Abstract
In this paper, a combination of Pixel based Seed points and textural Back Propagation Neural Networks is proposed for segmenting the Region of Interest (ROI) from the medical images. Medical images such as Fundus and Skin images are used to test the proposed algorithm. To develop the proposed algorithm, Pixel based Seed points are combined with the Texture based Back Propagation Neural Network (TBP-NN) by a trained knowledge of textural properties for segmenting the medical images which can be used in early Diabetic Retinopathy (DR) detection and Skin lesion detection. The proposed algorithm is tested with a total of 200 fundus and skin images each which is stored in a database used for further testing. The medical images were processed such that a knowledge in form of texture features such as Energy. Homogeneity, Contrast and Correlation were automatically obtained. The proposed algorithms efficiency was compared with traditional BP-NN methods and Support Vector Machine for segmenting medical images. The results obtained from the proposed methodology reveals that the accuracy of proposed algorithm is higher. It indicates that the proposed algorithm could achieve a better result in medical image segmentation more effectively.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Segmenting medical image is an important task in image analysis. Segmentation of a medical imaging is used to extract the required information regarding the presence of different abnormalities present in the respective image. Artificial Neural network (ANN) is a machine learning model which is used by simulating the anatomic neuron connections present in it. Its effects have been validated in scientific studies for various applications, such as image processing, pattern recognition, system control, and medical image diagnosis [4,5,6,7, 13, 14, 16]. It has some limitations such as false segmentation and region growing faults, when it is applied in some type of medical images. In this paper, improvements in this Neural Networks has been obtained when a trained knowledge as features was incorporated into it which is used to train it. Therefore, we aimed to combine the trained knowledge as features from medical images with a doctor’s trained knowledge in the segmentation of Retinal vessels and Skin lesions, for the application of early finding of diabetic retinopathy in eye fundus images and lesions in the Skin images. Important aim of this proposed methodology is to segment the region of interest in eye fundus images for finding the Diabetic Retinopathy and Skin lesions in medical images more effectively which is applied for training and testing as samples. Many methodologies have been given in the literature for the medical image segmentation. An automatic detection methodology was framed by Gardner et al. [8] for diabetic retinopathy using a neural network. The artifacts which are present in the medical images can be easily identified more effectively from the images of grey level. An automated back propagation type neural network is adopted to examine the eye fundus images. It does not work more effectively since the images are of low contrast.
Thresholding based technique proposed on eye fundus images by Sinthanayothin et al. in [9] The performance of their method is validated o using a 10 × 10 pixels instead of using the whole image. Usher et al. [10] endowed the various region of candidate exudates which are present in the eye fundus images by using a combination of Random Graph and intensity based adaptive thresholding methods. In their methodology, the regions of various candidate are removed and those are further used as an input for an artificial based neural network technology. Due to their algorithm the images with poor quality was affected and then all these images are extracted due to the bright and dark type of lesions.
Zheng et al. in [11] extracted the various types of artifacts which are present in the eye fundus images by combining the algorithms called as thresholding and region growing. The combination of Color normalization and local contrast enhancement methodology along with the clustering based on fuzzy C-means and neural network methodologies was proposed by Osareh et al. [12]. Their techniqua works more effectively only on Luv based colour space images even if it has an illumination of non-uniform in nature. Hence, the accuracy of detection less because of this dis advantage. Stoecker et al. [15] presented a new textural based segmentation methodology in skin images adopting the gray-level co-occurrence matrix which is a statistical based approach. Jeffrey et al. [1] adapted a novel methodology segmentation and classification of lesion present in the skin images. In their methodology, various lesions which are present in the skin images are segmented using a Distinctiveness based Joint Statistical Texture method. It results in an overall accuracy of 93%.
Menzies et al. in [5] proposed a novel algorithm based on a fusion of Semi-automatic and manual based methodologies A regression-based classifier is used for the purpose of classifying the further segmented results. They applied their algorithm. Their algorithm is applied for a set of 2430 lesion images of skin. A 65% average specificity and 91% average sensitivity is obtained to their algorithm.
2 Methodology
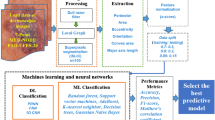
This section presents the overall process present in segmentation of region of interest in medical images. In this work, Skin and Retinal images are taken as testing datasets. Figure 1 gives the overall architecture of the proposed methodology for segmenting the medical images.
3 Image Pre-processing
All the obtained medical images were normalized for illumination, size, and color using an image processing process. In this work, the trained knowledge is defined as the texture features of the Region of Interest. This work is done after consulting with various experienced ophthalmologists and doctors for drawing the ground truth images.
-
A.
Computerized seed points extraction
In the segmentation algorithm, usually all the seed points were formed by manual process. To remove each and every seed points in all the images is a big-time process. The seed points which are corresponding to each and every pixel present in the image should be given prior to the segmentation of entire image. The further segmented image should be different from the previously segmented image. For this process, all the regions present in the current image is merged with one another for the process of splitting and then merging. More number of seed points should be given since it should be merged with all the pixels present in the original image. Since the given put is a medical set of data, all the seed points should be merged with all the pixels present in the entire image so that the entire image is considered. The overall method is as follows: The medical image which is to be segmented is considered as s binary image.
Each pixel present in the original binary image which is considered as the target region is considered as 1, and all the others are considered as 0. The proposed algorithm is as follows
-
(1)
Define the similarity threshold for color as \( a \).
-
(2)
Choose the pixel number manually which is present in the predefined target region, and also define them as an initial seed pixel as \( (x_{0} ,y_{0} ) \);
-
(3)
Define the initial seed pixel in center, in order to get the eight pixels in neighborhood as \( (x_{i} ,y_{i} )\left( {i = 0,1, \ldots .,10} \right) \)
-
(4)
Calculate the similarity in color between the predefined pixels \( (x_{i} ,y_{i} ) \) and \( (x_{0} ,y_{0} ) \)
-
(5)
If the similarity in color in between the predefined pixel is larger than the set similarity threshold \( a \), those pixels can be clustered towards a common region. Hence, the difference Pixel \( (x_{i} ,y_{i} ) \) should stored in a stack.
-
(6)
Repeat the above process until the stack become empty.
-
B.
Seed points based TBPNN
The seed point based Textural back propagation neural network (STBP-NN) [17, 18] is trained with pixel values as knowledge present in Fig. 2 has three layers such as an input layer, a hidden layer \( H_{in} (j) \) and an output layer. Initial Seed point is assumed as Xi and final seed point is assumed as X2. the pixel values as seed points and a Trained knowledge in the form of textural features are given as the input towards the input layer.
The framework of our STBPNN is as follows. The hidden layer input \( H_{in} \left( j \right) \) was defined as:
where \( x_{i} \) is the input features, \( \omega_{ij} \) is the weights between the neurons from the input layer and hidden layer, and \( a_{j} \) represents the threshold for hidden layer neurons.
The estimates of RBP-NN is as follows:
where, λ is a non-negative regularization parameter, x is the Blood vessel width and blood vessel tortuosity of retinal images, Y is the average accuracy and βs is the regression coefficient.
The number of neurons in the hidden layer is as follows:
where \( n \) is the number of input layer neurons, h is the number of neurons in the hidden layer, \( m \) was the number of neurons present in the output layer, and \( a \) is a threshold between 0 and 20. In this work, \( n \) is assumed as 80, m is set as 3, and his from 21 to 30. The texture features such as Energy.
Homogeneity, Contrast and Correlation were separately calculated for each training and testing. The results obtained from the traditional segmentation methods are compared with the proposed STBP-ANN. The same features used in this algorithm were used as the input towards SVM algorithm inorder to compare its performance with our proposed methodology.
4 Results and Discussion
In this research, the SVM, traditional BP-NN, and the proposed STBP-NN were each used to segment the Region of Interest from medical images. The objective of this comparison is to measure the accuracy of proposed algorithm based on sensitivity, specificity, and accuracy using Eqs. (5), (6) and (7) as shown in Table 1.
The images shown in Figs. 3 and 4 are the original. Ground truth and the segmented results of Skin and Retina images. The segmented images are very close to the ground truth image. In this approach, the ophthalmologists and doctors specified images are considered as ground truth images for the calculation of segmentation accuracy.
The results showed that the accuracy of this proposed methodology is higher than the SVM, BP-NN and STBP-NN was 94.39%, 94.69% and 95.71% respectively as shown in Table 1 for retinal images. This indicates that the Proposed STBP-NN with a priori knowledge can achieve better segmentation results for retinal images.
Table 2 presents the accuracy of this proposed methodology is higher than the SVM, BP-NN and STBP-NN was 94.75%, 94.84% and 95.61% respectively for skin images indicating that the Proposed methodology can achieve better segmentation results for segmenting skin images.
5 Conclusion
In this paper, a new method for segmenting Region of Interest present in the medical images using the combination of Seed point and supervised Texture based Back Propagation Neural Networks method is proposed. Medical images such as Skin and Fundus images are used to test the proposed algorithm in which the region of interests such as the skin lesion and the retinal vessels were segmented effectively. The main purpose of this approach is to improve the DR in Retinal images and Lesion in skin image detection accuracy by segmenting the region of interest in it. In this method, the obtained seed points are given as input for training and testing the proposed TBP-ANN. Compared with other segmentation method, our method can better for processing the fundus image in its vessel and lesion in skin image segmentation. This STBP-ANN could segment the region of interest in medical images better than the traditional Neural Networks and SVM methods and could be a promising measure for early DR and Lesion detection more effectively.
References
Glaister, J., Wong, A., Clausi, D.A.: Segmentation of skin lesions from digital images using joint statistical texture distinctiveness. IEEE Trans. Biomed. Eng. 61(4) (2014)
Stolz, W., Riemann, A., Cognetta, A.B., et al.: ABCD rule of dermatoscopy: a new practical method for early recognition of malignant melanoma. Eur. J. Dermatol. 4, 521–527 (1994)
Nachbar, F., Stolz, W., Merkle, T., et al.: The ABCD rule of dermatoscopy high prospective value in the diagnosis of doubtful melanocytic skin lesions. J. Am. Acad. Dermatol. 30, 551–559 (1994)
Lapuerta, P., L’Italien, G.J., Paul, S., et al.: Neural network assessment of perioperative cardiac risk in vascular surgery patients. Med. Decis. Making 18, 70–75 (1998)
Menzies, S.W., Bischof, L., Talbot, H., Gutenev, A., Avramidis, M., Wong, L.: The performance of solarscan: an automated dermoscopy image analysis instrument for the diagnosis of primary melanoma. Arch. Dermatol. 141(11), 1388–1396 (2005)
Argenziano, G., Soyer, H.P., De Giorgi, V., Piccolo, D., Carli, P., Delfino, M., et al.: Dermoscopy: a Tutorial. EDRA Medical Publishing & NewMedia, Milan (2002)
Salvi, M., Dazzi, D., Pellistri, I.: Classification and prediction of the progression of thyroid-associated ophthalmopathy by an artificial neural network. Ophthalmol. 109, 1703–1708 (2002)
Gardner, G.G., Keating, D., Williamson, T.H., Elliott, A.T.: Automatic detection of diabetic retinopathy using an artificial neural network: a screening tool. Br. J. Ophthalmol. (1996)
Sinthanayothin, C., Boyce, J.F., Williamson, T.H., Cook, H.L., Mensah, E., Lal, S.: Automated detection of diabetic retinopathy on digital fundus image. J. Diabet. Med. 19, 105–112 (2002)
Usher, D., Dumskyj, M., Himaga, M., Williamson, T.H., Nussey, S., Boyce, J.: Automated detection of diabetic retinopathy in digital retinal images: a tool for diabetic retinopathy screening. Diabet. Med. 21, 84–90 (2004)
Liu, Z., Opas, C., Krishnan, S.M.: Automatic image analysis of fundus photograph. In: Proceedings of the International Conference on Engineering in Medicine and Biology, vol. 2, pp. 524–525 (1997)
Osareh, A., Mirmehdi, M., Thomas, B., Markham, R.: Automated identification of diabetic retinal exudates in digital colour images. Br. J. Ophthalmol. 87, 1220–1223 (2003)
Mitra, S.K., Lee, T.-W., Goldbaum, M.: Bayesian network based sequential inference for diagnosis of diseases from retinal images. Pattern Recogn. Lett. 26, 459–470 (2005)
Dupas, B., Walter, T., Erginay, A.: Evaluation of automated fundus photograph analysis algorithms for detecting microaneurysms haemorrhages, and exudates, and of a computer- assisted diagnostic system for grading diabetic retinopathy. Diabet. Metab. 36, 213–220 (2010)
Stoecker, W.V., Chiang, C.-S., Moss, R.H.: Texture in skin images: comparison of three methods to determine smoothness. Comput. Med. Imag. Graph. 16(3), 179–190 (1992)
Faizal Khan, Z., Nalini Priya, G., Anwar, M.K.: Texture based back propagation neural networks for segmentation of arteriole and venule in fundus images. In: IEEE International Conference on Power, Control, Signals and Instrumentation Engineering (ICPCSI), pp. 84–89 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Faizal Khan, Z. (2018). Automated Seed Points and Texture Based Back Propagation Neural Networks for Segmentation of Medical Images. In: Zelinka, I., Senkerik, R., Panda, G., Lekshmi Kanthan, P. (eds) Soft Computing Systems. ICSCS 2018. Communications in Computer and Information Science, vol 837. Springer, Singapore. https://doi.org/10.1007/978-981-13-1936-5_29
Download citation
DOI: https://doi.org/10.1007/978-981-13-1936-5_29
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1935-8
Online ISBN: 978-981-13-1936-5
eBook Packages: Computer ScienceComputer Science (R0)