal pathways and neural stem cell differentiation. Neurodegener Dis 3:76–86
Lee GY, Kenny PA, Lee EH et al (2007) Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods 4:359–365
Lee-Liu D, Edwards-Faret G, Tapia VS et al (2013) Spinal cord regeneration: lessons for mammals from non-mammalian vertebrates. Genesis 51:529–544
Li X, Xiao Z, Han J et al (2013) Promotion of neuronal differentiation of neural progenitor cells by using EGFR antibody functionalized collagen scaffolds for spinal cord injury repair. Biomaterials 34:5107–5116
Lie DC, Colamarino SA, Song HJ et al (2005) Wnt signalling regulates adult hippocampal neurogenesis. Nature 437:1370–1375
Lim DA, Alvarez-Buylla A (1999) Interaction between astrocytes and adult subventricular zone precursors stimulates neurogenesis. Proc Natl Acad Sci USA 96:7526–7531
Lim DA, Tramontin AD, Trevejo JM et al (2000) Noggin antagonizes BMP signaling to create a niche for adult neurogenesis. Neuron 28:713–726
Lin C, Jiang X, Dai Z et al (2009) Sclerostin mediates bone response to mechanical unloading through antagonizing Wnt/beta-catenin signaling. J Bone Miner Res 24:1651–1661
Lledo PM, Alonso M, Grubb MS (2006) Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci 7:179–193
Louis SA, Rietze RL, Deleyrolle L et al (2008) Enumeration of neural stem and progenitor cells in the neural colony-forming cell assay. Stem Cells 26:988–996
Luca AC, Mersch S, Deenen R et al (2013) Impact of the 3D microenvironment on phenotype, gene expression, and EGFR inhibition of colorectal cancer cell lines. PLoS ONE 8:e59689
Ma DK, Marchetto MC, Guo JU et al (2010) Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci 13:1338–1344
Mao S, Li X, Wang J et al (2016) miR-17-92 facilitates neuronal differentiation of transplanted neural stem/precursor cells under neuroinflammatory conditions. J Neuroinflammation 13:208
Matigian N, Abrahamsen G, Sutharsan R et al (2010) Disease-specific, neurosphere-derived cells as models for brain disorders. Dis Model Mech 3:785–798
Mc Garrigle MJ, Mullen CA, Haugh MG et al (2016) Osteocyte differentiation and the formation of an interconnected cellular network in vitro. Eur Cell Mater 31:323–340
Mcconnell SK (1995) Strategies for the generation of neuronal diversity in the developing central nervous system. J Neurosci 15:6987–6998
Mcpherson A, Hormozdiari F, Zayed A et al (2011) deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 7:e1001138
Ming GL, Song H (2005) Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci 28:223–250
Monticone M, Liu Y, Pujic N et al (2010) Activation of nervous system development genes in bone marrow derived mesenchymal stem cells following spaceflight exposure. J Cell Biochem 111:442–452
Nelson LJ, Walker SW, Hayes PC et al (2010) Low-shear modelled microgravity environment maintains morphology and differentiated functionality of primary porcine hepatocyte cultures. Cells Tissues Organs 192:125–140
Nickerson CA, Ott CM, Wilson JW et al (2004) Microbial responses to microgravity and other low-shear environments. Microbiol Mol Biol Rev 68:345–361
Noctor SC, Martinez-Cerdeno V, Ivic L et al (2004) Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 7:136–144
Nomura T, Goritz C, Catchpole T et al (2010) EphB signaling controls lineage plasticity of adult neural stem cell niche cells. Cell Stem Cell 7:730–743
Nori S, Okada Y, Yasuda A et al (2011) Grafted human-induced pluripotent stem-cell-derived neurospheres promote motor functional recovery after spinal cord injury in mice. Proc Natl Acad Sci USA 108:16825–16830
Novosel EC, Kleinhans C, Kluger PJ (2011) Vascularization is the key challenge in tissue engineering. Adv Drug Deliv Rev 63:300–311
Okano H, Temple S (2009) Cell types to order: temporal specification of CNS stem cells. Curr Opin Neurobiol 19:112–119
Platel JC, Dave KA, Gordon V et al (2010) NMDA receptors activated by subventricular zone astrocytic glutamate are critical for neuroblast survival prior to entering a synaptic network. Neuron 65:859–872
Prewitz M, Seib FP, Pompe T et al (2012) Polymeric biomaterials for stem cell bioengineering. Macromol Rapid Commun 33:1420–1431
Puca A, Russo G, Giordano A (2012) Properties of mechano-transduction via simulated microgravity and its effects on intracellular trafficking of VEGFR’s. Oncotarget 3:426–434
Ravi M, Paramesh V, Kaviya SR et al (2015) 3D cell culture systems: advantages and applications. J Cell Physiol 230:16–26
Reynolds BA, Weiss S (1992) Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255:1707–1710
Riquelme PA, Drapeau E, Doetsch F (2008) Brain micro-ecologies: neural stem cell niches in the adult mammalian brain. Philos Trans R Soc Lond B Biol Sci 363:123–137
Schofield R (1978) The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 4:7–25
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Sher F, Boddeke E, Olah M et al (2012) Dynamic changes in Ezh2 gene occupancy underlie its involvement in neural stem cell self-renewal and differentiation towards oligodendrocytes. PLoS ONE 7:e40399
Shihabuddin LS (2008) Adult rodent spinal cord-derived neural stem cells: isolation and characterization. Methods Mol Biol 438:55–66
Song H, Stevens CF, Gage FH (2002) Astroglia induce neurogenesis from adult neural stem cells. Nature 417:39–44
Souza GR, Molina JR, Raphael RM et al (2010) Three-dimensional tissue culture based on magnetic cell levitation. Nat Nanotechnol 5:291–296
Stamenkovic V, Keller G, Nesic D et al (2010) Neocartilage formation in 1 g, simulated, and microgravity environments: implications for tissue engineering. Tissue Eng Part A 16:1729–1736
Sun G, Fu C, Shen C et al (2011) Histone deacetylases in neural stem cells and induced pluripotent stem cells. J Biomed Biotechnol 2011:835968
Szklarczyk D, Franceschini A, Wyder S et al (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–452
Tamura Y, Takahashi K, Takata K et al (2016) Noninvasive evaluation of cellular proliferative activity in brain neurogenic regions in rats under depression and treatment by enhanced [18F]FLT-PET imaging. J Neurosci 36:8123–8131
Tashiro A, Sandler VM, Toni N et al (2006) NMDA-receptor-mediated, cell-specific integration of new neurons in adult dentate gyrus. Nature 442:929–933
Trapnell C, Roberts A, Goff L et al (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Ueki T, Tanaka M, Yamashita K et al (2003) A novel secretory factor, Neurogenesin-1, provides neurogenic environmental cues for neural stem cells in the adult hippocampus. J Neurosci 23:11732–11740
Wang H, Zhang Z, Xin W (2007) Microgravity resulted from 3D dynamic culture induces compounding of bone morrow-derived mesenchymal stem cells with Pluronic F-127 scaffold used for repairing of cartilage defects. Chin Tissue Eng Clin Recov 11:2609–2613
Wang Y, An L, Jiang Y et al (2011) Effects of simulated microgravity on embryonic stem cells. PLoS ONE 6:e29214
Xu W, Li P, Qin K et al (2012) miR-124 regulates neural stem cells in the treatment of spinal cord injury. Neurosci Lett 529:12–17
Yamada KM, Cukierman E (2007) Modeling tissue morphogenesis and cancer in 3D. Cell 130:601–610
Yang K, Lee JS, Kim J et al (2012) Polydopamine-mediated surface modification of scaffold materials for human neural stem cell engineering. Biomaterials 33:6952–6964
Yu G, Wang LG, Han Y et al (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287
Yuge L, Kajiume T, Tahara H et al (2006) Microgravity potentiates stem cell proliferation while sustaining the capability of differentiation. Stem Cells Dev 15:921–929
Zhao C, Sun G, Li S et al (2010) MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci USA 107:1876–1881
Ziv Y, Ron N, Butovsky O et al (2006) Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat Neurosci 9:268–275
Acknowledgements
This work was supported by the “Strategic Priority Research Program of the Chinese Academy of Sciences” (XDA04020000) and National Science Foundation of China (U1738109 and 81601084).
We are greatly thankful to everyone who helped us to complete the SJ-10 satellite project. We thank Prof. Zhang Tao and his staff of the Shanghai Institute of Technical Physics (Chinese Academy of Sciences, CAS) for designing and manufacturing the fully automated bioreactors for 3D-cultured NSC survival in space. We thank Prof. Suo Guangli and his graduate student Zhou Yuanshuai of the Suzhou Institute of Nano-tech and Nano-Bionics (CAS) and Prof. Wu Xianming of the Institute of Genetics and Developmental Biology (CAS) for processing the bioinformatics data.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Science Press and Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Han, J., Cui, Y., Xu, B., Xue, W., Liu, S., Dai, J. (2019). Three-Dimensional Cell Culture and Tissue Restoration of Neural Stem Cells Under Microgravity. In: Duan, E., Long, M. (eds) Life Science in Space: Experiments on Board the SJ-10 Recoverable Satellite. Research for Development. Springer, Singapore. https://doi.org/10.1007/978-981-13-6325-2_10
Download citation
DOI: https://doi.org/10.1007/978-981-13-6325-2_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-6324-5
Online ISBN: 978-981-13-6325-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)