Abstract
Cultivated rice (Oryza sativa L.) was domesticated from the Asian wild species, Oryza rufipogon Griff. Among morphological differences between them, one of the striking traits specific to cultivated rice is loss of seed shattering. In the early stage of rice domestication, the related traits of this character have been desirable for the ancient seed gatherers because it enhances the efficiency of seed collection. In this chapter, we propose that three morphological traits, closed panicle shape, non-seed shattering, and seed awning, played important roles in controlling the degree of seed dispersal. First, we reviewed domestication loci controlling the three traits. We then evaluated allele effects at these loci using reciprocal backcross populations between O. sativa Nipponbare and our standard wild accession of O. rufipogon W630. In the genetic background of cultivated rice, all the wild functional alleles were responsible for these domestication traits. On the other hand, cultivated non-functional alleles were not always associated with the drastic morphological changes in the genetic background of wild rice. Since ancient humans have selected cultivated-type mutants in natural wild populations, possible domestication process for the emergence of cultivated rice is discussed based on the effects of cultivated non-functional alleles.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
12.1 Introduction
Cultivated rice (Oryza sativa L.) was domesticated from the Asian wild species, Oryza rufipogon Griff (Fig. 12.1) (Oka 1988). Several distinct morphological differences are observed between them, of which one of the striking traits specific to cultivated rice is loss of seed shattering. The related traits of this character have been desirable for the ancient seed gatherers because they enhance the efficiency of seed collection. When planting what they had harvested started, artificial selection was imposed on the shattering-related traits of rice plants. Therefore, it is considered that reduction of the seed-shattering degree is required in the early phase of domestication (Flannery 1973; Harlan et al. 1973; Fuller 2007).
Compared with cultivated rice, wild rice produces spikelets with long awns (bristle-like organs in the tip of the lemma) on spreading panicles (Fig. 12.2), and the seeds mature from top to bottom of the panicle. About 2 weeks after flowering, an abscission layer is completely formed between the pedicel and spikelet, and the seeds shed by themselves. Open panicle shape and long awns also enhance dispersal of mature seeds. Recently, we found a simple morphological change in panicle shape had a large impact on the seed-shedding behavior of wild rice (Fig. 12.3) (Ishii et al. 2013). In the maturing stage, wild rice plants with a cultivated-type closed panicle had significantly reduced seed shedding. There was a tendency to retain the upper mature seeds on the panicles through support from long awns in the lower immature seeds. In addition, the long awns in closed panicles inhibited the free exposure of anthers and stigmas on the flowering spikelets, resulting in a significant reduction in the outcrossing rate. These observations suggest that closed panicle shape and long awns played important roles in seed-shedding and pollinating behaviors during the early stage of rice domestication. We do not know the exact historical sequence of rice domestication, but three traits, i.e., closed panicle shape, non-seed shattering, and seed awning are closely associated with the emergence of cultivated rice. In this chapter, the genetic mechanisms of these three domestication traits are reviewed and discussed.
12.2 A Locus Responsible for Panicle Spreading
Panicles of cultivated rice are closed, whereas most Asian wild rice have spreading panicles. The morphological differences in panicles between cultivated and wild rice are mainly explained by a single locus, SPR3 (spreading panicle-3), where wild rice has the dominant allele for spreading panicles (Eiguchi and Sano 1990).
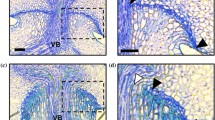
To examine morphological and genetic mechanisms of panicle structure, we previously investigated two plant materials, a cultivar of O. sativa Nipponbare (with closed panicles) and a wild accession of O. rufipogon W630 (with spreading panicles) (Ishii et al. 2013). The panicle shape of the cultivated and wild accessions is formed at the basal structure in the primary branches. At the cellular level, O. rufipogon W630 has tissue resembling a bump between the main and primary branches (Fig. 12.4). To confirm the chromosomal location of the panicle-spreading locus, we performed quantitative trait locus (QTL) analysis using 161 BC2F8 plants between O. sativa Nipponbare (recurrent parent) and O. rufipogon W630 (donor parent). One strong QTL corresponding to SPR3 was detected on the long arm of chromosome 4, which explained 80.1% of the phenotypic variance. Further fine mapping narrowed down the location of SPR3 to a 9.3-kb region. Although no coding sequences were predicted in this region, complementation tests showed that the region regulates the rice LIGULELESS1 gene (OsLG1), Locus ID Os04g0656500 in the Rice Annotation Project (RAP) Database (Fig. 12.5, Table 12.1). OsLG1 is located 10 kb away from the SPR3 locus and encodes a SQUAMOSA promoter binding protein (SBP) domain (Lee et al. 2007). Furthermore, it controls laminar joint and ligule development. The physical position is about 33.49 Mb on chromosome 4 according to the RAP Database. This gene was also named OsSPL8, one of the SBP-box genes in rice (Yang et al. 2008). In the basal parts of primary branches, higher levels of OsLG1 expression were observed in O. rufipogon W630 than in O. sativa Nipponbare (Ishii et al. 2013). In addition, overexpression of OsLG1 has been reported to induce the spreading panicle phenotype in transgenic lines (Zhu et al. 2013). Since the panicle phenotypes corresponded to the expression levels of OsLG1, we concluded that the SPR3 region contained the regulatory sequences of OsLG1 expression in the basal parts of the primary branches.
Panicles of O. sativa Nipponbare (a–c) and O. rufipogon W630 (d–f) (Modified from Ishii et al. 2013). (a, d) Panicles in the heading stage. Scale bar, 5 cm. (b, e) Basal structure of the primary branches indicated by white boxes in (a, d). Scale bar, 1 mm. (c, f) Longitudinal sections of the basal parts of primary branches indicated by white boxes in (b, e). Arrows show the bump structure tissue. Scale bar, 100 μm
12.3 Allele Effects at Panicle-Spreading Locus
Among the 161 BC2F8 plants between O. sativa Nipponbare and O. rufipogon W630 used in the above QTL analysis, all the plants having wild W630 alleles at SPR3 had spreading panicles. This demonstrates that wild alleles are responsible for panicle spreading in the genetic background of cultivated rice. We developed separately an introgression line that contained a chromosomal segment from O. sativa Nipponbare at the SPR3 region in the genetic background of O. rufipogon W630. This line had closed panicles with no bump structure tissue at the base of the primary branches, indicating that closed panicle shape is simply generated by a loss-of-function mutation at SPR3. The effects of the non-functional cultivated and functional wild alleles at SPR3 are shown in Fig. 12.6. We therefore infer that ancient humans may have selected closed panicle plants with the non-functional allele at SPR3 in natural wild populations.
12.4 Closed Panicle Influences Seed-Shedding and Outcrossing Behaviors in Wild Rice
To evaluate the effects of the closed panicle on seed-shedding and outcrossing behaviors in wild rice, the introgression line containing the Nipponbare allele in the genetic background of O. rufipogon W630 was produced (Ishii et al. 2013). For seed-shedding behavior, a seed-gathering experiment using the introgression line and O. rufipogon W630 was carried out in the field. Since both plants display a seed-shattering habit, their seeds were collected directly from the panicles by hand in the maturing stage, and seed-gathering rates were calculated. Significantly, more seeds (mean gathering rate = 30.6%) were collected from the introgression line than the wild parental accession of O. rufipogon W630 (19.8%), indicating that seeds could be gathered more efficiently from the closed panicles (Ishii et al. 2013). Furthermore, to confirm that the plants with closed panicles can retain mature seeds longer than those with open panicles, days from flowering to seed shedding were evaluated with the introgression line and the wild parent. A significant difference was observed for the average days between the introgression line (14.8 days) and O. rufipogon W630 (13.8 days). This indicates that plants with closed panicles could retain mature seeds for about 1 day longer than those with open panicles and explains why mature seeds were efficiently collected from the plants with closed panicles.
The outcrossing rates between the introgression line and two wild lines of O. rufipogon W630 were also examined (Ishii et al. 2013). All plant material displayed a similar wild morphology, except for the closed panicles observed in the introgression line. Outcrossing rates of the two wild lines were 10.24% and 11.71%, whereas that for the introgression line was 2.82%. A significant reduction in outcrossing rate was caused by a single non-functional allele at SPR3 which changed panicle structure from open to closed. This morphological change may have a big impact on pollination behavior during rice domestication.
12.5 Loci Responsible for Seed Shattering
Wild rice, O. rufipogon, has strong seed-shattering behavior that guarantees propagation success through seed dispersal under natural condition. A wide variation in seed-shattering degree is also observed among cultivated rice, O. sativa, suggesting that seed shattering is a quantitatively regulated trait. Two loci, sh4 and qSH1, were reported to have a strong influence on the shattering habit (Konishi et al. 2006; Li et al. 2006). The sh4 is located on the long arm of chromosome 4 with a physical position of about 34.23 Mb (just 0.74 Mb distal to SPR3 locus) (Fig. 12.5, Table 12.1). It was detected as a major QTL for seed shattering between O. rufipogon and O. sativa Indica and explains 69% of the total phenotypic variance in their segregating populations. A gene responsible for sh4 encodes a Myb-type transcription factor (Locus ID Os04g0670900). An allele with a loss-of-function mutation was found to inhibit the establishment of the abscission layer at flower development stage (Li et al. 2006). Since the loss-of-function mutation is commonly observed in cultivated rice, it is widely accepted that the mutation played an important role in rice domestication (Lin et al. 2007; Onishi et al. 2007; Zhang et al. 2009).
The other major QTL is qSH1, which was identified on chromosome 1 (ca. 36.45 Mb) using a segregating population between O. sativa Indica Kasalath and Japonica Nipponbare (Konishi et al. 2006) (Fig. 12.4, Table 12.1). A causative mutation at qSH1 was shown to be an SNP located 12 kb upstream of the rice homolog of the Arabidopsis REPLUMLESS (RPL) gene for dehiscence zone formation in the Arabidopsis silique. It belongs to the BEL1-type homeobox family (Locus ID Os01g0848400). The Nipponbare allele causes the reduction of rice RPL gene expression, resulting in the complete absence of abscission layer formation. This causative mutation is attributed to selection for the strong non-shattering behavior of Japonica cultivars.
12.6 Allele Effects at Seed-Shattering Loci
In both studies on seed-shattering loci, the functional alleles were found to be responsible for the formation and development of an abscission layer between the pedicel and spikelet in the genetic background of the cultivars (Konishi et al. 2006; Li et al. 2006).
We evaluated allele effects at these two loci using reciprocal backcross populations between O. sativa Japonica Nipponbare (with non-functional alleles) and our standard wild accession of O. rufipogon W630 (with functional alleles) (Ishikawa et al. 2010). We define the degree of seed shattering into three categories: non-seed shattering, weak seed shattering (mature seeds can be detached by hand gripping), and seed shattering (no mature seeds remained on the panicles). In the genetic background of Nipponbare, both wild alleles at sh4 and qSH1 generated plants with weak seed-shattering behavior (Fig. 12.7). These plants retain mature seeds on the panicles unless external force, such as hand gripping, is added. Strong seed shattering was observed in plants having wild alleles at both qSH1 and sh4. In the genetic background of wild rice, effects of the Nipponbare alleles were also examined (Fig. 12.7). It was serendipitous that the backcross plants having the Nipponbare alleles at either shattering locus (qSH1 or sh4) shed all seeds. This suggests that a non-functional allele at qSH1 or sh4 does not contribute to non-seed shattering in the genetic background of wild rice. The plants having the Nipponbare alleles at both qSH1 and sh4 retained mature seeds on the panicles; however, the seeds easily detached by hand tapping. This also confirms that non-shattering behavior is not obtained by a single mutation at these loci in the genetic background of wild rice. Probably, some other loci are still associated with non-shattering behavior of cultivated rice.
12.7 Loci Responsible for Seed Awning
Of the wild propagation-related traits, seed awning directly enhances seed dispersal together with seed-shattering behavior. The major seed-shattering loci have already been identified (Konishi et al. 2006; Li et al. 2006), whereas those for seed awning have been well-characterized only in the past few years. The first reported locus was Awn-1 (An-1) on chromosome 4 with the physical position of about 16.73 Mb in the RAP Database (Luo et al. 2013) (Fig. 12.5, Table 12.1). It was identified as a major QTL explaining 52% of the total phenotypic variance in the segregating population between O. rufipogon (long awn) and O. sativa (awnless). The responsible gene encodes a basic helix-loop-helix transcription factor (Locus ID Os04g0350700). In general, two types of mutations at the locus were observed among cultivars. Most Japonica cultivars have a transposon insertion in the promoter region, whereas most Indica cultivars possess a 1-bp deletion in the second exon which leads to a premature stop codon.
Another identified locus is long and barbed awn1 (LABA1) being located on chromosome 4 in the region of about 25.96 Mb, which encodes a cytokinin-activating enzyme (Locus ID Os04g0518800) (Hua et al. 2015) (Fig. 12.5, Table 12.1). This locus is associated with the transition from long barbed awns in wild rice to short barbless in Indica rice. A frame-shift 1-bp deletion in the coding region causes a premature stop codon and reduces the cytokinin concentration in awn primordia. This loss-of-function mutation is mainly observed in barbless Indica and tropical Japonica cultivars.
A regulator of awn elongation 2 (RAE2) locus regulates awn elongation in association with the epidermal patterning factor-like protein (Bessho-Uehara et al. 2016) (Fig. 12.5, Table 12.1). The locus ID is Os08g0485500 being located in the region of about 23.99 Mb on chromosome 8. In the second exon, a highly variable GC-rich region was found to harbor multiple independent mutations in cultivated rice plants. The RAE2 proteins are mainly classified into three types according to the number of cysteine residues: 4C, 6C, and 7C types. Of these, 6C is a functional type in wild rice, and putatively dysfunctional 4C and 7C types are mainly observed in Japonica and Indica cultivars, respectively.
Two loci of Awn-2 (An-2) and grain number, grain length, and awn development 1 (GAD1) were independently reported to be responsible for awn development (Gu et al. 2015; Jin et al. 2016). The An-2 and GAD1 loci are identical to LABA1 and RAE2, respectively.
12.8 Allele Effects at Seed-Awning Loci
We first performed QTL analysis for seed awning using 161 BC2F8 plants between O. sativa Nipponbare (recurrent parent without awns) and our standard wild accession of O. rufipogon W630 (donor parent with long awns). Two strong QTLs were detected on chromosomes 4 and 8 (Ikemoto et al. 2017). Subsequent causal mutation survey and fine mapping confirmed that they are identical to An-1 and RAE2 loci, respectively. Regarding LABA1, no QTL was detected in the chromosomal region near the locus. This was expected because Nipponbare has a functional allele at LABA1 just like other wild rice accessions (Hua et al. 2015). We then developed two backcross populations with reciprocal genetic backgrounds of Nipponbare and W630 to examine the allele effects and interactions at major seed-awning loci, An-1 and RAE2.
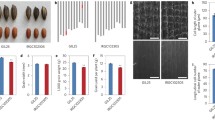
Awn length in wild rice varied among seeds even in the same plant. We noticed that the seed (or spikelet) position in a panicle affected awn length. Since wild rice exhibits seed-shattering behavior, awn lengths of O. rufipogon W630 were compared with the five upper spikelets on the top primary branch just after heading (Ikemoto et al. 2017). The results show that the second spikelet had a significantly shorter awn than the others, and the first and the third followed with similar values (Fig. 12.8). The fifth spikelet always gave a longer awn than the fourth on the same primary branches. Therefore, in this study, the gene effects and interactions were evaluated based on the awn length of the fifth spikelet which generates the longest awn among the five.
Panicle and spikelet morphology of O. rufipogon W630. (a) A panicle of O. rufipogon W630. A white box indicates the top primary branch in the panicle. (b) Spikelets from the top primary branch of O. rufipogon W630. They were arranged according to their positions from the top (first to fifth spikelets). Scale bar, 5 cm
A BC3F2 population segregated at An-1 and RAE2 was produced from the BC2F8 plants in the genetic background of Nipponbare to investigate gene interaction. In the BC3F2 population, we selected introgression plants with three combinations of genotypes on wild alleles, namely, wild homozygotes at single or both awning loci. Their awn lengths were measured based on spikelet position and compared with the parental accessions (Ikemoto et al. 2017). About half-length awns were observed in the plants having wild alleles at An-1 (40% in length compared to the wild) and RAE2 (51%). This suggests their wild alleles had awning effects in the genetic background of Nipponbare (Fig. 12.9). Additive effects were detected in the plants having wild alleles at both loci, and these produced awns whose length was about 78% of the W630 lengths.
In the genetic background of wild rice, allele effects of Nipponbare were examined using the other BC3F2 backcross population between O. sativa Nipponbare (donor parent) and O. rufipogon W630 (recurrent parent). Surprisingly, the plants having Nipponbare non-functional alleles at RAE2 showed almost the same awn lengths as those of W630 (Fig. 12.9). This suggests that the substitution of the non-functional alleles at RAE2 did not contribute to length reduction at all. But it was observed that the plants with Nipponbare alleles at An-1 had significantly shorter awns where the length reduction rate was approximately 13%. Even the plants having the Nipponbare non-functional alleles at both loci produced long awns (corresponding to about 74% of the W630 lengths). These results show that more loci other than An-1 and RAE2 are expected to contribute to awnlessness in the genetic background of wild rice.
12.9 Possible Domestication Process for the Emergence of Cultivated Rice
Loss of seed shattering is a key trait for the emergence of cultivated rice, because it enhanced the efficiency of seed collection by ancient seed gatherers. However, it is difficult to find non-shattering plants among wild rice populations. Non-shattering plants are easily eliminated under natural conditions due to their lower propagation ability. In addition, the appearance of non-shattering behavior probably requires independent mutations at multiple seed-shattering loci in the genetic background of wild rice. On the other hand, the temporary inhibition of seed shedding can be obtained through a simple morphological change in panicle shape. Wild rice has spikelets with long awns and spreading panicles, but a single mutation at SPR3 produces plants with closed panicles. They tend to retain mature seeds a little longer on the panicles with the support from the long awns of immature seeds. After maturity, all the seeds are shed without changing their wild propagation ability. Therefore, a closed panicle may be a first key trait in the development of cultivated rice (Fig. 12.10).
Furthermore, the long awns in closed panicles disturb the free exposure of anthers and stigmas on the flowering spikelets, resulting in a significant reduction of the outcrossing. This morphological change of the panicles promotes self-pollinating behavior and fixation of genes and may assist in the accumulation of homozygous alleles of recessive mutations on seed shattering. Once non-seed-shattering plants are generated in the early stage of rice domestication, seed awning may have been an undesirable trait during harvesting and rice-processing activities. Although seed awning in the genetic background of wild rice is under complex control, an awnless phenotype may be gradually generated by the accumulation of recessive alleles at seed-awning loci.
12.10 Perspective
Wild rice keeps many functional alleles at propagation-related loci to survive under natural condition, whereas cultivated rice has accumulated loss-of-function mutations at these loci. In rice domestication, the ancient humans selected desirable mutant plants in the wild natural populations. Therefore, to investigate the transition of the domestication traits, the effects of non-functional cultivated alleles should be examined in the wild genetic background. The non-functional alleles are not always associated with the drastic morphological changes, because many other wild genes are generally involved in the trait expression. Studies on gene interaction at multiple domestication loci in the wild genetic background will give good clues to elucidate the process of rice domestication.
References
Bessho-Uehara K, Wang DR, Furuta T et al (2016) Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc Natl Acad Sci USA 113:8969–8974
Eiguchi M, Sano Y (1990) A gene complex responsible for seed shattering and panicle spreading found in common wild rices. Rice Genet Newslett 7:105–107
Flannery KV (1973) The origins of agriculture. Annu Rev Anthropol 2:271–310
Fuller DQ (2007) Contrasting patterns in crop domestication and domestication rates, recent archaeobotanical insights from the Old World. Ann Bot 100:903–924
Gu B, Zhou T, Luo J et al (2015) An-2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice. Mol Plant 8:1635–1650
Harlan JR, de Wet JMJ, Price EG (1973) Comparative evolution of cereals. Evolution 27:311–325
Hua L, Wang DR, Tan L et al (2015) LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 27:1875–1888
Ikemoto M, Otsuka M, Thanh PT et al (2017) Gene interaction at seed-awning loci in the genetic background of wild rice. Genes Genet Syst 92:1–6
Ishii T, Numaguchi K, Miura K et al (2013) OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet 45:462–465
Ishikawa R, Thanh PT, Nimura N et al (2010) Allelic interaction at seed-shattering loci in the genetic backgrounds of wild and cultivated rice species. Genes Genet Syst 85:265–271
Jin J, Hua L, Zhu Z et al (2016) GAD1 encodes a secreted peptide that regulates grain number, grain length and awn development in rice domestication. Plant Cell 28:2453–2463
Konishi S, Izawa T, Lin SY et al (2006) An SNP caused loss of seed shattering during rice domestication. Science 312:1392–1396
Lee J, Park JJ, Kim SL et al (2007) Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65:487–499
Li C, Zhou A, Sang T (2006) Rice domestication by reducing shattering. Science 311:1936–1939
Lin Z, Griffith ME, Li X et al (2007) Origin of seed shattering in rice (Oryza sativa L.) Planta 226:11–20
Luo J, Liu H, Zhou T et al (2013) An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25:3360–3376
Oka HI (1988) Origin of cultivated rice. Elsevier, Amsterdam
Onishi K, Takagi K, Kontani M et al (2007) Different patterns of genealogical relationships found in the two major QTLs causing reduction of seed shattering during rice domestication. Genome 50:757–766
Yang Z, Wang X, Gu S et al (2008) Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 407:1–11
Zhang LB, Zhu Q, Wu ZQ et al (2009) Selection on grain shattering genes and rates of rice domestication. New Phytol 184:708–720
Zhu Z, Tan L, Fu Y et al (2013) Genetic control of inflorescence architecture during rice domestication. Nat Commun 4:2200
Acknowledgments
We thank Dr. Cristina Castillo, University College London, UK, for her critical reading and editing of the manuscript. The seeds of wild rice accession, O. rufipogon W630, were kindly provided by the National Institute of Genetics (National Bioresource Project), Japan. This work was supported in part by a Grant-in-Aid from Japanese Society for Promotion of Science (nos. 26292004 and 26450003).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Ishii, T., Ishikawa, R. (2018). Domestication Loci Controlling Panicle Shape, Seed Shattering, and Seed Awning. In: Sasaki, T., Ashikari, M. (eds) Rice Genomics, Genetics and Breeding. Springer, Singapore. https://doi.org/10.1007/978-981-10-7461-5_12
Download citation
DOI: https://doi.org/10.1007/978-981-10-7461-5_12
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-7460-8
Online ISBN: 978-981-10-7461-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)