Abstract
Solution small-angle neutron scattering (SANS) combined with contrast variation provides information about the size and shape of individual components of a multi-component biological assembly, as well as the spatial arrangements between the components. The large difference in the neutron scattering properties between hydrogen and deuterium is key to the method. Isotopic substitution of deuterium for some or all of the hydrogen in either the molecule or the solvent can greatly alter the scattering properties of the biological assembly, often with little or no change to its biochemical properties. Thus, SANS with contrast variation provides unique information not easily obtained using other experimental techniques.
If used correctly, SANS with contrast variation is a powerful tool for determining the solution structure of multi-component biological assemblies. This chapter discusses the principles of SANS theory that are important for contrast variation, essential considerations for experiment design and execution, and the proper approach to data analysis and structure modeling. As sample quality is extremely important for a successful contrast variation experiment, sample issues that can affect the outcome of the experiment are discussed as well as procedures used to verify the sample quality. The described methodology is focused on two-component biological complexes. However, examples of its use for multi-component assemblies are also discussed.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
5.1 Introduction
Small-angle neutron scattering (SANS) is able to provide the size, molecular mass and shape of a macromolecular complex in solution on length scales between approximately 10 Å to about 1,000 Å (Jacrot 1976; Svergun and Koch 2003; Svergun 2010; Jacques and Trewhella 2010; Zaccai 2012; Ankner et al. 2013). SANS combined with contrast variation allows for the unique retrieval of the internal structure and organization of chemically distinct components in a biological assembly. Recent reviews (Neylon 2008; Whitten and Trewhella 2009; Heller 2010; Gabel 2015; Zaccai et al. 2016) as well as classic papers (Engelman and Moore 1975; Ibel and Stuhrmann 1975; Jacrot 1976) describe the contrast variation technique as it applies to biological systems in detail.
Contrast variation takes advantage of the large difference in neutron scattering properties between hydrogen and deuterium. By systematically varying the ratio of H2O to D2O (H2O:D2O ratio) of the solvent, conditions are found in which one component in a multi-component assembly has the same scattering properties as the solvent. Under these conditions, the “matched” component is essentially invisible, much like a transparent material, e.g., a glass rod, can be optically invisible when in a solution that matches its index of refraction. In theory, a systematic series of measurements can be made allowing the scattering from each component to be determined as well as their positions with respect to each other in the complex. Substitution of deuterium for hydrogen in one or more of the components of the complex is another form of contrast variation.
This chapter introduces contrast variation experiments, outlining pertinent principles of SANS theory, essential considerations for experiment design and execution, and the proper approach to data analysis and structure modeling. The discussions on experiment planning and sample quality verification provide a guide for preparing samples that will lead to a successful contrast variation experiment. While the methodology described here is focused on two-component complexes, its use for multi-component assemblies is also discussed.
5.2 Theory
5.2.1 Scattering Intensity
The measured SANS intensity from a macromolecule consists of a coherent and an incoherent contribution such that
The coherent contribution is dependent on the scattering vector,\( \overrightarrow{q}, \) which has a magnitude defined as
where 2θ is the scattering angle (typically in degrees), measured from the axis of the incoming neutron beam, and λ is the neutron wavelength. The wavelength is usually expressed in nm or Å, such that q is stated in units of nm−1 or Å−1. The shape of the coherent scattering intensity profile depends on the shape of the molecule. On the other hand, the incoherent contribution is not q-dependent and contributes mainly to the noise level or “background”. Thus, the scattering intensity will refer to only the coherent term unless otherwise specified. The incoherent term will be further discussed in the Experimental Method, Data Analysis and Structure Modeling section.
Since SANS does not provide information on the length scale of atomic bonds, the strength of the scattering interaction can be described in terms of a uniform scattering length density of the entire molecule, ρ, within the molecular volume, V (in cm3 or Å3). The scattering length density is usually expressed in units of cm−2 or cm Å−3, but can be found stated in units of Å−2. The SANS intensity from the molecule can be written in terms of the scattering length density as
or
where \( {\left|F\left(\overrightarrow{q}\right)\right|}^2 \) is the form factor of the molecule.
5.2.2 Contrast
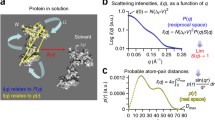
The coherent scattering from a biological molecule in a solvent can be thought of in terms of the cartoon shown in Fig. 5.1. Each fish represents one of N randomly-oriented molecules in the solvent. The molecules are identical in composition, size and shape and they are not interacting with each other. In other words, the solution is monodisperse, containing N identical particles and dilute, such that the particles do not sense one another.
The coherent scattering from any one of the molecules in the solution equals that from the molecule with scattering length density, ρ, in the molecular volume, V, plus that from the solvent with scattering length density, ρ s , in an effectively infinite volume, minus that from a “ghost” molecule with scattering length density of the solvent, ρ s , in the volume, V. The coherent scattering from a solvent of infinite extent is a delta function at q = 0 because the solvent has no long range-order; thus, it is not observed in practice. The scattering is therefore defined only in terms of the difference in the scattering length densities of the molecule and the solvent within the same molecular volume. Thus, Eq. 5.4 becomes
The difference in scattering length densities is known as the contrast,
Figure 5.2 shows the scattering length density of water as a function of the percent D2O (% D2O) in the solvent, along with scattering length densities of some typical biological molecules and compounds. The large slope of the water line as compared to the others is due to the large difference in scattering length density between hydrogen and deuterium. The lines are horizontal for CH2 and CD2 because there is no exchange of deuterium for hydrogen as the % D2O increases in the solvent. However, for proteins and nucleic acids, labile hydrogen atoms, i.e., those bound to nitrogen and oxygen, will exchange with deuterium in the solvent, causing an increase in scattering length densities for increased % D2O.
Neutron scattering length density as a function of percentage of D2O (% D2O) in the solvent for some typical biological molecules and compounds (Zaccai et al. 2016)
The vertical double-arrowed line at 10% D2O represents the contrast between protein and water at that % D2O value. A similar line can be drawn at any % D2O. Note that the protein and water lines cross near 40% D2O indicating that the scattering length densities of the molecule and solvent are the same, i.e., Δρ = 0. This is called a contrast match point and occurs at approximately 40% D2O for typical proteins. The contrast match points for DNA, RNA, lipid head groups, and CH2 can be found in the same manner. Note that the scattering length densities for perdeuterated molecules, in which all of the non-exchangeable hydrogen atoms, i.e., those bound to carbon, have been chemically replaced by deuterium, do not cross the water line. Thus, the theoretical contrast match points for these molecules would be greater than 100% D2O, which is not obtainable in practice.
The SANS intensity from all N monodisperse, randomly-oriented biological macromolecules in a dilute solution can be written in terms of the contrast as
where n is the number density (N per unit volume, v), of molecules (in cm−3). The brackets represent an averaging over all orientations of the molecule. The rotationally averaged form factor is sometimes also called P(q). It can be seen from Eq. 5.7 that the scattering intensity is zero at the contrast match point, Δρ = 0.
A typical contrast variation experiment involves measuring a complex consisting of two components that have different scattering length densities in solvent consisting of mainly water with perhaps a small amount of salts and buffering compounds. When water mixtures with different H2O:D2O ratios are used, the contrast of each component will change as a function of the concentration of D2O in the solvent. Thus, contrast match points exist for each of the components as well as the entire complex. As the H2O:D2O conditions in the solvent are varied, different parts of the complex scatter more strongly depending on their individual scattering length densities. By varying the amount of D2O in the solvent, one component can be essentially transparent at its contrast match point while the others are still visible. If enough different contrast conditions are measured, the scattering intensities for the individual components as well as their positions relative to each other are obtained. It is this feature of SANS that makes the method so powerful for selective measurement of individual components within a complex.
From Fig. 5.2, it is clear that proteins and nucleic acids have different contrast match points. The protein contrast match point is around 40% D2O, meaning that only the DNA or RNA is visible at this contrast. The DNA and RNA contrast match points are around 65% D2O such that only the protein is visible under these conditions. Therefore, complexes consisting of proteins and nucleic acids are ideal candidates for contrast variation experiments (Gabel 2015).
For a complex consisting of two proteins, replacement of some or all of the non-exchangeable hydrogen atoms with deuterium in one of the components is required in order for the two contrast match points to be different. Since the contrast match point of perdeuterated proteins is above 100% D2O, partially deuterated proteins are generally used for contrast variation experiments so that the contrast match point of the deuterated component is somewhere between 60% D2O and 100% D2O (Jacques et al. 2011). The exact contrast match point of a deuterated component is dependent on the amount of deuteration achieved. The contrast variation experiment can be used to verify this parameter, especially if a reliable determination cannot be made by other methods such as NMR or mass spectrometry. The method can be extended to larger assemblies consisting of multiple copies of the two different components (Appolaire et al. 2014).
Contrast variation methods also exist for solving the structures of multi-component assemblies. The triple isotopic substitution method (TISM) (Serdyuk and Zaccai 1996) allows for the determination of the scattering from one component at a time. Each determination requires three measurements with different deuteration levels of the component of interest. The label triangulation method (LTM) (Engelman and Moore 1972; Hoppe 1973) allows for the structure of a complex to be obtained by determining the distances between pairs of components. Each distance determination requires the selective labeling of the two components of interest. After determining a requisite number of distances, a 3D model of the structure can be determined by triangulation. The main applications of this method have been for the determination of the location of various components of the small (30S) and large (50S) subunits of the ribosome. These historic works are referenced in (May and Nowotny 1989).
Both the TISM and LTM methods require a large number of measurements. In practice, measurements of complexes consisting of three or more components are often made under solvent conditions that match each of the individual components. While some components still must be deuterated, this approach limits the number of measurements needed. An attempt is then made to find model structures that best fit all of the contrast variation data. This approach often works well when atomistic level starting structures of individual components are already available or can be built using homology modeling techniques, distance constraints are available from other techniques and/or docking software can be used to build structure models. The value of any such model is determined by its ability to reproduce the SANS data.
5.2.3 Radius of Gyration and Forward Scattering Intensity
The radius of gyration, R g , which is similar to the moment of inertia with respect to the scattering center of mass, and the forward scattering intensity, I(0), which is the scattering intensity at q = 0, are two important model-independent parameters that are obtained from SANS data. R g provides information about the size of the molecule whereas I(0) provides information about its molecular weight. Both parameters depend on the contrast, which can provide important clues about the spatial distribution of contrast within the molecule.
By definition, P(q) in Eq. 5.7 is equal to 1 at q = 0. Thus,
The number density, n (cm−3), can be written in terms of the concentration of the molecule, c (g cm−3), as
where M w is the molecular weight of the molecule (in Da, where 1 Da = 1 g mole−1) and N A is Avogadro’s number. In addition, the molecular volume, V, can be written in terms of the partial specific volume, \( \overline{v} \) (cm3 g−1), as
Equations 5.8, 5.9 and 5.10 can be used to relate I(0) to the M w of the molecule if the SANS data are on an absolute scale, usually in units of cm−1. While I(0) = 0 at the contrast match point, this is not true at larger angles that correspond to length scales on the order of the internal scattering length density fluctuations that were ignored by assuming a uniform scattering length density in Eq. 5.3.
The Guinier approximation (Guinier and Fournet 1955),
can be used on the low-q portions of the data to obtain values for R g and I(0). This low-q analysis is valid only in the region where qR g ≲ 1.3, so the valid q range depends on the size of the molecule. A shape must be assumed for the molecule to relate R g to the molecular dimensions. R g and I(0) are found by plotting the natural log of Eq. 5.11 such that
A linear fit of ln(I(q)) vs q 2 (Eq. 5.12) to the low-q portion of the data allows the determination of R g from the slope and I(0) from the intercept.
Another method to obtain R g and I(0), which makes use of all of the data rather than a limited data set at small q values, is to use the distance distribution function, P(r) vs r (Glatter and Kratky 1982). This function represents the probability distribution of distances, r, between all pairs of atoms in the molecule. The result is a smooth histogram-like plot with peaks at the most probable distances in the molecule. Thus, the shape of the P(r) vs r curve depends strongly on the shape of the molecule and can vary as a function of contrast.
P(r) is typically obtained from the SANS data using an indirect Fourier transformation method (Glatter 1977; Moore 1982; Semenyuk and Svergun 1991) using the relation
This analysis requires a stipulation by the user of a maximum dimension, D max , beyond which P(r) = 0. Typically, several values of D max are explored in order to find the range over which the P(r) function doesn’t change as a function of D max . Typically, the condition that P(r) = 0 at r = 0 is also assumed. R g and I(0) can be derived from P(r), as described in (Glatter 1982).
5.3 Experimental Method, Data Analysis and Structure Modeling
The successful SANS contrast variation experiment requires a number of steps from experiment planning and sample preparation to data collection and analysis, culminating with structure modeling. A flow chart showing these different steps is shown in Fig. 5.3 and each is described in detail in this section.
5.3.1 Sample Considerations
To obtain the most information possible from a contrast variation experiment, the main requirement is that the biological complex must be measured under dilute and monodisperse conditions in all H2O:D2O solvents. In other words, the sample must be of high purity. Ideally, this means that all the molecules have the same stoichiometry under all contrast conditions and there are no “free” components in the solution. Furthermore, all of the complexes are in identical conformations and there are no interactions between the complexes. The above equations describing the scattering intensity from macromolecules in solution assume these conditions are met.
In practice, there are many reasons why a sample might not meet all of these conditions. Sample preparation for contrast variation experiments involves making biological complexes in solvents containing D2O. In some cases, the complexes themselves must be made using deuterated components, which is typically accomplished by expressing one of the protein components using bacteria grown in deuterium-enriched media.
Whenever deuterium is introduced into the molecule or solvent, there is an increased chance for undesirable aggregation, even under conditions where the complex would not aggregate were deuterium absent. SANS is a volume-weighted technique. Since larger aggregates have larger volumes, a small amount of larger aggregates contribute more to the scattering intensity than a larger amount of small aggregates. While the signature of aggregation is most obvious at lower q values, the contribution to the total scattering persists over a much wider q range. Another issue when using deuterated components is that the complex might not form as readily or stably. This can lead to a situation for which the complex is associating and dissociating at a constant rate. In such situations, it may not always be possible to avoid having an excess of the constituent components in the solution.
The amount of sample needed for a contrast variation experiment is on the order of 0.5 mL at a concentration of 1–5 mg mL−1 for complexes with a typical M w between 50 and 100 kDa. Under solvent conditions where the contrast, and thus I(q) is low, higher concentrations must sometimes be used. This may introduce interparticle interference or aggregation effects that need to be mitigated as much as possible, e.g., by adding salts to the solvent to screen electrostatic interactions or by adding compounds that inhibit hydrophobic interactions. The mitigation of interparticle interference and aggregation effects is a difficult problem since every complex is different and measures that work in one case may not work in another.
Given the amount of material needed for a contrast variation experiment, the care that must be taken to prepare the samples, and the cost and time involved in getting access to a SANS beamline, it is important that such experiments are well planned and that these issues are considered and tested well before committing to making samples for a complete set of contrast variation measurements. Steps that can be taken to insure sample purity are discussed in detail in (Jacques and Trewhella 2010). Thorough sample characterization under the conditions that are being used for SANS should be performed prior to the SANS experiments using not only biochemical assays such as SDS-PAGE and gel filtration, but also complementary physical characterization techniques such as size exclusion chromatography with multi-angle light scattering (SEC-MALS), dynamic light scattering (DLS), or analytical ultracentrifugation (AUC). The amount of deuteration in a deuterated component can be assessed by mass spectroscopy or deuterium NMR. Small-angle X-ray scattering (SAXS) measurements can also be very helpful to detect concentration or D2O-dependent aggregation as will be discussed in more detail below.
5.3.2 Experiment Planning
For two component systems, the contrast of each component as well as that for the complex can be calculated from the chemical composition of each component and the stoichiometry of the complex as a function of the solvent composition. Two applications for performing this calculation are the MULCh (Whitten et al. 2008) software and the Contrast Calculator (Sarachan et al. 2013) module of the SASSIE (Curtis et al. 2012) software. For proteins and nucleic acids, the chemical composition is defined in terms of the amino acids or nucleotide bases, respectively, including the amount of H-D exchange and deuteration for each component. Non-protein or nucleic acid components can be entered based on their chemical formulas. Non-water solvent components such as salts and buffering compounds can be included in the calculation of the solvent scattering length densities to determine if they significantly alter the contrast match points. This is usually not the case if they are present in typical mmoles L−1 (mM) quantities.
Both contrast calculating programs provide a web interface for easy access and use. While MULCh is limited to two-component complexes, it also provides options for data analysis that will be described further below. The Contrast Calculator module allows for an unlimited number of components in the complex and additionally calculates expected I(0) values as a function of solvent composition. The predicted I(0) values are invaluable in determining the concentration needed to obtain a measurable signal for each solvent condition. If atomic coordinates are available, such as from X-ray crystallography or NMR structures, model scattering intensity curves can be calculated at each contrast to perform the SANS experiment in silico prior to the actual experiment. Software such as CRYSON (Svergun et al. 1998) or the SasCalc (Watson and Curtis 2013) module of SASSIE can be used for this purpose. The experimental SANS data may not match that calculated from the model structure if, for instance, disordered residues are missing or if the structure doesn’t accurately represent the complex in solution. Model structures must be complete if they are to be used as starting points for further modeling once SANS data are obtained. Thus, missing N or C terminal residues, internal loops, domains and/or disordered linkers must be added if necessary.
The calculations of the scattering length densities, contrast, I(0) and model scattering intensity curves used to plan an experiment apply only to the coherent scattering component in Eq. 5.1. The hydrogen and deuterium in the biological complex also have an incoherent scattering component, which contributes a small amount to the total scattering in dilute solutions. On the other hand, the incoherent scattering from a solvent containing mostly H2O:D2O is significant. In fact, it can often be greater than the calculated coherent intensity of a typical complex in a dilute solution. For example, the calculated coherent I(0) value for a 1 mg mL−1 protein-protein complex consisting of a trimer of the chaperone, Skp, bound to 50% deuterated outer membrane protein, OmpA, is 0.086 cm−1 in H2O (i.e., 0% D2O) and 0.04 cm−1 in 98% D2O (Sarachan et al. 2013). However, the incoherent scattering intensity for 98% D2O is ≈ 0.07 cm−1 and that of H2O is ≈ 1.0 cm−1 (Rubinson et al. 2008). Thus, the incoherent component is the dominant contributor to the total scattering in H2O, and remains a significant contributor in 98% D2O.
The calculated scattering curves for the Skp-OmpA complex are shown in Fig. 5.4 with and without the addition of the incoherent scattering component. The scattering that is actually measured is that shown in Fig. 5.4b. Under ideal conditions where there are no error bars on the data points in Fig. 5.4b and the incoherent background is measured equally well, the scattering curves in Fig. 5.4a could be recovered. However, this is not the case in practice. Features such as that shown at q ≈ 0.2 Å−1 for the complex in H2O (Fig. 5.4a) generally are not observable in practice since the incoherent component already significantly influences the total scattering at q ≈ 0.1 Å−1 (Fig. 5.4b). On the other hand, features such as those shown at 0.15 Å−1 ≤ q ≤ 0.25 Å−1 for the complex in 98% D2O typically are observable after subtraction of the incoherent scattering component. This subtraction is accomplished by measuring the incoherent scattering from a solvent that matches that of the sample as closely as possible. Thus, dialysis is an ideal method for exchanging the biological complex into buffers with different H2O:D2O ratios, as the dialysate solution can then be used to measure the incoherent background scattering.
The incoherent scattering from the solvent must be considered when planning data collection times. For a given calculated coherent I(0) value, it will take longer to obtain data in H2O than in D2O if the data are to have similar statistics after the incoherent background is subtracted. Furthermore, since the incoherent background is much higher in H2O, the maximum q value obtained after buffer subtraction will be lower for data obtained in H2O. Thus, when planning an experiment, samples prepared in solvents containing less than 50% D2O often must be measured a higher concentration so that the coherent I(0) is higher to make up for the higher incoherent background under these conditions. It should be noted that the scattering from water can contain both incoherent multiple scattering and inelastic scattering contributions and thus can differ depending on instrument conditions and sample geometry (Carsughi et al. 2000; Rubinson et al. 2008; Do et al. 2014). As the instrument scientists are most familiar with their instruments, it is important to rely on their advice for the I(0) values that can reasonably be measured at a given contrast at their facility.
Prior to performing a series of contrast variation measurements, two series of sample concentration measurements should be performed, one in H2O and one in >95% D2O, to determine the maximum concentration at which the sample is dilute and monodisperse. These measurements also identify whether there are any effects due to the presence of D2O in the solvent. Discussions with instrument scientists, who are familiar with the range of I(0) values that are measurable at their facility, provide the best resource in planning the range of sample concentrations to examine. This will avoid wasting time measuring the complex at sample concentrations that would result in I(0) values that are too low to be measured. It should also be noted that the accuracy of the concentration is extremely important, as it is the main source of error for determining the M w from I(0) (Eqs. 5.8 and 5.9).
The SANS curves from a complex often are significantly different in H2O and D2O solvents. Indeed, this is the reason for performing a contrast variation experiment in the first place. Therefore, the measurements described above are for comparison of the behavior of the complex as a function of concentration separately in H2O and D2O. The concentration needed to satisfy the dilute and monodisperse conditions may be lower in D2O than in H2O. If a SAXS instrument is available, SAXS measurements are a good option to determine whether there are any effects from either deuterium in the complex (when it consists of a deuterated and non-deuterated protein, for example) or D2O in the solvent. Since X-rays are not sensitive to the difference between hydrogen and deuterium, the scattering curves should ideally be the same regardless of sample or solvent deuteration. Another advantage is that SAXS measurements require smaller samples, on the order of 50 μL, requiring less sample preparation time for these important feasibility measurements. The SAXS measurement can also serve as another contrast point that can assist in modeling the structure of the complex.
Once optimal concentrations in H2O and D2O solvents have been found, then the feasibility of performing the contrast variation experiment can be further assessed by referring back to the predicted I(0) values obtained for the complex as a function of D2O in the solvent. For instance, is there a conflict between the concentration needed to obtain a reasonable signal and that needed to obtain a dilute and monodisperse sample at a given solvent condition? Is there a way to mitigate the issue, perhaps by altering the solvent conditions slightly? Again, an experienced instrument scientist can be a valuable resource when considering these types of issues.
5.3.3 Data Collection and Reduction to I(q) vs q
Data collection consists of measuring both the transmission of neutrons through the sample and the scattering of neutrons from the sample. Typically, these two different types of measurements require different instrument configurations and are performed separately. Since the sample consists of a biological complex in a solvent, these same measurements are also made for the solvent alone. Recall from Fig. 5.1 that the coherent scattering from the solvent is not observable. However, Fig. 5.4b shows that the incoherent scattering from a solvent containing mostly H2O:D2O is significant and therefore must be subtracted from the total sample scattering, which also contains this incoherent component from the solvent. Since both the sample and solvent alone are measured in a sample holder, often a quartz cuvette or a demountable cell with quartz windows, the transmission and scattering from the empty holder (i.e, the empty cell) should also be measured.
Typical windows used for SANS such as quartz, aluminum and titanium have significant scattering in the forward direction. Subtracting the scattering of the empty cell from both the sample and the solvent alone will eliminate the scattering contribution from the windows so that the shape of the scattering from the sample and solvent can be accurately observed. The solvent scattering should then be approximately flat since it is mainly incoherent and, hence, not q-dependent (Eq. 5.1). Furthermore, the scattering observed from the sample is then attributed to the biological complex of interest plus the solvent (as in Fig. 5.4b) with no contribution from the sample holder. The transmission measurement is important for proper subtraction of the empty cell and for placing the data on an absolute scale as described, for example, in (Glinka et al. 1998).
An additional consideration when collecting data is the time spent measuring each sample. Counting times for transmission measurements from the sample, solvent and empty cell are typically just a few minutes. On the other hand, counting times for scattering measurements from the sample (complex plus solvent) are concentration, contrast and instrument dependent and should be determined in consultation with the instrument scientist. For dilute samples, the counting statistics on the sample and solvent alone should be equivalent since the incoherent scattering from the solvent dominates the total scattering at most q values measured (Fig. 5.4). Thus, counting times for the sample and solvent alone should be the same. This is especially critical for measurements made under low contrast conditions. An exception can sometimes be made for a high M w complex, or for measurements made under high contrast conditions, if the scattering is significantly higher than that of the solvent alone in the low-q region of the I(q) vs q curve. However, the counting statistics of the sample and solvent scattering should still be equal in the higher-q portion of the curves, where the signal from the complex is weak. Since the empty cell scatters predominately in the forward direction, the counting times for these measurements can be shorter, matching only the statistics of the sample in the low-q region.
In a dilute solution, the incoherent scattering from the hydrogen and deuterium in the biological complex itself is negligible compared to that from the solvent. While the background levels of both sample and solvent alone should be the same in this case, mismatches can occur for a variety of reasons. Some possible sources for these mismatches include statistical differences in transmission values, the influence of incoherent multiple scattering, micro- or macroscopic bubbles in the solution, and mismatch in H2O:D2O ratio between the sample and solvent alone. However, under dilute conditions, the incoherent scattering from the solvent alone can be removed by scaling it to match the high-q region of the total sample scattering and then subtracting it. The acceptability of the solvent subtraction can be assessed by not requiring that P(r) = 0 at r = 0 when calculating P(r) from the SANS data (Jacques and Trewhella 2010). If P(0) is positive or negative, then the solvent has been under- or over-subtracted, respectively.
This approximation, used to remove the solvent scattering by scaling it to match the sample scattering, does not apply to concentrated solutions (concentrations ≈ 10 mg mL−1 and above for samples in a D2O buffer), where the incoherent scattering from the non-exchangeable hydrogen atoms in the complex itself is significant. As an alternative, it is tempting to simply subtract a constant representing the incoherent scattering from the solvent rather than subtract the measured scattering from the solvent alone, but this only works if the scattering from the solvent is truly flat. Typically the solvent scattering is not exactly flat due to incoherent multiple scattering, inelastic scattering, and other contributions(Carsughi et al. 2000; Rubinson et al. 2008; Do et al. 2014), with the shape depending on the H2O:D2O conditions of the solvent. Given these issues, in practice it is quite difficult to accurately subtract the solvent scattering from the sample scattering. Thus, it is not uncommon to find a systematic background mismatch when comparing the calculated SANS curves to the measured SANS data during the structure modeling stage.
Methods of reducing data from the raw data collected on a 2D position-sensitive detector to the 1D I(q) vs q data vary depending on the facility. However, if the end result is a set of separate I(q) vs q curves on an absolute scale for both the sample and the solvent alone, the scattering from the solvent can be successfully subtracted. If the solvent subtraction is performed as part of the 2D data reduction, then it is more difficult to diagnose problems that often arise during the solvent subtraction step.
5.3.4 Data Quality Checks
5.3.4.1 R g and I(0)
Once contrast variation data have been reduced and the contribution from the solvent has been subtracted, the basic parameters, R g and I(0) should be obtained at all contrasts using both the Guinier and P(r) analyses (Eqs. 5.11–5.13). Guinier analysis is usually provided by scattering facilities as part of the data reduction software. If P(r) analysis isn’t also included, a program such as GNOM (Semenyuk and Svergun 1991) or BayesApp (Hansen 2014) can be used. The experimental values of I(0) are then compared to the calculated values at each contrast in order to confirm the integrity of the samples. The number density, n, is defined as in Eq. 5.9, but it is now in terms of the concentration and M w of the entire complex. Similarly, Δρ refers to the mean contrast of the entire complex and V is the volume of the complex. The M w values should match the calculated values to within 10% if the data are on an absolute scale, I(0) has an error of 1‑2%, and the concentrations are measured to within 5%.
If the solution consists of a mixture of monomers and dimers of the complex or another combination of low-order oligomers, a valid Guinier region may still exist, but the I(0) value will be larger than that of the monomeric complex. If there are larger aggregates of the molecule in the solution, the Guinier region will occur at smaller q values than expected and Eq. 5.12 will not be linear in the q-range related to the expected size of the complex. Rather it will have some curvature and the fit to a straight line in the expected q-range will be poor. The effects can be subtle or very obvious depending on the severity of the aggregation (Jacques and Trewhella 2010). If aggregation is present, whether subtle or severe, the R g and I(0) values no longer represent that of a monomer complex in a monodisperse solution. Rather, they are influenced by the larger aggregates present in the solution. The P(r) function also changes in response to aggregation, primarily in the high-r region. In cases of minor aggregation, it may still be possible to calculate a P(r) corresponding to the monomer but the more severe the aggregation, the more difficult it is to determine D max (Jacques and Trewhella 2010).
5.3.4.2 Contrast Match Point Analysis
To further confirm the integrity of the sample at all contrasts, the contrast match point of the of the complex can be determined from the experimental data. Expanding on Eqs. 5.8–5.10 I(0) can be written in terms of a two-component complex as,
where f 1 and f 2 are the respective mass fractions of the first and second components in the complex. Now, Δρ 1 and Δρ 2 are respectively the scattering contrasts of the first and second components and \( {\overline{v}}_1 \) and \( {\overline{v}}_2 \) are respectively the partial specific volumes of the first and second components (Sarachan et al. 2013).
Because I(0) is proportional to c and to (Δρ)2, which is in turn dependent on the fraction of D2O in the solvent, f D2O, the contrast match point of the complex can be determined from the x-intercept of a linear fit to \( \sqrt{\raisebox{1ex}{$I(0)$}\!\left/ \!\raisebox{-1ex}{$c$}\right.} \) vs \( {f}_{{\mathrm{D}}_2\mathrm{O}} \). An example of this analysis is shown in Fig. 5.5, where experimental values of \( \sqrt{\raisebox{1ex}{$I(0)$}\!\left/ \!\raisebox{-1ex}{$c$}\right.} \) vs \( {f}_{{\mathrm{D}}_2\mathrm{O}} \) are plotted for the aforementioned Skp-OmpA complex that was measured in 0%, 15%, 30% and 98% D2O buffer. An unweighted linear fit to the data resulted in a line with a slope of −0.51 ± 0.01 and a y-intercept of 0.294 ± 0.006, where the errors represent one standard deviation. The x-intercept corresponding to the match point of the complex was calculated to be 0.576 ± 0.014, or 57.6% ± 1.4% D2O. This analysis can also be performed using MULCh (Whitten et al. 2008) and requires the experimental I(0) values, the concentration of the complex and the fraction of D2O in the buffer at each contrast.
Experimental \( \sqrt{\raisebox{1ex}{$I(0)$}\!\left/ \!\raisebox{-1ex}{$c$}\right.} \) values as a function of \( {f}_{{\mathrm{D}}_2\mathrm{O}} \) for a Skp-OmpA complex measured in 0%, 15%, 30% and 98% D2O buffers (red squares). The blue dashed line is the linear fit to the data while the black solid line is the calculated curve assuming 50% deuteration for OmpA
Comparison of this experimentally determined contrast match point to that calculated with Δρ 1 and Δρ 2 determined from the chemical composition of each component provides another quality assurance test on the data in that the calculated and experimentally determined contrast match points should agree with each other. If that is the case, then there is confidence that the calculated match points of the individual components are correct. The match point calculated from the chemical composition of the Skp-OmpA complex assuming 50% deuteration for OmpA using the Contrast Calculator module of SASSIE (Sarachan et al. 2013) was 59.8% D2O, in excellent agreement with the experimental result. It should be noted that the experimental and calculated complex match points can agree even if the slopes of the experimental and theoretical lines are quite different. The fit to the experimental data points should be compared to the theoretical curve as well. In this case, the data point for the 98% D2O sample falls slightly below the calculated line, which could mean that the sample concentration was lower than the value used in the analysis. Thus, even if the concentration is measured to a precision of ±5% prior to the experiment, the actual concentration of material in the beam can be affected by micro- or macro-bubbles in the sample, a small amount of precipitate in the sample (not uncommon for samples in D2O buffers), pipetting errors while diluting the sample and other such issues. Therefore, it is a good idea to recheck the sample concentrations after the SANS experiment if possible.
5.3.5 Component Analysis
5.3.5.1 Component R g and Center of Mass (CM) Distance
The R g values obtained at each contrast are related by the relationship (Ibel and Stuhrmann 1975),
where R m is the R g value of the equivalent complex with a homogeneous scattering length density, α is the second moment of the density fluctuations and β is the square of the first moment of the density fluctuations. For two component systems with different scattering length densities, the term α relates to the distribution of scattering length densities relative to the center of mass (CM) of the complex, and the term β provides the separation of the scattering CM of the two components (Moore 1982). A Stuhrmann plot (Ibel and Stuhrmann 1975) of \( {R}_g^2 \) vs (Δρ)−1 (Eq. 5.15) is used to determine R m , α, and β. If the plot is linear, then β = 0 and the CM of the two components are concentric. In this case, the sign of the slope of the line, α, is an indication of whether the component with the higher scattering length density is on the interior (negative) or exterior (positive) of the complex. Examples of parabolic and linear Stuhrmann plots are shown in Fig. 5.6. In practice, it is not always easy to distinguish between a parabolic and linear Stuhrmann plot, especially if R g doesn’t change appreciably as a function of contrast (α is close to zero) or if R g values are not available close to the individual contrast match points of the two components.
Similar information can be obtained from the parallel axis theorem
where R 1 and R 2 are the radii of gyration of the components and D CM is the distance between the scattering CM of the two components (Engelman and Moore 1975). Here, Δρ 1, Δρ 2, V 1 and V 2 refer to the individual components and Δρ and V refer to the complex. The parallel axis theorem provides the radii of gyration of the components and the CM distance between them directly, whereas they are calculated from the definitions of α and β when the Stuhrmann analysis is used. In both cases, R 1, R 2 and D CM are contrast-independent values that can be used in the structure modeling process. The MULCh software provides both of these analyses (Whitten et al. 2008). The inputs are the measured R g values as a function of f D2O in the solvent. The contrasts, Δρ, Δρ 1 and Δρ 2, are determined from the chemical composition of each component based on the sequence information input earlier in the analysis.
5.3.5.2 Component Scattering Intensities and Cross-Term
The scattering intensity from a two-component system with different scattering length densities can be written as (Whitten et al. 2008)
where I 1(q) and I 2(q) are the scattering intensities of components 1 and 2, respectively, and I 12(q) is the scattering intensity due to the interference between the two components. I 1(q) and I 2(q) are related to the shapes of the two components and I 12(q) is related to their spatial distribution. For a given set of measured contrast variation intensities, I(q), and known values for the contrasts, Δρ 1 and Δρ 2, the three unknowns, I 1(q), I 2(q) and I 12(q), are found by solving the resultant set of linear equations at each q value. Thus, data must be obtained at a minimum of three contrasts to solve for the three unknowns. The MULCh software (Whitten et al. 2008) provides this analysis. The measured I(q) vs q SANS curves and complex concentrations are required for each contrast and Δρ 1 and Δρ 2 are determined from the chemical composition of each component based on the sequence information input earlier in the analysis.
In practice, successful contrast variation studies that have resulted in the determination of I 1(q), I 2(q) and I 12(q) have employed at least five strategically chosen contrast points including the contrast match points of the individual components, where high quality data were obtained (Whitten et al. 2008). An example of a successful application of this method can be found in a study of a kinase, KinA, in complex with an inhibitor, Sda (Whitten et al. 2007). Based on this study and an analysis of the corresponding theoretical scattering curves with noise added, a data collection strategy was recommended that includes a minimum of two contrast points on either side of the average match point of the entire complex (Whitten et al. 2008). This results in a well-spread range of contrast points that allows the individual R 1, R 2 and D CM to be obtained with good accuracy and precision.
For a complex consisting of a protein and nucleic acid or a protein and a deuterated protein (with a deuteration level such that the match point is between 60% D2O and 80% D2O), a typical contrast variation data set would include 0% D2O and 100% D2O, along with 20% D2O, 40% D2O (the protein match point) and a fifth contrast point between 70% D2O and 80% D2O (near the match point of the second component). To obtain accurate scattering intensities of the components (Eq. 5.17), high quality data are needed at all measured contrasts. This analysis can be a useful tool to model the components separately and then arrange them with respect to each other in their proper position within the complex. However, structure modeling can proceed on the basis of the individual R 1, R 2 and D CM information alone, as is shown by example below.
5.3.6 Structure Modeling
Both SAXS and SANS are being used for structural determination of large biological complexes and for complexes containing flexible regions in solution. Many options are available for modeling multimeric biological complexes using a combination of rigid body and atomistic approaches, as described in recent reviews (Putnam et al. 2007; Rambo and Tainer 2010; Schneidman-Duhovny et al. 2012; Boldon et al. 2015). The SASSIE software suite (Curtis et al. 2012) is one tool that is available to assist in the atomistic and rigid body structure modeling of biological molecules for comparison to SAXS and SANS data. SASSIE provides users access to molecular dynamics, Monte Carlo, docking and rigid body modeling methods to assist in generating structure models and assessing how well models match the data. Constraints can be incorporated from other techniques such as NMR and AUC. SASSIE has been used for the structure modeling of many biological systems, including intrinsically disordered monomeric proteins (Curtis et al. 2012), large protein complexes (Krueger et al. 2011, 2014) and single-stranded nucleic acids (Peng et al. 2014). It has also been applied to the study of monoclonal antibodies using free energy analysis (Clark et al. 2013). A web version is available (https://sassie-web.chem.utk.edu/sassie2/) for ease of access and to handle the intensive computational requirements of the structural modeling and data analysis.
For a two-component complex, SANS and contrast variation experiments provide the added structural information from the individual components as constraints for modeling the entire complex. If obtainable, the scattering intensities of the separate components (Eq. 5.17) can be helpful for the modeling of the individual components and for construction of the model structure for the entire complex (Whitten et al. 2008). However, the contrast-independent R 1, R 2 and D CM distance constraints found by the Stuhrmann (Eq. 5.15) and parallel axis theorem (Eq. 5.16) analyses add unique information that can be used in the modeling process even in the absence of the component scattering intensities. Often, structural information for one or both of the components alone in solution is used as a starting point for their structures in the complex. Whether or not models are constructed from the scattering intensities of the separate components, the model SANS curves should always be judged against the entire contrast variation data set.
The first step in the structure modeling process is to construct starting models that satisfy the R 1, R 2 and D CM (as well as I 1(q), I 2(q) and I 12(q), if available) constraints found from the data analysis. Since these parameters have errors associated with them, several starting model structures that encompass the range of these parameters may be needed. Model structures that are consistent with the SANS data at all contrast conditions take full advantage of the information content of the contrast variation data set and provide the most robust representation of the data. Working model structures are first tested against the data by calculating both R g and the theoretical SANS curves from the model structures at each contrast measured. The theoretical SANS curves are then compared to the measured SANS curves, usually using a goodness of fit criterion such as the reduced χ 2 equation
where I exp(q) is the experimentally determined SANS intensity curve, I calc(q) is the calculated intensity curve from the model structure and σ exp (q) is the q-dependent error of the I exp(q) values. The sum is taken over N p independent data points.
The model structures producing the best-fit curves to the SANS data are then evaluated to verify that the R g values at each contrast match the values obtained from the experimental data. This process can be partially automated by plotting χ 2 vs R g at each contrast to identify the structures that match the experimental R g values as well as give the best fits to the data. The chi-square filter module in SASSIE (Curtis et al. 2012) provides this analysis. The inputs are the SANS data at a given contrast and the calculated SANS curves from the model structures at that same contrast.
The subset of best-fit model structures at each contrast are then further filtered to obtain the best overall agreement with the entire SANS contrast variation data set. The global best-fit structures can be evaluated using a parameter such as the average reduced χ 2 value
where N c is the number of contrast variation I(q) vs q scattering curves and \( {\chi}_i^2 \) is the reduced χ 2 value for the i th scattering curve. An example of this procedure is found in a recent methods paper (Zaccai et al. 2016), in which complexes of the Skp chaperone with two different unfolded Omp proteins (uOmps) were studied, Skp-OmpA and Skp-OmpW.
Structure modeling is an iterative process, especially for complexes since the structures of two or more components and their spatial arrangement need to be determined. This can be illustrated by examining the recent Skp-OmpA and Skp-OmpW study (Zaccai et al. 2016) more closely. For both complexes, the uOmp component was determined to be 50% ± 5% deuterated from the contrast match point analysis. First, hybrid model structures for Skp-OmpW were tested in which the Skp component was modeled by an all-atom structure and OmpW was modeled by an ellipsoid of revolution. Once a working model structure was found that agreed with all of the contrast variation data, the structure of the Skp component was adopted for the Skp-OmpA complex as well.
In the Skp-OmpA complex, the OmpA component contained a periplasmic domain connected to the transmembrane domain by a flexible linker. The Complex Monte Carlo module of SASSIE (Curtis et al. 2012) was used to create an ensemble of possible conformations of the periplasmic domain that best match the entire contrast variation data set. In this case, both all-atom and ellipsoid models were used for the OmpA component. However, in both cases, there was a mismatch in the 98% D2O data at higher q values, as shown in Fig. 5.7a. Since the scattering from the deuterated OmpA is weak at this contrast, the mismatch in the data was attributed to the Skp component, which was not varied from that found for Skp-OmpW (Zaccai et al. 2016). In fact, the 98% D2O scattering curves from Skp-OmpW and Skp-OmpA clearly differed in shape at the higher q values. It was evident that the Skp trimer is more symmetric, with respect to the position of the three monomers relative to each other, in the Skp-OmpA complex than in the Skp-OmpW complex. This was an important finding that provided support to the notion that Skp changes its conformation to accommodate different uOmps (Zaccai et al. 2016).
SANS contrast variation data for a Skp-OmpA complex. Solid lines represent the calculated SANS curves from the model structures in the inset assuming 60% deuteration for OmpA. Skp is shown in blue and OmpA is shown in red. a The model curves were calculated from the Skp-OmpA structure described in (Zaccai et al. 2016). The dashed line for the 98% D2O contrast was calculated assuming 50% deuteration for OmpA for comparison. b Best-matched model curve calculated using the same OmpA structure as in a paired with a more symmetric Skp structure. Error bars represent the standard error of the mean with respect to the number of pixels used in the data averaging
To determine if a more symmetric Skp structure could be found to better agree with the 98% D2O data for the Skp-OmpA complex, additional structures of Skp were further examined. The R g values were calculated for 60 Skp structures that were recorded during a biased MD simulation (Zaccai et al. 2016) in which the Skp monomers were splayed out to specific separations constrained to specific R g values. The ten structures that were in the best agreement with the R g value of Skp from the Stuhrmann analysis of Skp-OmpA were paired with the OmpA component from the best-matched Skp-OmpA structure. The new Skp-OmpA structures were checked for overlap between the basis CA atoms and then energy minimized using NAMD (Phillips et al. 2005). The SasCalc (Watson and Curtis 2013) module of SASSIE (Curtis et al. 2012) was then used to calculate their scattering curves. Two sets of calculations were performed assuming both 50% and 60% deuteration for the OmpA component. The model SANS curves were then compared to the SANS data at all contrasts using the Chi-Square Filter module of SASSIE (Curtis et al. 2012) to identify four structures that were in better agreement with the 98% D2O data. The model SANS curve from the overall best-matched structure is shown in Fig. 5.7b for each contrast. The Skp structures from the complexes shown in Fig. 5.7 have been superimposed in Fig. 5.8 for comparison. While the Skp structure in Fig. 5.7b is more symmetric with respect to the location of the three monomers as expected, there is still a mismatch in the range 0.06 Å−1 ≤ q ≤ 0.1 Å−1.
Given the flexibility of the monomers in the Skp trimer, it is not surprising that a single structure does not match the 98% D2O data. Mismatches of the type observed in Fig. 5.7b often are an indication of polydispersity. Since a large part of the OmpA structure is disordered, including the entire transmembrane (TM) domain that is encapsulated by Skp, it is also likely that the TM domain of OmpA exists in many different forms. Therefore, Skp likely takes on different structures to encapsulate these different OmpA TM structures. This is in addition to the conformations that can be assumed by the periplasmic domain of OmpA. While Skp and OmpA presumably exist in multiple different conformations in the Skp-OmpA complex, the SANS data reveal that an overall symmetry with respect to the configuration of the Skp monomers exists in the best-matched structural ensemble.
5.4 Concluding Remarks
Contrast variation combined with small-angle neutron scattering is a powerful tool for determining the structure of biological assemblies in solution. Contrast variation can be easily applied using neutrons due to the different scattering properties between hydrogen and deuterium. Through isotopic substitution of deuterium for hydrogen in both the molecule and/or solvent, the structures of individual components in a complex can be determined as well as their spatial arrangement. This represents unique information that cannot be easily obtained using other experimental techniques.
Experiments should be well-planned and the quality of the samples verified in advance to make the best use of beam time at neutron scattering facilities. Software tools are available to assist in experiment planning and facilities employ experienced instrument scientists who can offer assistance as well. Sample quality is extremely important for a successful contrast variation experiment. The integrity of the sample must be verified under all contrast conditions to have confidence in the data analysis and structure modeling.
After quality samples have been measured and the data reduced, model structures can provide valuable insight into the experimental system. Calculated SANS curves from model structures should be consistent with the SANS data at all contrasts. A wide variety of structure modeling software is becoming available to assist in developing and testing models to identify those that agree with SANS data.
Tips for Performing a Successful SANS Contrast Variation Experiment
Sample Preparation
-
Prepare highly pure samples
-
SDS-PAGE, gel filtration to remove larger M w species
-
A280:260 nm to detect nucleic acid
-
DLS, AUC to assess aggregation
-
SEC-MALS for monodispersity
-
-
Buffer and sample should match as closely as possible
-
Dialyze into final buffer
-
Measure sample concentration as accurately as possible
-
Re-check after experiment
-
Measure amount of deuteration in deuterated components as accurately as possible
-
Mass spectroscopy
-
Deuterium NMR
-
Preliminary Calculations and Measurements
-
Measure sample at multiple concentrations in 0% and 100% D2O buffers before contrast variation experiment
-
Assess concentration effects and D2O-dependent aggregation
-
Use SAXS if possible to also assess more subtle D2O effects
-
Plan the experiment ahead of time
-
Calculate contrast match points of complex and components
-
Calculate expected I(0) values vs fraction of D2O in the solvent
-
Determine contrast conditions for measurement, sample concentrations and counting times (with help from beam line instrument scientist)
-
Data Collection
-
Measure data on an absolute scale
-
Measure the buffers for the same counting time as the samples
Data Reduction and Analysis
-
Calculate I(q) vs q for samples and buffers
-
Subtract buffer scattering
-
Perform preliminary Guinier and P(r) analysis to obtain R g and I(0) at each contrast
-
Verify calculated complex match point from experimental data
-
Perform Stuhrmann and Parallel Axis Theorem analyses to obtain R 1, R 2, and D CM
-
Perform component analysis to obtain I 1(q), I 2(q) and I 12(q) (if possible)
Structure Modeling
-
Verify that model structures are complete (no missing residues, loops, domains, linkers)
-
Verify that starting model structures satisfy R 1, R 2, D CM (and I 1(q), I 2(q) and I 12(q))
-
Choose modeling method(s) appropriate for complex
-
Rigid body
-
Monte Carlo for flexible regions
-
Molecular Dynamics
-
-
Best-fit structures must fit the entire contrast variation data set
-
Narrow choices at each contrast and find subset that matches entire data set
References
Ankner JF, Heller WT, Herwig KW et al (2013) Neutron scattering techniques and applications in structural biology. In: Coligan JE, Dunn BM, Speicher DW, Wingfield PT (eds) Current protocols in protein science. Wiley, Hoboken
Appolaire A, Girard E, Colombo M et al (2014) Small-angle neutron scattering reveals the assembly mode and oligomeric architecture of TET, a large, dodecameric aminopeptidase. Acta Crystallogr D Biol Crystallogr 70:2983–2993. doi:10.1107/S1399004714018446
Boldon L, Laliberte F, Liu L (2015) Review of the fundamental theories behind small angle X-ray scattering, molecular dynamics simulations, and relevant integrated application. Nano Rev. doi:10.3402/nano.v6.25661
Carsughi F, May RP, Plenteda R, Saroun J (2000) Sample geometry effects on incoherent small-angle scattering of light water. J Appl Crystallogr 33:112–117. doi:10.1107/S0021889899013643
Clark NJ, Zhang H, Krueger S et al (2013) Small-angle neutron scattering study of a monoclonal antibody using free-energy constraints. J Phys Chem B 117:14029–14038. doi:10.1021/jp408710r
Curtis JE, Raghunandan S, Nanda H, Krueger S (2012) SASSIE: a program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints. Comput Phys Commun 183:382–389. doi:10.1016/j.cpc.2011.09.010
Do C, Heller WT, Stanley C et al (2014) Understanding inelastically scattered neutrons from water on a time-of-flight small-angle neutron scattering (SANS) instrument. Nucl Instrum Methods Phys Res Sect Accel Spectrometers Detect Assoc Equip 737:42–46. doi:10.1016/j.nima.2013.11.030
Engelman DM, Moore PB (1972) A new method for the determination of biological quarternary structure by neutron scattering. Proc Natl Acad Sci U S A 69:1997–1999
Engelman D, Moore PB (1975) Determination of quaternary structure by small-angle neutron scattering. Q Rev Biophys 4:219–241
Gabel F (2015) Small-angle neutron scattering for structural biology of protein–RNA complexes. Methods in enzymology. Elsevier, 391–415
Glatter O (1977) A new method for the evaluation of small-angle scattering data. J Appl Crystallogr 10:415–421. doi:10.1107/S0021889877013879
Glatter O (1982) Data treatment. In: Small-angle x-ray scattering. Academic Press, New York, pp 119–165
Glatter O, Kratky O (1982) Small-angle x-ray scattering. Academic Press, New York
Glinka CJ, Barker JG, Hammouda B et al (1998) The 30 m small-angle neutron scattering instruments at the national institute of standards and technology. J Appl Crystallogr 31:430–445. doi:10.1107/S0021889897017020
Guinier A, Fournet G (1955) Small-angle Scattering of X-rays. Wiley, New York
Hansen S (2014) Update for BayesApp : a web site for analysis of small-angle scattering data. J Appl Crystallogr 47:1469–1471. doi:10.1107/S1600576714013156
Heller WT (2010) Small-angle neutron scattering and contrast variation: a powerful combination for studying biological structures. Acta Crystallogr D Biol Crystallogr 66:1213–1217. doi:10.1107/S0907444910017658
Hoppe W (1973) The label triangulation method and the mixed isomorphous replacement principle. J Mol Biol 78:581–585. doi:10.1016/0022-2836(73)90480-4
Ibel K, Stuhrmann HB (1975) Comparison of neutron and x-ray scattering of dilute myoglobin solutions. J Mol Biol 93:255–265
Jacques DA, Trewhella J (2010) Small-angle scattering for structural biology – expanding the frontier while avoiding the pitfalls. Protein Sci Publ Protein Soc 19:642–657. doi:10.1002/pro.351
Jacques DA, Langley DB, Hynson RMG et al (2011) A novel structure of an antikinase and its inhibitor. J Mol Biol 405:214–226. doi:10.1016/j.jmb.2010.10.047
Jacrot B (1976) The study of biological structures by neutron scattering from solution. Rep Prog Phys 39:911–953. doi:10.1088/0034-4885/39/10/001
Krueger S, Shin J-H, Raghunandan S et al (2011) Atomistic ensemble modeling and small-angle neutron scattering of intrinsically disordered protein complexes: applied to minichromosome maintenance protein. Biophys J 101:2999–3007. doi:10.1016/j.bpj.2011.11.006
Krueger S, Shin J-H, Curtis JE et al (2014) The solution structure of full-length dodecameric MCM by SANS and molecular modeling: structure of dodecameric MCM helicase. Proteins Struct Funct Bioinf 82:2364–2374. doi:10.1002/prot.24598
May RP, Nowotny V (1989) Distance information derived from neutron low– Q scattering. J Appl Crystallogr 22:231–237. doi:10.1107/S0021889888014281
Moore PB (1982) Small-angle scattering techniques for the study of biological macromolecules and macromolecular aggregates. In: Ehrenstein G, Lecar H (eds) Methods of experimental physics. Academic, New York, pp 337–390
Neylon C (2008) Small angle neutron and x-ray scattering in structural biology: recent examples from the literature. Eur Biophys J 37:531–541. doi:10.1007/s00249-008-0259-2
Peng Y, Curtis JE, Fang X, Woodson SA (2014) Structural model of an mRNA in complex with the bacterial chaperone Hfq. Proc Natl Acad Sci 111:17134–17139. doi:10.1073/pnas.1410114111
Phillips JC, Braun R, Wang W et al (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802. doi:10.1002/jcc.20289
Putnam CD, Hammel M, Hura GL, Tainer JA (2007) X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40:191–285. doi:10.1017/S0033583507004635
Rambo RP, Tainer JA (2010) Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle x-ray scattering. Curr Opin Struct Biol 20:128–137. doi:10.1016/j.sbi.2009.12.015
Rubinson KA, Stanley C, Krueger S (2008) Small-angle neutron scattering and the errors in protein structures that arise from uncorrected background and intermolecular interactions. J Appl Crystallogr 41:456–465. doi:10.1107/S0021889808004950
Sarachan KL, Curtis JE, Krueger S (2013) Small-angle scattering contrast calculator for protein and nucleic acid complexes in solution. J Appl Crystallogr 46:1889–1893. doi:10.1107/S0021889813025727
Schneidman-Duhovny D, Kim S, Sali A (2012) Integrative structural modeling with small angle X-ray scattering profiles. BMC Struct Biol 12:17. doi:10.1186/1472-6807-12-17
Semenyuk AV, Svergun DI (1991) GNOM – a program package for small-angle scattering data processing. J Appl Crystallogr 24:537–540. doi:10.1107/S002188989100081X
Serdyuk IN, Zaccai G (1996) The triple isotopic substitution method in small-angle neutron scattering: application to studying macromolecular complexes. J Mol Struct 383:197–200. doi:10.1016/S0022-2860(96)09286-1
Svergun DI (2010) Small-angle X-ray and neutron scattering as a tool for structural systems biology. Biol Chem. doi:10.1515/bc.2010.093
Svergun DI, Koch MHJ (2003) Small-angle scattering studies of biological macromolecules in solution. Rep Prog Phys 66:1735–1782. doi:10.1088/0034-4885/66/10/R05
Svergun DI, Richard S, Koch MHJ et al (1998) Protein hydration in solution: experimental observation by x-ray and neutron scattering. Proc Natl Acad Sci 95:2267–2272
Watson MC, Curtis JE (2013) Rapid and accurate calculation of small-angle scattering profiles using the golden ratio. J Appl Crystallogr 46:1171–1177. doi:10.1107/S002188981301666X
Whitten AE, Trewhella J (2009) Small-angle scattering and neutron contrast variation for studying bio-molecular complexes. In: Foote RS, Lee JW (eds) Micro and nano technologies in bioanalysis. Humana Press, Totowa, pp 307–323
Whitten AE, Jacques DA, Hammouda B et al (2007) The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition. J Mol Biol 368:407–420. doi:10.1016/j.jmb.2007.01.064
Whitten AE, Cai S, Trewhella J (2008) MULCh : modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J Appl Crystallogr 41:222–226. doi:10.1107/S0021889807055136
Zaccai G (2012) Straight lines of neutron scattering in biology: a review of basic controls in SANS and EINS. Eur Biophys J EBJ 41:781–787. doi:10.1007/s00249-012-0825-5
Zaccai NR, Sandlin CW, Hoopes JT, et al (2016) Deuterium labeling together with contrast variation small-angle neutron scattering suggests how Skp captures and releases unfolded outer membrane proteins. In: Kelman Z (ed) Methods in enzymology. Elsevier, pp 159–210
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Krueger, S. (2017). Designing and Performing Biological Solution Small-Angle Neutron Scattering Contrast Variation Experiments on Multi-component Assemblies. In: Chaudhuri, B., Muñoz, I., Qian, S., Urban, V. (eds) Biological Small Angle Scattering: Techniques, Strategies and Tips. Advances in Experimental Medicine and Biology, vol 1009. Springer, Singapore. https://doi.org/10.1007/978-981-10-6038-0_5
Download citation
DOI: https://doi.org/10.1007/978-981-10-6038-0_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-6037-3
Online ISBN: 978-981-10-6038-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)