Abstract
Medical concept extraction was the part of i2b2 challenge 2010 in which three concepts like problem, treatment, and test were targeted. This paper presents a rule-based method for automatic concept extraction from clinical notes. The method is compound of two modules that are text preprocessing and automatic rules creation. Rules creation module generates rules to recognize single and composite words for concept identification and mapping these concepts with their semantic types using medical dictionary UMLS. The method is applied to two different training datasets by Beth Medical Center with 73 annotated clinical notes and by Partners Healthcare with 97 annotated clinical notes, and then evaluated its performance using a test dataset with 256 annotated notes. The method achieved an average precision of 70% and average recall of 60%.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
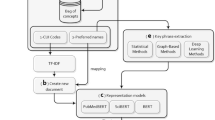
Medical concept extraction is divided into two sequential subtasks: first one is identification of medical entities, and other is classification of the semantic category for each detected medical entity. I2b2 had organized an NLP challenge for clinical data in 2010. Extracting clinical concepts from natural language text, to include medical problems, tests, and treatments was one of the three tasks of that challenge [1]. In this paper, a rule-based method is proposed for the automatic medical concept extraction. The method contains two modules text preprocessing and rule template creation. Rule template creation module generates some rules for single and composite word identification and rules for concept mapping with their semantic types using UMLS. Performance of the method is evaluated using precision and recall and comparison with MetaMap results.
2 Background
Many NLP challenges such as the i2b2 Challenge Shared Tasks [2] and the ShARe/CLEF eHealth Shared Task [3, 4] had focused on medical concept extraction. In numerous previous works, rule-based approaches were used for natural language processing research for clinical notes. MetaMap was developed to recognize Metathesaurus concepts from biomedical texts by utilizing the UMLS [5]. Experimentation of MetaMap 2013v2 on i2b2 2010 clinical data with the 2013AB NLM relaxed database is performed in [6] and gives low score of precision (47.3%) and recall (36%). Other than rule-based, some machine learning approaches, ensemble-based approach [7], and hybrid approaches [8] have been used for concept extraction. We still identified some issues related to classification and entity boundary identification which has some scope to improve recall of the system. For text preprocessing, many natural language tools have been used like openNLP, tree tagger, Stanford parser [9], Lingpipe, Splitta, SPECIALIST, c-TAKES, and Stanford CoreNLP. Evaluation of sentence boundary detection using these tools is performed in [10]. For semantic types mapping, many systems have used UMLS [11]. Unsupervised biomedical named entity recognition is performed on GENIA corpus and i2b2 2010 dataset [12]. Various issues are still present in the identification and classification of clinical concepts because of unstructured nature of clinical notes. Common challenges like boundary identification of single and multi-adjacent words for designing and developing clinical decision support systems had focused in [13].
3 Proposed Method and Dataset
The proposed method has used clinical records provided by I2b2 National Center in 2010 NLP challenge. The dataset consisted of discharge summaries from Partners Healthcare and Beth Israel Deaconess Medical Center. All these records had been manually annotated for three types of concepts (medical problems, tests, and treatments), according to guidelines provided by the i2b2/VA challenge organizers [2]. Gold dataset of Beth Center contains 73 annotated notes, Partners Healthcare contains 97 annotated notes, and for system evaluation, they have provided test dataset which contains 256 annotated notes. For the experiment, the method has used both training and test annotated notes and for evaluation, gold dataset is used. The proposed rule-based method is divided into two sub-modules: text preprocessing and rules template creation.
3.1 Text Preprocessing
I2b2 clinical notes require text preprocessing because of theirs unstructured and semi-structured nature of the text. These notes contain some sections such as discharge date, admission date, allergies, history of present illness, past medical history, etc. Every section contains some information related to every patient with some special characters, colons, semicolons, punctuations, hyphens, etc. In this method, Natural Language Toolkit (NLTK) is used for line tokenization, word tokenization, and POS tagging. Special characters identification is performed by some regular expressions, which is used for word tokenization. After text preprocessing, numerous single words with their POS tags have been identified, which is applied as input for concepts generation.
3.2 Rules Template Creation
After getting words with POS tags, concepts have been identified. Concepts can be single word or composite word. For concepts identification, some rule templates are created, which are some common patterns and takes different values for different conditions. Rule template creation is divided into two subparts: (1) rules for composite words identification, and (2) then rules for map these words as concepts with UMLS using their semantic types. For multiword or composite word identification, some features like word, previous word, next word, previous word POS, and next word POS are used. For POS feature, same rules as word feature are used for rule template generation, only few POS tag combinations such as noun, pronoun, adjective, and determiner have considered for concept extraction. Rules using word feature are defined below: Suppose w = “single word” and cw = “composite word”:
-
Rule 1: if w is middle word, then y2 = w, cw = y1 + y2 + y3, where y1 = previous one word and y3 = next one word. Rule 2: if w is middle word, then y3 = w, cw = y1 + y2 + y3 + y4 + y5, where y1 = previous two words from y3, y2 = previous one word, y4 = next one word, y5 = next two words from y3. Rule 3: if w is the first word, then y1 = w, cw = y1 + y2 + y3, where y2 = next one word and y3 = next two words from y1. Rule 4: if w is the last word, then y3 = w, cw = y1 + y2 + y3, where y2 = previous one word and y1 = previous two words from y3. Rule 5: if w is the first word, then y1 = w, cw = y1 + y2, where y2 = next one word. Rule 6: if w is the last word, then y2 = w, cw = y1 + y2, where y1 = previous one word.
Rule template for concept mapping with UMLS
Some rule templates have defined below in which Semantic type is one attribute of medical information which is defined in UMLS Metathesaurus database. Table 1 show some categories of semantic type which is used for three medical concepts.
-
Rule 7: (Semantic type = x1) ˅ (Semantic type = x2) ˅ (Semantic type = x3) ˅ (Semantic type = x4) ˅ (Semantic type = x5) → Class = X, where X = 1 to n are semantic types corresponding to their concept category which is used for classification such as if x1 = “Disease or Syndrome”, x2 = “Sign or Symptom”, x3 = “Finding”, x4 = “Pathologic Function”, then X = Problem; if x1 = “Tissue”, x2 = “Cell”, x3 = “Laboratory Procedure”, x4 = “Laboratory or Test Result”, x5 = “Clinical Attribute” then X = Test; and if x1 = “Antibiotic”, x2 = “Organic Chemical”, x3 = “Therapeutic or Preventive Procedure”, x4 = “Pharmacologic substance”, x5 = “Diagnostic Procedure”, then X = Treatment. If word is matched with any of semantic type of categories (given in Table 1), then it can be correctly mapped with appropriate concept class. In the proposed method, exact matching is performed.
4 Results and Discussions
The proposed method has performed experiments on I2b2 2010 clinical notes. It contains 73 annotated clinical notes provided by Beth Medical Center and 97 annotated clinical notes provided by Partners Healthcare and test dataset with 256 annotated notes. Rules created for these clinical notes and evaluated using gold dataset. The system has achieved an average precision of 70% and average recall of 60%, average of all concepts problem, test, and treatment. The system has performed better than MetaMap 2013v2. MetaMap gave a low score of precision (47.3%) and recall (36%). Figure 1a shows a comparison of performance of proposed method with MetaMap 2013v2 for all concepts based on precision and recall. Performance is measured for every concept individually also and compare Beth and Partners results concept wise (see Fig. 1b). The precision of Beth data for problem and treatment concept is more than Partners data, but for the test is equal. Recall of Partners data is more than Beth data for every concept.
After error analysis, it has been found that recall still has some scope for improvement. Few concepts have been missed because of incorrect boundary identification of composite words. Boundary identification issue is more observed in problem concept because gold standard contains some composite words of problem like “burst of atrial fibrillation” and found in treatment concept also like “saphaneous vein graft -> posterior descending artery”. The method has performed strict matching but not used partial matching; large composite words are not correctly or matched in UMLS database as medical concept. This issue can be resolved using relaxed or partial matching. Rules for composite words creation are designed in such a way which considers word features for maximum five words. Some concepts are combination of 4–5 or more words such “mild postoperative widening of the cardiomediastinal silhouette”, which are not identified in the proposed method. Text preprocessing included some regular expressions for special character identification, which are used as word splitter, but few words identified in gold standard which contains these characters in between composite words such as “severe 3 vessel disease, “heel/shin”, “leg, emg &apos”, etc., which are incorrectly recognized as concept using proposed method. POS feature has also used for composite word identification; few combinations of POS patterns have been defined in rules like “NN, NNP, NST”, “DT, NNP”, “DT, JJ, NNP”. These errors can be resolved in future work by designing some other rules or using hybrid approaches.
5 Conclusions
In the proposed rule-based method, medical concepts have been recognized using rules template generation for multiword identification and concept mapping with UMLS. It has been found that the performance of the system is better than MetaMap 2013v2 in terms of precision and recall. Still recall can be improved by more accurate multiword boundary identification; for this, rules with more features like stemming, prefix, and suffix can be added. Rules are generalized not domain-dependent and regardless of the semantics of the sentences. In future work, this method can be applied to other corpuses for entity extraction with different regular expressions for text preprocessing, it will give better results. The method has not used any machine learning approach, that is why performance is not dependent on the size of dataset. Rules can be applied to small and large dataset in a similar way, but system processing time will be increased for the large dataset. In future work, this method can be implemented in a distributed environment for fast processing of large dataset.
References
Jiang, M., Chen, Y., Liu, M., Rosenbloom, S.T., Mani, S., Denny, J.C., Xu, H.: A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J. Am. Med. Inform. Assoc. JAMIA 18, 601–606 (2011)
Uzuner, Ö., South, B.R., Shen, S., DuVall, S.L.: 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J. Am. Med. Inform. Assoc. JAMIA 18, 552–556 (2011)
Suominen, H., Salanterä, S., Velupillai, S., Chapman, W.W., Savova, G., Elhadad, N., Pradhan, S., South, B.R., Mowery, D.L., Jones, G.J.F., Leveling, J., Kelly, L., Goeuriot, L., Martinez, D., Zuccon, G.: Overview of the ShARe/CLEF eHealth Evaluation Lab 2013. In: Forner, P., Müller, H., Paredes, R., Rosso, P., Stein, B. (eds.) Information Access Evaluation. Multilinguality, Multimodality, and Visualization. Proceedings of 4th International Conference of the CLEF Initiative, CLEF 2013, Valencia, Spain, pp. 212–231. Springer Berlin Heidelberg, Berlin, Heidelberg, 23–26 Sept 2013
Kelly, L., Goeuriot, L., Suominen, H., Schreck, T., Leroy, G., Mowery, D.L., Velupillai, S., Chapman, W.W., Martinez, D., Zuccon, G., Palotti, J.: Overview of the ShARe/CLEF eHealth evaluation lab 2014. In: Kanoulas, E., Lupu, M., Clough, P., Sanderson, M., Hall, M., Hanbury, A., Toms, E. (eds.) Information Access Evaluation. Multilinguality, Multimodality, and Interaction. Proceedings of 5th International Conference of the CLEF Initiative, CLEF 2014, Sheffield, UK, pp. 172–191. Springer International Publishing, Cham, 15–18 Sept 2014
Aronson, A.R., Lang, F.-M.: An overview of MetaMap: historical perspective and recent advances. J. Am. Med. Inform. Assoc. JAMIA 17, 229–236 (2010)
Kim, Y., Riloff, E., Hurdle, J.F.: A study of concept extraction across different types of clinical notes. In: AMIA Annual Symposium Proceedings 2015, pp. 737–746 (2015)
Kang, N., Afzal, Z., Singh, B., van Mulligen, E.M., Kors, J.A.: Using an ensemble system to improve concept extraction from clinical records. J. Biomed. Inform. 45, 423–428 (2012)
Minard, A.-L., Ligozat, A.-L., Ben Abacha, A., Bernhard, D., Cartoni, B., Deléger, L., Grau, B., Rosset, S., Zweigenbaum, P., Grouin, C.: Hybrid methods for improving information access in clinical documents: concept, assertion, and relation identification. J. Am. Med. Inform. Assoc. 18, 588 (2011)
Xu, H., AbdelRahman, S., Jiang, M., Fan, J.W., Huang, Y.: An initial study of full parsing of clinical text using the Stanford Parser. In: 2011 IEEE International Conference on Bioinformatics and Biomedicine Workshops (BIBMW), pp. 607–614 (2011)
Griffis, D., Shivade, C., Fosler-Lussier, E., Lai, A.M.: A quantitative and qualitative evaluation of sentence boundary detection for the clinical domain. AMIA Summits Transl. Sci. Proc. 2016, 88–97 (2016)
Bodenreider, O.: The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 32, D267–D270 (2004)
Zhang, S., Elhadad, N.: Unsupervised biomedical named entity recognition: experiments with clinical and biological texts. J. Biomed. Inform. 46, 1088–1098 (2013)
Dehghan, A., Keane, J.A., Nenadic, G.: Challenges in clinical named entity recognition for decision support. In: 2013 IEEE International Conference on Systems, Man, and Cybernetics, pp. 947–951 (2013)
Acknowledgements
I would like to thank the 2010 i2b2/VA challenge organizers for the development of training and test corpora. I also thank U.S. National Library of Medicine for providing UMLS for the research work.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Sahu, R. (2018). Rule-Based Method for Automatic Medical Concept Extraction from Unstructured Clinical Text. In: Sa, P., Bakshi, S., Hatzilygeroudis, I., Sahoo, M. (eds) Recent Findings in Intelligent Computing Techniques . Advances in Intelligent Systems and Computing, vol 709. Springer, Singapore. https://doi.org/10.1007/978-981-10-8633-5_27
Download citation
DOI: https://doi.org/10.1007/978-981-10-8633-5_27
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-8632-8
Online ISBN: 978-981-10-8633-5
eBook Packages: EngineeringEngineering (R0)