Abstract
Majority of RNAs expressed in animal cells lack protein-coding ability. Unlike other cellular RNAs, circular (circ)RNAs include a large family of noncoding (nc)RNAs that lack the 5′ or 3′ ends. The improvements in high-throughput RNA sequencing and novel bioinformatics tools have led to the identification of thousands of circRNAs in various organisms. CircRNAs can regulate gene expression by influencing the transcription, the mRNA turnover, and translation by sponging RNA-binding proteins and microRNAs. Given the broad impact of circRNA on miRNA activity, there is huge interest in understanding the impact of miRNA sponging by circRNA on gene regulation. In this review, we summarize our current knowledge of the miRNA-circRNA interaction and mechanisms that influence gene expression.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
RNA molecules were conventionally believed to transfer the genetic information coded in the genomic DNA into specific proteins [1]. However, the protein-coding mRNAs represent only ~5% of the human transcriptome, while the rest of the transcriptome is noncoding (nc)RNAs [2]. The vast majority are ribosomal (r)RNA and transfer (t)RNA, both involved in translation [1, 3]. The other categories of ncRNAs include microRNAs (miRNAs), pseudogenes, long (l)ncRNAs, and circular (circ)RNAs [4,5,6]. In 1976, electron microscopy of plant viroid discovered covalently closed single-stranded RNA molecules for the first time [7]. Later, the hepatitis delta virus (HDV) was found to have a circRNA genome [8]. Another report suggested the expression of scrambled exon RNA from tumor suppressor gene DCC in human cells [9]. Due to lack of RNA-sequencing technologies and inability to map the circRNAs to the genome, circRNAs were completely neglected for last two decades. Traditional molecular biology techniques used for RNA analysis cannot differentiate circRNAs from linear RNAs [10, 11]. Interestingly, the innovations in next-generation sequencing associated with new computational pipelines to map circRNAs to the genome have moved circRNA to the forefront of RNA research [12,13,14,15]. Most of the circRNAs are found to be abundant, conserved across species, and often show tissue-specific expression pattern [13, 16]. Recent studies established that covalently closed circRNAs are generated from the canonical splicing machinery by a process called backsplicing [17]. CircRNAs are categorized as exonic (E), intronic (I), and exon-intron (EI) circRNAs based on the primary transcript sequence they are generated from [15, 17,18,19]. circRNAs are very stable due to lack of the 5′-3′ ends which makes them resistant to exonucleases [20, 21]. Recent studies reported that some may act as sponges for miRNAs, sponges for RBPs, compete with linear splicing, and translated into peptides [4, 18, 22]. Growing evidence indicated that circRNAs involved in various cellular events including proliferation, differentiation, apoptosis, and metastasis [23]. This review briefly discusses the impact of circRNAs on key cellular processes by acting as a competing endogenous RNA (ceRNA) for target miRNAs.
2 MiRNA Sponging by circRNA
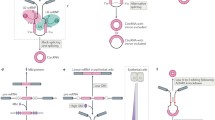
MicroRNAs are small, evolutionarily conserved ncRNA molecules predicted to regulate ~30% of the protein-coding genes [24, 25]. There are ~1800 miRNAs which have been identified in humans [26]. The miRNA genes are transcribed into primary miRNA (pri-miRNA) by RNA polymerase II followed by processing with Drosha and DGCR8 to generate pre-miRNA [27]. The pre-miRNA is exported to the cytoplasm and processed by Dicer to produce ~19–22 nt mature miRNA. The functional miRNA is incorporated in the effector RNA-induced silencing complex (RISC) and activates the RISC complex to bind the target mRNA that has sequence complementarity with the loaded miRNA. The miRNAs usually target the 3′ untranslated regions (UTRs) of specific mRNA targets and regulate their stability and/or translation [28]. The level of complementarity between the mRNA and miRNA determines the mechanism of miRNA action on target mRNA. As each miRNAs can have complementarity with many mRNAs, they have the potential to regulate multiple genes [29]. miRNAs are shown to be involved in posttranscriptional regulation of gene expression in nearly all cellular events including cell proliferation, migration, differentiation, and apoptosis [29,30,31,32,33]. Given their importance in gene expression regulation, there is enormous interest in understanding the regulatory mechanisms that can regulate miRNA function. Accumulating evidence indicates that circRNAs play a crucial role in gene expression regulation partly by inhibiting miRNA activity (Fig. 6.1). In this review, several circRNA-miRNA interactions and their function are listed as follows (Table 6.1).
Schematic representation of circRNA biogenesis and their impact on gene expression by sponging miRNA
(a) The canonical splicing machinery generates mature linear RNA from the primary transcript by removing intervening introns. (b) The circRNA is generated by the “head-to-tail” backsplicing of the circularizing exons. (c) Most circRNA sponges are enriched in miRNA-binding sites. Overexpression of circRNA leads to inactivation of miRNAs, thereby upregulating miRNA target gene expression. (d) The decrease in circRNA expression allows higher levels of functional miRNAs to suppress the expression of mRNAs containing miRNA-binding sites
CDR1as
The CDR1as is generated from the antisense transcript of CDR1 gene (also termed as ciRS-7). CDR1as was the first miRNA sponge reported to negatively regulate miR-7 and found to be expressed in brain tissue, neuroblastoma, astrocytoma, HeLa cells, and lung carcinoma [14, 34,35,36]. The CDR1as has more than 60 binding sites for miR-7, and its resistance to miRNA-mediated RNA degradation makes this a perfect ceRNA for miR-7 [12, 14]. Expression of CDR1as inhibits miR-7 activity that leads to increase in expression of miR-7 targets. Coexpression of CDR1as and miR-7 is necessary for sponging activity of circRNA. The sponging of miR-7 by ciRS-7 is reported to affect the expression of ubiquitin protein ligase A (UBE2A) in Alzheimer’s disease [37]; Myrip and Pax6 in insulin secretion and synthesis, respectively [38]; and epidermal growth factor receptor (EGFR) in cancer [36, 14, 39]. Upregulation of CDR1as in gastric cancer suppresses miR-7 activity which leads to more aggressive oncogenic phenotype mediated by PTEN/PI3K/AKT pathway [40]. Another study reported the upregulation of CDR1as and downregulation of miR-7 in hepatocellular carcinoma (HCC) tissue compared with the adjacent non-tumor tissues [34]. The overexpression of miR-7 or silencing of CDR1as inhibited the HCC cell proliferation and invasion by inhibiting the expression of target genes CCNE1 and PIK3CD. Together, CDR1as act as an oncogene in HCC through sponging miR-7 [34].
cir-SRY
The sex-determining region Y gene produces a circRNA known as cir-SRY. The cir-SRY is highly expressed in adult mouse testis [41]. A recent study reported that there are 16 putative miR-138-binding sites present in cir-SRY which can act as ceRNA and thus potentially regulate expression of miR-138 target genes [14, 42].
cir-ITCH
Cir-ITCH (itchy E3 ubiquitin protein ligase) is generated from the ICTH gene. Cir-ICTH was reported to enrich in miRNA regulatory elements (MREs) for miR-7, miR-17, and miR-214 and act as a sponge for these miRNAs, leading to upregulation of miRNA target gene ITCH. Cir-ITCH was reported to have antitumor activity by suppressing the Wnt/β-catenin signaling by regulating ICTH expression in various cancers including esophageal squamous cell carcinoma and bladder, lung, and colorectal cancer [43,44,45,46].
circHIPK3
The second exon of homeodomain-interacting protein kinase 3 (HIPK3) gene generates a circRNA called circHIPK3 that can act as a sponge for nine miRNAs including miR-124. Silencing of CircHIPK3 reduced cell growth through suppressing miR-124 activity which directly interacts with circHIPK3 [47]. CircHIPK3 was upregulated in HCC tissues [48]. CircHIPK3 acted as a sponge for miR-124 and suppressed miR-124 activity in HCC, leading to upregulation of miR-124 target gene aquaporin 3 (AQP3). Further, increase in AQP3 expression promoted cell proliferation and migration in HCC cells. Together, these results suggested that circHIPK3 regulated HCC growth through the miR-124-AQP3 axis [48]. CircHIPK3 was also found to be downregulated in bladder cancer tissues compared with normal bladder tissues, and the level of circHIPK3 negatively correlates with bladder cancer grade [49]. Increase in circHIPK3 level led to inhibition of migration, invasion, and angiogenesis of bladder cancer cells in vitro. CircHIPK3 acted as a ceRNA for miR-558 and inhibit miR-558 activity, thereby regulating the expression of heparanase (HPSE) in bladder cancer cells [49]. Another study reported the expression pattern of circHIPK3 in six non-small cell lung cancer (NSCLC) cell lines [50]. The NSCLC cell lines NCI-H2170 and NCI-H1299 had the highest and lowest expression level, respectively. The circHIPK3 overexpression in NCI-H1299 promoted cell proliferation and circHIPK3 silencing in NCI-H2170-inhibited cell proliferation. CircHIPK3 was found to sequester miR-379 and increase the expression levels of miR-379 target IGF1, leading to increase in cell proliferation [50].
circPVT1
Circular RNA expression pattern in proliferating (early-passage) and senescent (late-passage) human diploid WI-38 fibroblasts were analyzed and identified hundreds of differentially expressed senescence-associated circRNAs (SAC-RNAs) [51]. One of the SAC-RNA called circPVT1 was significantly downregulated in senescent fibroblasts. Further, circPVT1 silencing in proliferating fibroblasts promoted cellular senescence. CircPVT1 selectively sponged let-7 and promoted the expression of let-7 target genes including IGF2BP1, KRAS, and HMGA2. Together, these data suggested that the SAC-RNA circPVT1, elevated in the proliferating cells, inhibits endogenous let-7 activity to enable a proliferative phenotype [51]. Another report suggested that the circPVT1 is often upregulated in gastric cancer (GC) tissues compared with matched normal tissues [52]. The circPVT1 acted as a sponge for the members of the miR-125 family and promoted cell proliferation. Further, circPVT1 expression level was correlated with the survival of patients with gastric cancer. In sum, circPVT1 acts as a proliferative factor and prognostic marker in gastric cancer [52].
circRNA-CER
A recent study explored the circRNA expression pattern and function of chondrocyte extracellular matrix (ECM)-related circRNAs (circRNA-CER) in cartilage. Several circRNAs were differentially expressed in osteoarthritis samples compared to normal cartilage. The circRNA-CER expression was upregulated in chondrocytes upon interleukin-1 (IL-1) and tumor necrosis factor α (TNFα) treatment [53]. The circRNA-CER was found to have five putative MREs for miR-636, miR-665, miR-217, miR-646, and miR-136. The MMP13 gene was regulated by miR-136 which is sponged by circRNA-CER. The silencing of circRNA-CER increased chondrocyte extracellular matrix formation by inhibiting MMP13 expression [53].
circRNA-MYLK
A recent study found that the circRNA-MYLK and VEGFA were significantly upregulated in bladder carcinoma. The circRNA-MYLK directly binds to miR-29a that leads to increase in expression of miR-29a target VEGFA. Overexpression of circRNA-MYLK activated VEGFA/VEGFR2 and downstream Ras/ERK signaling pathway by acting as a ceRNA for miR-29a [54].
circTCF25
The circRNA circTCF25 was predicted to sponge miR-103a-3p and miR-107. The overexpression of circTCF25 sequestered miR-103a-3p and miR-107, leading to upregulation of their target CDK6 which in turn enhanced the proliferation and migration of bladder cancer cells [55].
circHIAT1
The expression of circHIAT1 was downregulated in clear cell renal cell carcinoma (ccRCC) compared to the adjacent normal tissues. Androgen receptor (AR) downregulated the expression of circHIAT1 by suppressing transcription of its host gene, hippocampus abundant transcript 1 (HIAT1). The circHIAT1 may act as a sponge for miR-195-5p/29a-3p/29c-3p to modulate CDC42 expression which is linked to ccRCC cell migration and invasion [56].
HRCR
A recent work suggested that a heart-related circRNA (HRCR) could act as a sponge for miR-223 to suppress cardiac hypertrophy. ARC was identified to be the downstream target to mediate the function of miR-223 in cardiac hypertrophy. The circRNA HRCR was found to sequester and inhibit the function miR-223, leading to upregulation of ARC expression which is involved in cardiac hypertrophy and heart failure [57].
cir-ZNF609
In Hirschsprung disease (HSCR) the expression of cir-ZNF609 was lower as compared with normal bowel tissues. The silencing of cir-ZNF609 led to suppression of proliferation and migration of cells. Further, cir-ZNF609 was found to regulate the expression of AKT3 by acting as a decoy for miR-150-5p. These findings suggest that the cir-ZNF609-miR-150-5p-AKT3 axis plays a critical role in the onset of HSCR [58].
has_circ_001569
The expression of hsa_circ_001569 was upregulated in colorectal cancer tissues and predicted to sponge miR-145. The miR-145 can modulate the expression of BAG4, E2F5, and FMNL2 transcripts in colorectal cancer cells. Expression of circ_001569 upregulated the expression of BAG4, E2F5, and FMNL2 by sponging miR-145, leading to colorectal cancer cell proliferation and invasion [59].
hsa_circ_001564
The circRNA hsa_circ_0001564 is derived from the gene called CANX and significantly upregulated in osteosarcoma cell lines. Silencing of hsa_circ_0001564 reduced the proliferation by inducing cell cycle arrest and apoptosis in HOS and MG-63 cells. Further, hsa_circ_0001564 was found to be a sponge for miR-29c-3p which could reverse the tumorigenic effect of circ_0001564. These data suggested that hsa_circ_0001564 plays a critical role in osteosarcoma by acting as a ceRNA for miR-29c-3p [60].
circVMA21
The role of circVMA21 was explored in nucleus pulposus (NP) cells and degenerative NP tissues from intervertebral disc degeneration (IVDD) patients. The circVMA21 was found to directly interact with miR-200c which inhibits the expression of the target gene X-linked inhibitor-of-apoptosis protein (XIAP). The NP cell function and viability was regulated by miR-200c through suppression of XIAP. Together, circVMA21 could inhibit NP cell apoptosis through miR-200c-XIAP axis [61].
circRNA_Atp9b
In osteoarthritis, circRNA_Atp9b was overexpressed in mouse chondrocytes upon interleukin-1 beta (IL-1β) treatment. Further, circRNA_Atp9b silencing upregulated type II collagen expression and suppressed the expression of MMP13, COX-2, and IL-6. miR-138-5p was found to be sponged by circRNA_Atp9b, and their expression levels are negatively correlated. Moreover, the effects of circRNA_Atp9b on extracellular matrix catabolism and inflammation were partly reversed by inhibition of miR-138-5p. In sum, these data suggested that the extracellular matrix in chondrocytes is regulated by circRNA_Atp9b through sponging miR-138-5p [62].
circDOCK1
A TNF-α-induced apoptotic model was developed for oral squamous cell carcinoma (OSCC) to study the impact of circDOCK1 on apoptosis. The silencing of circDOCK1 increased the apoptosis in OSCC cells. Bioinformatics analysis predicted the interaction of circDOCK1 with miR-196a-5p which targets BIRC3. Interestingly, both overexpression of miR-196a-5p or knockdown of circDOCK1 led to suppression of BIRC3 which is a negative regulator of apoptosis. Together, these results suggested that the apoptosis of OSCC cells is regulated by circDOCK1 through sponging miR-196a-5p [63].
circMTO1
A recent study reported the circRNA expression profile in HCC and identified circMTO1, generated from the gene mitochondrial translation optimization 1 (MTO1). The level of circMTO1 was found to be downregulated in HCC tissues and positively correlated with survival of HCC patients. Biochemical assays revealed the interaction of miR-9 with circMTO1in HCC cells. The circMTO1 silencing led to the suppression of p21 which is a target of miR-9, leading to increase in proliferation and invasion of HCC cells. Taken together, these data suggest that circMTO1 inhibits HCC progression by acting as ceRNA for miR-9 to upregulate the expression of p21 [64].
hsa_circ_0005986
The circRNA hsa_circ_0005986 was found to be downregulated in HCC tissue samples compared with adjacent non-tumorous tissues [65]. Furthermore, hsa_circ_0005986 expressions were significantly downregulated in HCC cell lines, HepG2, SMMC7721, and HCCLM3 compared to normal hepatic cell line L02. Interestingly, the level of hsa_circ_0005986 downregulation was found to be correlated with Barcelona clinic liver cancer (BCLC) stage, chronic hepatitis B family history, and tumor diameters. miR-129-5p was one of the miRNA that could be sponged by hsa_circ_0005986 and regulated the target gene Notch1. Silencing of hsa_circ_0005986 increased miR-129-5p activity and downregulated Notch1 expression, leading to increase in cell proliferation and tumorigenesis in HCC [65].
hsa_circ_000984
The circular RNA hsa_circ_000984 generated from the CDK6 gene was significantly overexpressed in colorectal cancer (CRC) tissues from patients as well as in the CRC cell lines [66]. Further, the expression level of hsa_circ_000984 was positively associated with CRC advancement. The knockdown of hsa_circ_000984 expression led to inhibition of cell proliferation, migration, and invasion in CRC cell lines. Hsa_circ_000984 could act as a sponge for miR-106b, leading to upregulation of miR-106b target CDK6. Together, these data suggest that the hsa_circ_000984 could upregulate CDK6 by inhibiting miR-106b activity, thereby promoting colon cancer growth and metastasis [66].
hsa_circ_0020397
Another study reported that the circRNA hsa_circ_0020397 was upregulated, while its target miR-138 was downregulated in CRC cells. Furthermore, overexpression of hsa_circ_0020397 could inhibit the miR-138 activity indicating that hsa_circ_0020397 act as a ceRNA for miR-138. The hsa_circ_0020397 promoted the expression of miR-138 targets telomerase reverse transcriptase (TERT) and programmed death-ligand 1 (PD-L1) by sponging miR-138, thereby promoting cell viability and invasion of CRC cells. Together, these data suggest that hsa_circ_0020397 plays a crucial role in CRC pathogenesis by acting as a sponge for miR-138 [67].
hsa_circ_0009910
In osteosarcoma, the hsa_circ_0009910 expression was found to be upregulated and silencing of circ_0009910 promoted cell cycle arrest and apoptosis. The miR-449a expression was found to be suppressed in osteosarcoma cells and predicted to be sponged by circ_0009910. Further, miR-449a could target and downregulate the expression of IL6R in osteosarcoma cells, thereby promoting inhibition of cell proliferation, cell cycle arrest, and apoptosis. The transcript level of IL6R was also found to be negatively correlated with the level of miR-449a in osteosarcoma cells. Taken together, these data suggested that the carcinogenesis of osteosarcoma cells was induced by the circ_0009910/miR-449a/IL6R axis [68].
circGFRA1
A little is known about the role of circRNA in triple negative breast cancer (TNBC). A recent study reported that the circGFRA1 was upregulated in TNBC cell lines and tissues. Kaplan-Meier survival analysis suggested that the level of circGFRA1 was negatively correlated with survival. CircGFRA1 silencing led to the suppression of proliferation and induced apoptosis in TNBC cells. Further, biochemical assays found that circGFRA1 can directly bind and inhibit miR-34a activity, leading to increase in miR-34a target gene GFRA1. In sum, circGFRA1 act as a sponge for miR-34a to regulate GFRA1 expression in TNBC [69].
hsa-circ-0016347
Hao J et al. reported that the circ-0016347 acted as a ceRNA for miR-214 in osteosarcoma cell leading to upregulation of miR-24 target caspase-1. Moreover, circ-0016347 was found to induce proliferation and invasion of osteosarcoma cells. These data suggested that the circ-0016347 plays a crucial role in osteosarcoma progression by acting as a sponge for miR-124, which could be used as a potential target for development of therapy for osteosarcoma [70].
circWDR77
The circRNA expression profiling in glucose-induced vascular smooth muscle cells (VSMCs) discovered hundreds of differentially expressed circRNAs. CircWDR77 is one of the upregulated circRNAs whose silencing led to inhibition of proliferation and migration of VSMCs. Computational prediction suggested the interaction of circWDR77 with miR-124. Furthermore, circWDR77 inhibited miR-124 activity and upregulated the expression of miR-124 target fibroblast growth factor 2 (FGF2) in VSMCs. Together, these results indicated that the proliferation and migration of VSMCs were regulated through circWDR77/miR-124/FGF2 axis [71].
circACTA2
Neuregulin-1 (NRG-1) was found to be upregulated and cleaved in response to transforming growth factor-β1 in VSMCs. NRG-1 was also known to promote the expression of an extracellular epidermal growth factor-like domain and intracellular domain (NRG-1-ICD) which induced circular ACTA2 (alpha-actin-2; circACTA2) expression. Further, circACTA2 acted as a sponge for miR-548f-5p which upregulated α-SMA expression, leading to increase in stress fiber formation and cell contraction in VSMCs. Together, these data indicated that circACTA2 fine-tunes the α-SMA expression and VSMC contraction through the NRG-1-ICD/circACTA2/miR-548f-5p axis [72].
hsa_circ_0012673
The hsa_circ_0012673 expression was upregulated in lung adenocarcinoma (LAC) tissues compared with adjacent non-tumor tissues. Further, the expression level of circ_0012673 was positively correlated with tumor size. Biochemical assays revealed that hsa_circ_0012673 promoted LAC proliferation by acting as a sponge for miR-22, which inhibits erb-b2 receptor tyrosine kinase 3 (ErbB3) [73].
circRNA_LARP4
The large tumor suppressor kinase 1 (LATS1) acts as a tumor suppressor in gastric cancer by regulating the Hippo signaling pathway. The circRNA_ LARP4 was downregulated in gastric cancer and predicted to sponge miR-424 computationally. The direct interaction between miR-424 and LATS1 or circLARP4 was verified by various biochemical assays. Overexpression of miR-424 suppressed the expression of miR-424 target gene LATS1 which led to increase in proliferation and invasion of gastric cancer cells. In sum, circLARP4 act as a novel tumor suppressive factor by sponging miR-424-5p which modulate the expression of LATS1 in gastric cancer cells [74].
circ-ABCB10
The circ-ABCB10 was significantly overexpressed in breast cancer tissue. Further experiments suggested that silencing of circ-ABCB10 inhibited the proliferation and promoted apoptosis of breast cancer cells. Bioinformatics and biochemical analysis found that miR-1271 can be sponged by circ-ABCB10. Furthermore, the effect of circ-ABCB10 on breast cancer cells was rescued by miR-1271. These data indicated that circ-ABCB10 promotes breast cancer pathogenesis via sponging miR-1271 [75].
3 Web Tools for Analysis of miRNA-circRNA Interaction
Besides in-depth studies on functional circRNAs, several circRNA databases have been developed to explore the interaction of circRNAs with miRNAs. The databases such as CircInteractome, Circ2Traits, CircNet, and StarBase v2.0 provide excellent platforms to predict the miRNA-circRNA interactions (Table 6.2).
circInteractome
The CircInteractome (http://circinteractome.nia.nih.gov) database is the first web tool developed to predict the miRNAs-circRNA interactions for circRNAs listed in CircBase [76,77,78]. Furthermore, this is the only web tool available to date for designing divergent primer and siRNA against circRNAs. Together, the CircInteractome web tool provides bioinformatic analyses of miRNA-binding sites on circRNAs and predicts potential miRNA sponge circRNAs that are crucial for posttranscriptional gene regulation [76].
circ2Trait
The circ2Traits (http://gyanxet-beta.com/circdb/) is a database of 1951 human circRNAs and their association with 105 human diseases [79]. The circRNAs were categorized based on number of disease-associated SNPs, AGO interaction sites, and interaction with disease-associated miRNA. Circ2Trait also checks the enrichment of sets of genes in the miRNA-circRNA interactome that is associated with particular diseases. Circ2Trait also provides the complete information of miRNA-circRNA-mRNA-lncRNA interaction networks for the human diseases.
CircNet
The CircNet (http://circnet.mbc.nctu.edu.tw/) is a database to explore circRNA expression in specific tissues, circRNAs isoforms, circRNA sequences, and circRNA-miRNA interactions. The CircNet database was the first database to report the expression of tissue-specific circRNAs and circRNA-miRNA-gene interaction network. In sum, the CircNet is an interactive web interface for visualization and analysis of the regulatory network of circRNA, miRNA, and genes [80].
starBase v2.0
The starBase v2.0 (http://starbase.sysu.edu.cn/) is the first database to systematically identify the regulatory RNA-RNA and protein-RNA interaction networks using the experimentally supported CLIP-Seq data. This study identified ∼9000 miRNA-circRNA regulatory interactions. Moreover, starBase v2.0 provides CLIP-supported miRNA target sites for interacting circRNAs. This web server predicts the functional interaction of miRNA-circRNA and their coordinated regulatory networks [81].
4 Conclusion and Future Directions
With the advancement in circRNA enrichment and sequencing technology, a huge number of circRNAs have been identified in various organisms, and the number will most likely increase. The circRNAs are currently one of the focus areas in biological research, and the field is still in its early stages. There is huge interest in how circRNAs are generated and what are their biological functions. Although thousands of circRNAs have been identified, only a handful of circRNAs has been reported to have a biological function. Almost all studies on circRNAs largely focused on their interactions with miRNAs and RBPs. The expression of vast number and types of circular RNAs increases the difficulty level for understanding their regulatory mechanisms. Further intensive studies are required to understand the biogenesis of circRNAs, functional interaction of circRNAs with genome and mRNA transcripts, and their translatability into proteins. With ever-increasing evidence of functional circRNA and discovery of novel molecular mechanisms of action, there is no doubt that circRNAs will be used for disease diagnosis and treatment in near future.
References
Crick F (1970) Central dogma of molecular biology. Nature 227(5258):561–563
Derrien T, Johnson R, Bussotti G et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22(9):1775–1789
Noller HF (1991) Ribosomal RNA and translation. Annu Rev Biochem 60:191–227
Hsiao KY, Sun HS, Tsai SJ (2017) Circular RNA – new member of noncoding RNA with novel functions. Exp Biol Med (Maywood) 242(11):1136–1141
Wery M, Kwapisz M, Morillon A (2011) Noncoding RNAs in gene regulation. Wiley Interdiscip Rev Syst Biol Med 3(6):728–738
Yoon JH, Abdelmohsen K, Gorospe M (2013) Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol 425(19):3723–3730
Sanger HL, Klotz G, Riesner D et al (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 73(11):3852–3856
Kos A, Dijkema R, Arnberg AC et al (1986) The hepatitis delta (delta) virus possesses a circular RNA. Nature 323(6088):558–560
Nigro JM, Cho KR, Fearon ER et al (1991) Scrambled exons. Cell 64(3):607–613
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32(5):453–461
Panda AC, Abdelmohsen K, Gorospe M (2017) RT-qPCR detection of senescence-associated circular RNAs. Methods Mol Biol 1534:79–87
Memczak S, Jens M, Elefsinioti A et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338
Jeck WR, Sorrentino JA, Wang K et al (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19(2):141–157
Hansen TB, Jensen TI, Clausen BH et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Panda AC, De S, Grammatikakis I et al (2017) High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res 45(12):e116
Xia S, Feng J, Lei L et al (2017) Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform 18(6):984–992
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12(4):381–388
Panda AC, Grammatikakis I, Munk R et al (2017) Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA 8(2):e1413
Li Z, Huang C, Bao C et al (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22(3):256–264
Chen LL (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17(4):205–211
Zhang Y, Xue W, Li X et al (2016) The biogenesis of nascent circular RNAs. Cell Rep 15(3):611–624
Abdelmohsen K, Panda AC, Munk R et al (2017) Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14(3):361–369
Haque S, Harries LW (2017) Circular RNAs (circRNAs) in health and disease. Genes (Basel) 8(12):353
Berezikov E, Guryev V, van de Belt J et al (2005) Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120(1):21–24
Rajewsky N (2006) microRNA target predictions in animals. Nat Genet 38(Suppl):S8–S13
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39(Database issue):D152–D157
Siomi H, Siomi MC (2010) Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell 38(3):323–332
Pillai RS (2005) MicroRNA function: multiple mechanisms for a tiny RNA? RNA 11(12):1753–1761
Zhou X, Yang PC (2012) MicroRNA: a small molecule with a big biological impact. Microrna 1(1):1
Lee KP, Shin YJ, Panda AC et al (2015) miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4. Genes Dev 29(15):1605–1617
Panda AC, Abdelmohsen K, Gorospe M (2017) SASP regulation by noncoding RNA. Mech Ageing Dev 168:37–43
Panda AC, Sahu I, Kulkarni SD et al (2014) miR-196b-mediated translation regulation of mouse insulin2 via the 5′UTR. PLoS One 9(7):e101084
Munk R, Panda AC, Grammatikakis I et al (2017) Senescence-associated microRNAs. Int Rev Cell Mol Biol 334:177–205
Yu L, Gong X, Sun L et al (2016) The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One 11(7):e0158347
Tang W, Ji M, He G et al (2017) Silencing CDR1as inhibits colorectal cancer progression through regulating microRNA-7. Onco Targets Ther 10:2045–2056
Peng L, Yuan XQ, Li GC (2015) The emerging landscape of circular RNA ciRS-7 in cancer (review). Oncol Rep 33(6):2669–2674
Lukiw WJ (2013) Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet 4:307
Xu H, Guo S, Li W et al (2015) The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 5:12453
Kefas B, Godlewski J, Comeau L et al (2008) microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res 68(10):3566–3572
Pan H, Li T, Jiang Y et al (2018) Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem 119(1):440–446
Capel B, Swain A, Nicolis S et al (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73(5):1019–1030
Long L, Huang G, Zhu H et al (2013) Down-regulation of miR-138 promotes colorectal cancer metastasis via directly targeting TWIST2. J Transl Med 11:275
Huang G, Zhu H, Shi Y et al (2015) cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One 10(6):e0131225
Yang C, Yuan W, Yang X et al (2018) Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer 17(1):19
Li F, Zhang L, Li W et al (2015) Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 6(8):6001–6013
Wan L, Zhang L, Fan K et al (2016) Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta-catenin pathway. Biomed Res Int 2016:1579490
Zheng Q, Bao C, Guo W et al (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215
Chen G, Shi Y, Liu M et al (2018) circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis 9(2):175
Li Y, Zheng F, Xiao X et al (2017) Circ HIPK3 sponges mi R-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep 18(9):1646–1659
Tian F, Wang Y, Xiao Z et al (2017) Circular RNA circHIPK3 promotes NCI-H1299 and NCI-H2170 cell proliferation through miR-379 and its target IGF1. Zhongguo Fei Ai Za Zhi 20(7):459–467
Panda AC, Grammatikakis I, Kim KM et al (2017) Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res 45(7):4021–4035
Chen J, Li Y, Zheng Q et al (2017) Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett 388:208–219
Liu Q, Zhang X, Hu X et al (2016) Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation. Sci Rep 6:22572
Zhong Z, Huang M, Lv M et al (2017) Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett 403:305–317
Zhong Z, Lv M, Chen J (2016) Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep 6:30919
Wang K, Sun Y, Tao W et al (2017) Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett 394:1–12
Wang K, Long B, Liu F et al (2016) A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 37(33):2602–2611
Peng L, Chen G, Zhu Z et al (2017) Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget 8(1):808–818
Xie H, Ren X, Xin S et al (2016) Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 7(18):26680–26691
Song YZ, Li JF (2018) Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun 495(3):2369–2375
Cheng X, Zhang L, Zhang K et al (2018) Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis 77(5):770–779
Zhou ZB, Du D, Huang GX et al (2018) Circular RNA Atp9b, a competing endogenous RNA, regulates the progression of osteoarthritis by targeting miR-138-5p. Gene 646:203–209
Wang L, Wei Y, Yan Y et al (2018) CircDOCK1 suppresses cell apoptosis via inhibition of miR196a5p by targeting BIRC3 in OSCC. Oncol Rep 39(3):951–966
Han D, Li J, Wang H et al (2017) Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 66(4):1151–1164
Fu L, Chen Q, Yao T et al (2017) Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget 8(27):43878–43888
Xu XW, Zheng BA, Hu ZM et al (2017) Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget 8(53):91674–91683
Zhang XL, Xu LL, Wang F (2017) Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int 41(9):1056–1064
Deng N, Li L, Gao J et al (2018) Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun 495(1):189–196
He R, Liu P, Xie X et al (2017) circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res 36(1):145
Jin H, Jin X, Zhang H et al (2017) Circular RNA hsa-circ-0016347 promotes proliferation, invasion and metastasis of osteosarcoma cells. Oncotarget 8(15):25571–25581
Chen J, Cui L, Yuan J et al (2017) Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem Biophys Res Commun 494(1–2):126–132
Sun Y, Yang Z, Zheng B et al (2017) A novel regulatory mechanism of smooth muscle alpha-actin expression by NRG-1/circACTA2/miR-548f-5p axis. Circ Res 121(6):628–635
Wang X, Zhu X, Zhang H et al (2018) Increased circular RNA hsa_circ_0012673 acts as a sponge of miR-22 to promote lung adenocarcinoma proliferation. Biochem Biophys Res Commun 496(4):1069–1075
Zhang J, Liu H, Hou L et al (2017) Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer 16(1):151
Liang HF, Zhang XZ, Liu BG et al (2017) Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res 7(7):1566–1576
Dudekula DB, Panda AC, Grammatikakis I et al (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13(1):34–42
Panda AC, Dudekula DB, Abdelmohsen K et al (2018) Analysis of circular RNAs using the web tool CircInteractome. Methods Mol Biol 1724:43–56
Glazar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. RNA 20(11):1666–1670
Ghosal S, Das S, Sen R et al (2013) Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 4:283
Liu YC, Li JR, Sun CH et al (2016) CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 44(D1):D209–D215
Li JH, Liu S, Zhou H et al (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42(Database issue):D92–D97
Acknowledgments
This work was supported by the Science and Engineering Research Board, a statutory body of the Department of Science and Technology (DST), Government of India (SERB/F/6890/2017-18).
Conflicts of Interest
The authors have no conflicts of interest to declare.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Panda, A.C. (2018). Circular RNAs Act as miRNA Sponges. In: Xiao, J. (eds) Circular RNAs. Advances in Experimental Medicine and Biology, vol 1087. Springer, Singapore. https://doi.org/10.1007/978-981-13-1426-1_6
Download citation
DOI: https://doi.org/10.1007/978-981-13-1426-1_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1425-4
Online ISBN: 978-981-13-1426-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)