Abstract
In this book chapter, a practical approach for conducting small angle X-ray scattering (SAXS) experiments is given. Our aim is to guide SAXS users through a three-step process of planning, preparing and performing a basic SAXS measurement. The minimal requirements necessary to prepare samples are described specifically for protein and other macromolecular samples in solution. We address the very important aspects in terms of sample characterization using additional techniques as well as the essential role of accurately subtracting background scattering contributions. At the end of the chapter some advice is given for trouble-shooting problems that may occur during the course of the SAXS measurements. Automated pipelines for data processing are described which are useful in allowing users to evaluate the quality of the data ‘on the spot’ and consequently react to events such as radiation damage, the presence of unwanted sample aggregates or miss-matched buffers.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
2.1 Planning a SAXS Experiment
Many researchers are becoming increasingly inspired by the growing number of success stories that use SAXS to analyze the structures of macromolecules in solution (see reviews such as Vestergaard 2016; Trewhella 2016; Graewert and Svergun 2013). They ask themselves: SAXS is clearly beneficial, but what are the minimal requirements needed to prepare samples for solution scattering experiments? Figure 2.1 lists the fundamental points to consider when planning SAXS measurements that encompasses:
-
Sample purity and polydispersity.
-
Sample quantities.
-
Sample stability.
-
Sample handling and transport.
-
Obtaining an exactly-matched buffer for accurate background subtraction.
-
Measuring dilution series.
-
Trouble shooting at the beam line, e.g., overcoming radiation damage.
2.1.1 Sample Purity and Polydispersity
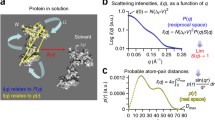
A biological solution SAXS experiment is very straightforward. A sample containing a macromolecule of interest is held directly in an X-ray beam with a defined energy (i.e., at a specific wavelength, λ) and the intensities of the scattered X-rays, I, are recorded as a function of the angle, q, to ultimately produce a plot of I(q) vs q; where q = 4πsinθ /λ (2θ is the scattering angle). In a second step, the scattering of an identical solution that does not contain the sample, i.e., the supporting solvent, is collected and the scattering intensities are subtracted from the sample scattering to yield the scattering contributions from the macromolecule of interest. However, as easy as these two measurements may sound, it is just as easy to collect meaningless scattering data as any material placed into the beam will scatter X-rays (Jacques and Trewhella 2010). Therefore, it is important that the composition of both the sample and the matched buffer are known and well characterized.
With respect to sample quality, the degree to which known and unknown contamination affects the outcome of the experiment depends on a number of factors. Of particular importance is the size, or more specifically the volume of contaminating species. For example, a protein has to be purified to at least 95% as assessed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE; Jacques et al. 2012a). In some cases a higher degree of purity is required depending on the size of the contaminating proteins. As the total scattering is proportional to the square of the particle volume (V 2) even trace amounts of species present in a sample that are larger than the macromolecule of interest can ‘swamp’ the scattering signal rendering the data (often) uninterpretable. Producing samples that are free of higher molecular weight species and free of aggregates represents the biggest challenge when preparing SAXS samples. Figure 2.2 summarizes a number of techniques that are especially suited for detecting and quantifying higher oligomeric species (described in more detail below).
Sample polydispersity can also significantly impact the interpretation of SAXS data and, if possible, should be minimized as data interpretation and modeling is greatly simplified for monodisperse samples. However, for many biological systems polydispersity is often an intrinsic property of the sample such as monomer-oligomer equilibrium or the formation of complexes with low affinity constants. Such samples will generate scattering profiles representing the summed, volume-fraction weighted contribution of each species in the mixture. Recording SAXS data from a dilution series and evaluating changes in the scattering profiles (that includes determining the molecular weight, MW) is one way to evaluate whether a sample forms a concentration dependent mixture. In addition, the application of size exclusion chromatography (SEC) immediately prior to SAXS allows for the separation and sequential measurement of the separated mixture components. Furthermore, many recent methodological advances have made the analysis of mixtures relatively straightforward (Petoukhov et al. 2013). These include the analysis of intrinsically-disordered or flexible macromolecular systems that are by definition, polydisperse (Kikhney and Svergun 2015). Yet, for both monodisperse and polydisperse samples the general over-arching rule of sample preparation remains the same: it is crucial to prepare samples that are as pure as possible and free of unwanted contaminants.
2.1.2 Sample Quantities and Concentrations
Once the question of purity has been addressed, the next question becomes: how much sample is needed? Approximate guidelines on the quantity of material required for SAXS experiments are listed in Table 2.1. New developments in sample-delivery robotics and their installation at many biological SAXS beam lines not only prevent human errors but also ensure precise and efficient sample loading (David and Perez 2009; Round et al. 2015; Blanchet et al. 2015). Thus, usually 5–25 μl of protein sample are sufficient to fill the measuring cell at concentrations between 0.5 and 8 mg/ml. As a rule-of-thumb, a suitable protein concentration for synchrotron-based SAXS can be described by:
For example, a protein concentration of 2 mg/ml is most likely adequate for studying a monodisperse 50 kDa protein. However, for polydisperse samples that undergo concentration dependent oligomerization, it may be necessary to increase or decrease the sample concentration to either assemble or disassemble the oligomers, respectively. For more advanced measurements such as SEC-SAXS, more sample is likely required. Finally, it is always prudent to have some additional material on-hand in cases where a sample is sensitive to radiation damage so ‘at the beam line’ adjustments can be made to reduce the effects of this damage to the sample (see below).
2.1.3 Sample Environment (Buffer)
A solution SAXS measurement comprises two essential steps: (i) the measurement of the scattering data from the sample and; (ii) the measurement of scattering data from an identical, exactly matched buffer that does not contain the macromolecule of interest which is used for background subtraction. Imprecise buffer matching is a frequent stumbling block for first time SAXS users. Only after data collection and processing does it become apparent just how sensitive the method is to small discrepancies between correct and incorrectly-matched buffers. While planning a SAXS measurement the practical question that has to be addressed is: How is a suitable buffer obtained? For example, if the final purification step for a protein is ion-exchange chromatography, then it becomes difficult to evaluate the exact salt concentration at which the sample elutes from the column and then to prepare an exact replica of this buffer for SAXS. Or, in other words, as X-rays scatter from electrons, it is difficult to manually prepare a buffer with matched X-ray scattering and absorption properties as the supporting solvent of the sample. In such circumstances, a dialysis step is strongly recommended to obtain a good matching buffer, or to use buffer-exchange using SEC.
The atomic composition, or more precisely, the electron density of a chosen buffer is indeed a crucial aspect to consider when preparing samples for SAXS. As it happens, the average electron density of water (0.33 electrons/Å3) is not that much lower than the average electron density of protein (~0.43 electrons/Å3) and it is this very small difference that, after background subtraction, gives rise to a coherent SAXS pattern at low angle that can be used to extract structural information. However, the small difference in electron density decreases even further with the addition of components to the buffer, for example high-salt, glycerol, sucrose, etc. If the concentration of these buffer components becomes too great the X-ray contrast will limit to zero and the net scattering from the macromolecule will be effectively negated. For example the addition of either ~35% v/v glycerol, ~3 M NaCl, or ~1.2 M sucrose to a buffer will result in an approximate 50% reduction in the net scattering intensity measured from proteins in solution (see Box 2.1).
Box 2.1: Calculating the Contrast, Δρ, of a Sample
One of the advantages of solution SAXS is that a diverse range of solution conditions can be screened to assess the effects of changing sample environment on the structures of macromolecules. However, the addition of high concentrations of small molecules (e.g., 2 M NaCl) or the addition of electron-dense molecules to the supporting solvent will reduce the difference in electron density between the solvent and the macromolecules of a sample. As a result, the net scattering intensities derived from the macromolecules of a sample, i.e., the scattering contributions after the buffer scattering has been subtracted, will decrease. This could become important to consider, for example, when adding small molecules to a sample that limit the effects of radiation damage (e.g., electron-dense polyols). Adding too much will eventually result in the ‘matching out’ of the scattering signal.
The effect of changing the electron density of a buffer on the overall magnitude of the scattering intensities can be assessed in advance by calculating the contrast of a sample (Δρ), for example using the program MULCh (modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies (Whitten et al. 2008)). The Δρ is the difference between the average scattering length density of a macromolecule and the average scattering length density of the buffer which relates to the difference in electron density between a macromolecule and the buffer. The magnitude of the net small-angle scattering intensities from the macromolecules of the sample will be proportionate to Δρ2. The CONTRAST module of MULCh is specifically tailored for calculating X-ray (and neutron) scattering contrasts of a macromolecular system. For this calculation, the scattering data is not required. CONTRAST simply uses protein, RNA or DNA sequences in combination with the atomic formulae and concentrations of small molecules in the solvent. Using this information, CONTRAST calculates the X-ray and neutron-scattering-length densities of the macromolecule and solvent (ρ) and subtracts these values to obtain Δρ of the sample. The entire MULCh package, which includes CONTRAST, can be downloaded as an off-line tool (with instructions) or used interactively online via http://smb-research.smb.usyd.edu.au/NCVWeb/. All you need as input is: (i) the list of solvent/buffer components (atomic formulae) and their molar concentrations: (ii) the one-letter amino-acid code or one-letter DNA/RNA code of the macromolecules; (iii) the atomic formulae of any small molecules bound to the macromolecule of interest— e.g., metal ions, cofactors.
2.1.4 Sample Stability
Further considerations have to include the assessment of sample stability during the measurement. SAXS experiments performed ‘in house’ using a laboratory X-ray source may require higher sample concentrations combined with longer exposure times. Thus, sample conditions may have to be found where the sample is both concentration and time-stable, specifically in regard to the formation of aggregates, during potentially prolonged measurements (e.g., up to 1 h). The brilliance afforded by synchrotron based SAXS means that samples can be measured using very short exposure times (in the order of milliseconds to seconds) and at low concentration. However, synchrotron SAXS poses a different set of challenges. Although radiation damage is a universal problem for both lab-based and synchrotron SAXS, the rate of damage using a synchrotron X-ray source may be more apparent even during very short exposure periods. Predicting whether a sample might be prone to radiation damage prior to a SAXS experiment is difficult and has to be treated on a case-by-case basis. For example, some proteins such as biomolecules with metal centers, may be particularly sensitive to radiation damage (e.g., cytochrome C, that binds Fe-heme), then again others are not (e.g., glucose isomerase, that binds Mg2+ or Mn2+). In Box 2.2 and 2.3 the means of dealing with radiation damage at the beam line are listed.
Box 2.2: Addition of Small Molecules to Limit X-Ray Radiation Damage
There a few ‘tricks’ that can be used to limit the effects of X-ray radiation damage by adding small-molecule free radical scavengers or polyols to a sample. Unfortunately, there is no single ‘tried-and-true’ method that can be applied and, somewhat annoyingly, it is impossible to predict before a SAXS experiment whether such measures will be effective. There a few considerations that can be helpful to determine which (if any) scavenger might be compatible to the system being studied. As a reminder, care has to be taken when adding accurate and equal measures of additive to both the sample and to the corresponding solvent blank. Ideally, a dialysis of the sample should be performed against the buffer with the added scavenger. However, dialysis might not be feasible as beam-time and sample quantities might be limited. In such cases, well calibrated pipettes or a microbalance should be used to add an equal volume or mass of concentrated additive stock solutions. Extreme care has to be applied, especially when adding viscous polyol solutions such as glycerol or sucrose. The main disadvantage of the solution additive approach is the increased risk of altering the chemical or physical properties of a macromolecule.
-
DTT, @1–5 mM:
-
Dithiotheritol has been often described as a useful scavenger, as it is not overly expensive and available in most molecular biology/structural biology laboratories. However, one must keep in mind that DTT is a reducing agent and is therefore not suitable for systems in which disulfide bonds play an essential role. Reduction of disulfide bonds can resulting in undesirable changes in structure. One must also remember that DTT has a short shelf life (just up to a few hours) and should first be added directly before the measurement. In this sense, it is not suitable for SEC-SAXS. In addition, DTT undergoes oxidation and changes its ultraviolet (280 nm) absorption properties that may affect protein concentration estimates.
-
Ascorbic acid, @ 1–2 mM
-
Ascorbic acid is a ‘classic’ free radical scavenger that can be added to a sample to limit radiation damage. As the name suggests, ascorbic acid is acidic and thus one must be acutely aware that adding ‘neat’ ascorbic acid to a sample can significantly lower the pH which may then induce chemical alterations to a sample. Ascorbic acid also changes the UV absorbance properties so that concentration determination using UV methods may be hindered.
-
Glycerol, @ 3–5% v/v
-
Glycerol is not scavenger per se but is very good at limiting X-ray induced aggregation in solution. The addition of glycerol will increase the electron-density of the solvent, i.e., reduce the contrast of a sample, which needs to be considered with respect to maintaining the SAXS signal intensities (see Box 2.1). Glycerol can also influence protein–solvent/protein–proteins interactions that may affect concentration dependent oligomerization. In addition, due to its high viscosity, glycerol is difficult to add in exactly-equivalent amounts to sample and to the corresponding solvent/buffer blank needed for the SAXS measurements. Therefore, it is often preferable to make up a 10–20% v/v glycerol dilution in the buffer of choice (checking that the pH does not change) and then add the more diluted glycerol stock to the SAXS sample and buffer using a microbalance or a pipette (with the pipette tip-end clipped off). For SEC-SAXS, glycerol is often very effective in reducing radiation damage in the often slower sample flows through the X-ray beam line, but care must be taken that the SEC columns can withstand the increased pressure caused by the addition of glycerol to the mobile phase.
Box 2.3: Tips for Performing a SAXS Experiment at a Synchrotron Beam Line
-
Tip 1: Take time to think about and plan the experiment. Importantly ask yourself the question. What question do I want to focus on when using SAXS to probe the structure(s) of my samples?
-
Tip 2: Contact a local beam line responsible, or someone with experience, to coordinate the experiment in regard to sample handling, shipment, any additional equipment or necessary paperwork at a facility.
-
Tip 3: Thoroughly characterize the sample, this includes measuring small test batch-samples using SAXS to assess the susceptibility of samples to radiation damage prior to SEC-SAXS.
-
Tip 4: Determine the sample concentration immediately prior to the SAXS measurements to account for any sample loss during storage/transportation. If applicable, have some back-up material at hand for unforeseen problems (e.g., to add free radical scavengers to a sample if radiation damage is observed).
-
Tip 5: Remind yourself that it is crucial to have sufficient matching buffer for background subtraction. Make sure to set aside a large volume of exactly-matched buffer for each sample for all of the SAXS measurements (e.g., dilution series, SEC-SAXS, etc.).
-
Tip 6: Prioritize your experiments. It is often preferable to measure a smaller number of characterized samples well during a beam-line shift compared to collecting data from as many poorly-characterized samples as possible (‘garbage-in-garbage-out’).
Another challenge faced by synchrotron SAXS users concerns the storage and shipping of samples to large-scale facilities. The general shelf-life of a sample and the tendency to form aggregates over time needs to be assessed. Effects of long-term storage at 4 °C or freeze-thawing under different conditions should be inspected. The practical aspects of transporting or shipping the sample to the synchrotron facility must also be considered. The stability of a sample can easily be tested in advance. Analytical size exclusion chromatography (SEC) and dynamic light scattering (DLS; both described in more detail below) are convenient methods for such an assessment, especially for detecting the presence of time-dependent aggregates. Small aliquots of a test sample can be screened using different handling protocols, such as fast/slow freezing, fast/slow thawing, plus or minus salt, glycerol, etc. and then be compared with each other. The objective is to identify the condition that prevents the sample from aggregating.
2.1.5 Support SAXS with Sample Characterization Data During Sample Preparation
During the planning stage of a SAXS experiment that more-often-than-not involves optimizing sample conditions, it is recommended to gather as much information as possible to support the conclusions from the SAXS investigation. This includes but is not limited to: what is the estimated MW of my sample (e.g., determined using light scattering techniques, for example multi-angle, right-angle laser light or static light laser scattering, MALLS, RALLS, SLS); what is the oligomeric state and does it change with different pH values and/or salt concentration (e.g., using SEC)? How is the system influenced by small-molecule ligands, temperature, etc. (e.g., using DLS or thermofluor assays to assess stability/aggregation (Boivin et al. 2013))? How flexible/folded is the system (e.g., using circular dichronism CD)?
In summary, a basic SAXS experiment is conceptually simple, but can be demanding in terms of preparing quality samples and matched buffers; not necessarily in regard to obtaining the required amount of sample, but regarding the quality and stability of the sample. In the next section, protocols for preparing the sample and the buffer for a solution SAXS experiment are described in more detail.
2.2 Preparation for SAXS Measurement
Here, we discuss general options and techniques that can be performed in the laboratory to achieve the goal of producing SAXS-quality samples and matched sample buffers under the general concepts of:
-
Sample characterization using gel electrophoresis, SEC, light scattering techniques, analytical ultra-centrifugation and mass spectrometry.
-
Assessing sample concentration using spectrophotometry or refractive index.
-
Preparing the matched sample buffer.
-
Organizing SAXS experiments
2.2.1 Sample Characterization: Assessing Polydispersity
It is very important to set aside sufficient time to thoroughly characterize those samples that will be used for the SAXS measurements (Jacques et al. 2012b). Due to the often irrecoverable and deleterious influence on the scattering data by aggregates or other large MW species it is very important to determine the association processes of a sample in solution. The ultimate aim should be to present the results obtained from a SAXS investigation in such a manner that the quality of the samples used to obtain the data can be assessed. For this, the sample purification procedure must be documented and reported, along with an estimate of the final purity of the sample. Available methods for such assessments are summarized in Table 2.2 as well as Fig. 2.2 and are shortly outlined here.
Gel electrophoresis provides an invaluable tool to assess the purity of the native proteins and complexes. Denaturing SDS-PAGE (both reducing and non-reducing) is excellent to evaluate whether a sample contains additional higher MW contaminants or if the target protein is affected by non-specific disulphide cross links. Native PAGE (run without SDS and non-reducing/non-degrading conditions) is useful to assess whether higher oligomeric species and to some extent self-associated aggregates are present in the sample. A big advantage of PAGE is that only small volumes of sample are required and a number of samples can be analyzed in parallel. A drawback of native-PAGE is that the separation is dependent on the size as well as the overall net charge of the molecule. Thus, the success of the separation is dependent on the isoelectric point (pI) of the protein as well as the behavior of the protein in the somewhat limited choice of native PAGE buffer systems (that may be very different to the final buffer selected for SAXS). However, in general, both SDS- and native-PAGE are exceptionally useful for routinely checking the purity and the stability of a sample over time.
In Size Exclusion Chromatography (SEC) a solvent carrying the sample, or mobile phase, passes through porous particles (the matrix, or stationary phase)–typically supported in a column–in which smaller particles are trapped for a short time resulting in a shift in their retention time. Consequently, larger particles (e.g., particles with a larger molecular weight or hydrodynamic radius) migrate through the separation matrix faster and are separated from the smaller species, assuming that there are no significant interactions with the column matrix. The column resolution (separation of two individual peaks) depends on a number of controllable parameters such as the choice of column (material, pore size, length) and running conditions (flow rate, mobile phase, loading volume). A major advantage of SEC is that it can be performed in a number of buffer conditions that can be screened and optimized for maintaining a sample in a desired state. There are, however, some limitations. Unavoidable interactions between the macromolecules of the sample and the stationary phase can result in adsorption, shifts in retention time, elution peak tailing/asymmetry, or even to changes in the three dimensional conformation (Hong et al. 2012). These undesirable interactions can often be prevented through the addition of salts to the running buffer. In addition, the type of separation matrix can be chosen; silica-based SEC columns represent a good choice for samples that may interact with dextran-based matrices used in the most common sepharose columns.
Another useful advantage of SEC is that it can be used in combination with UV-spectroscopy to qualitatively evaluate the oligomerization or aggregation state(s) of a protein sample. Although quantitative MW estimates based on the retention volume are not reliable, UV-SEC provides a means through which to visualize and detect the presence of the aggregates and higher oligomeric species and how these species may change in the sample over time or in different buffer conditions. In some circumstances, the equilibrium driven self-association of the individual components within a sample, even after purification, may be unavoidable. If this is observed, one can profit from SEC-SAXS set-ups which are now offered at almost all biological SAXS beam lines whereby the SEC column outlet is directly connected in-line to the SAXS capillary so that scattering data can be collected from the freshly-separated components as they elute from the column (Mathew et al. 2004; David and Perez 2009; Round et al. 2013; Graewert et al. 2015). As a SEC run is often accompanied with a solvent exchange, collecting SAXS data from the buffer that has run through the column acts as a convenient means to obtain the scattering required for background subtraction. However, and once again, care maybe required when selecting the correct buffer for SEC-SAXS background subtraction. Unknowable buffer-matrix interactions may cause extremely subtle alterations to the electron density of the exchanged solvent flowing through the column that may have a slightly different X-ray scattering and absorption properties compared to the buffer of the eluted macromolecules.
Sample component separation using SEC is normally monitored with UV detectors. There are, however, advantages of adding other detectors such as static light scattering (SLS) to the system. Using RALLS or MALLS in combination with the concentration estimates derived from UV or refractive index (RI) measurements, the molecular mass of the samples can be determined, independent from the elution volume and can be used to validate the MW of the SAXS samples (Graewert et al. 2015). This approach is ideal to determine to exact oligomeric state(s) of the SEC-separated components of a protein sample. In the simplest case, light scattering is detected for just one angle (90° in right angle laser light scattering, RALLS, or <7° for low angle scattering, LALLS). However, using multiple of detectors (up to 18) placed at different angles (multi angle laser light scattering, MALLS) significantly increases the sensitivity for the high MW aggregates as well as accuracy of the MW estimations, especially for larger complexes (Ahrer et al. 2003).
In a different type of laser light scattering experiment, Dynamic Light scattering (DLS), the correlations between the fluctuations in the intensity of scattered light from macromolecules relating to their movement (Brownian motion) in solution are analyzed. As this is an exceptionally sensitive technique it can be employed to detect aggregates as well as to estimate polydispersity. Large globular particles not only scatter more strongly compared to smaller counterparts, but their Brownian motion is decreased due to their increased mass (as they dwell longer within the illuminated area). Consequently, relative to a starting time, t0, the fluctuations in the intensities for larger globular particles will be correlated relative to t0 for longer time periods compared to smaller particles, before exponentially decaying to zero, i.e., will eventually become uncorrelated relative to the initial time point. As the polydispersity and/or aggregation of a sample increases, the time taken for the auto-correlation to decay increases and it no longer smoothly decays towards zero. Accordingly, information can be obtained from the auto-correlation function by fitting the data (for example using a sphere model) from which hydrodynamic radius distributions of the particles and sample polydispersity can be estimated. An examination of the populations present within a sample using DLS, especially the presence of aggregates, is a good way to evaluate sample quality for SAXS.
There is no limitation for the size or type of particles that can be studied with DLS (peptides, proteins, polymers, micelles, carbohydrates, nanoparticles, etc.), however the resolution of the technique is quite low, i.e., it is not possible to distinguish between a sample consisting of monomers from a sample in monomer-dimer equilibrium. Its power as a characterization technique for SAXS samples is that DLS is exceptional for detecting trace amounts of aggregates (if aggregates are detected in DLS then they will also interfere with the SAXS) as well as for stability testing and condition screening. As DLS is a non-destructive method it is very suitable to examine the samples directly before they are loaded into the capillary of the beam line for example with a 96-well plate reader.
Analytical Ultracentrifugation is based on the sedimentation of macromolecules in their native state, often under extreme g-forces, is followed using an optical device (UV light absorption, fluorescent system or Rayleigh interferometer). The separation of the sample components within a mixture is dependent on the hydrodynamic and thermodynamic properties/association states of the macromolecules and enables the analysis of the shape, size distribution and molar masses of the sample components. Using a different approach, called a sedimentation equilibrium experiment, the final steady-state of the components are analyzed, where their sedimentation is balanced by diffusion opposing the concentration gradients. From this, one can retrieve information directly on the MW of the macromolecules as it is insensitive to shape. The amount of required sample is low (<0.5 mg). If the experiment is designed well molecules between 100 Da and 10 GDa and the size resolution can be chosen to detect even small mass changes. However, sophisticated equipment and technical know-how is required so that it is not typically accessible to the inexperienced user (Lebowitz et al. 2002). The experiment itself can be lengthy, depending on type of run (3–6 h for sedimentation velocity analysis, and even days for a sedimentation equilibrium experiment). However, ultracentrifugation can be exceptionally informative and a valuable asset in interpreting and supporting the conclusions reached from a SAXS experiment.
Structural biology studies can also profit from native mass spectrometry (nMS). Here, electrospray ionization techniques are commonly employed so that the tertiary and quaternary protein structures are preserved. Very accurate mass estimates of proteins and the stoichiometry of subsequent assemblies can be determined. In turn, information can be gained regarding quaternary structure stability, dynamical behavior, conformation(s), subunit interaction sites, glycosylation state(s) and the topological arrangement of the individual proteins within a complex (Sharon and Horovitz 2015). However, as with ultracentrifugation, sophisticated equipment and technical know-how are absolutely required.
2.2.2 Sample Characterization: Assessing Concentration
Along with assessing the polydispersity of a sample, it is also very important to experimentally determine the concentration of a sample used for a SAXS experiment. The method for concentration determination should be chosen such that the accuracy of the method is sufficient to derive the MW of the sample from the SAXS measurements, specifically from the forward scattering at zero angle, I(0). In this respect, the recent wwwPDB SAS task force has emphasized the importance on reporting which concentration technique was employed for a specific experiment (Jacques et al. 2012b). The concentration determination of proteins based on UV absorption at 280 nm is the most frequently used method as it is often the most understood and the fastest technique that has low sample consumption. However, and especially for proteins that completely lack or only have a few aromatic amino acid side chains (e.g., tryptophan) that consequently do not absorb strongly at 280 nm or for proteins that bind to non-protein ligands that absorb strongly in the UV region (e.g., heme) the concentration estimates using UV at 280 nm tend to become inaccurate. An alternative is differential refractometry (RI) which is far less dependent on amino acid sequence composition. Importantly, RI can be employed to measure the concentration of hetero complexes, or intrinsically disordered proteins (that often lack or are aromatic amino acid poor). With refractometry, the degree to which light bends as it passes through the interface between two substances is measured. The physical characteristic is dependent on the protein concentration and the protein’s refractive increment (dn/dc). However, for most proteins in standard aqueous solutions dn/dc is 0.187 mL/g, allowing one to determine the concentration of the sample.
In some cases, using colorimetric assays may produce the most reliable results. In these instances, it is always beneficial to correlate and/or standardize the results obtained from colorimetric assays to UV or RI measurements. Correlating the results to UV also means that a fast assessment of the sample concentration, for example after transport, can easily be performed directly at a SAXS beam line, as most facilities offer access to an UV spectrometer. For the determination of nucleotide concentration commonly three different methods can be used: (i) UV absorbance at 260 nm (specific absorption peak of purine and pyrimidine rings), (ii) fluorescence (the amount of binding of fluorescent dye to double stranded DNA is determined with a fluorimeter and compared to reference measurements) and (iii) lesser available method: diphenylamine reaction (Li et al. 2014). For the latter, DNA is heated under acidic conditions to obtain 2-deoxyribose, which after dehydration can react with diphenylamine to produce a blue substance with an absorption maximum at 595 nm.
2.2.3 Buffer Preparation
As mentioned above, buffer matching is crucial for any SAXS measurement. SAXS is sensitive to even small changes in the composition of a buffer, in particular the electron density and X-ray absorption properties so that even small discrepancies between a sample and its corresponding buffer can lead to an erroneous background subtraction. In our experience the best way to obtain the optimal buffer is to perform dialysis (Fig. 2.3a) and this should always be the first method of choice. There are, however, cases in which dialysis is not feasible (for example a sample is prone to time-induced self-association/aggregation) and alternative approaches are available (Fig. 2.3b). In the following section we discuss:
-
Dialysis.
-
How to accurately add small molecules or expensive ligands to a sample (sample ‘spiking’).
-
Diafiltration.
-
Desalting columns.
In dialysis small molecules are exchanged into or out of a macromolecular sample by size-restricted diffusion through a porous membrane. The sample is placed on one side of the membrane and a buffer solution, the so-called dialysate, on the other side. Usually the dialysate is 200–500 times the volume of the sample. Sample molecules that are larger than the molecular-weight cutoff (MWCO) of the semi-permeable membrane are retained on the sample side of the membrane while smaller components (specifically buffer components) freely diffuse through the membrane and approach an equilibrium concentration with the dialysate. The process of buffer exchange may require 4–24 h to complete depending on the dialysate viscosity and typically produces an optimal matched buffer for SAXS background subtraction. Aside from using regular dialysis tubing, a number of devices are now commercially available which are essentially ready to use and resist sample leakage (Fig. 2.3a). The dialysis of small sample volumes (~200 μl) can benefit from both commercial or home-made “cup devices” which can be placed inside a medium size reagent tube containing the dialysate (for instance a 50 ml Falcon® tube) which enables easy transport. To ensure complete buffer exchange, different aspects such as ratio of sample volume to dialysate volume as well as the surface area of the membrane and factors including temperature, viscosity, mixing, etc., have to be taken into account. When choosing the membrane for the dialysis one has to be aware that the MWCO is not a sharply defined value; in general, the MWCO should be 5× smaller than the expected MW of the macromolecule.
Preparing sufficient dialysate volume (200–500 times larger than the sample) is not always feasible especially when expensive ligands are required. In such cases, spiking the sample and buffer will be necessary. If the sample is stable in the absence of the ligand, then the apo, or unligated, variant of the sample should be prepared and dialysed against the ligand-free buffer. Afterwards the ligand can be added in small amounts from a stock solution to the sample as well as the buffer (dialysate) using accurately calibrated pipettes or an electronic micro-balance. If the ligand has to be added throughout the preparation or the sample is generally prone to time-dependent aggregation, then diafiltration might be an alternative option for obtaining a matched buffer. As in dialysis, a semi-permeable membrane is used to separate macromolecules from low molecular-weight compounds. However, instead of relying on passive diffusion, the solutions are forced through the membrane by pressure or centrifugation. As the water along with the low molecular-weight solutes collect on one side of the membrane, the macromolecules are concentrated on the opposite side. Thus, diafiltration devices are often employed as concentrators, but can–if used with caution–also be used for buffer exchange. For this, the sample undergoes successive rounds of dilution in a buffer of choice, concentration, followed by subsequent rounds of dilution and concentration. If this method is used to adjust sample solvent to the desired buffer for background subtraction it is very important to thoroughly rinse the membrane before use to remove any preservatives on that membrane that are included in the manufacturing process (e.g., glycerol, azide, etc.). This is done, by passing larger volumes of buffer through the membrane before adding the sample. In addition, it is preferable to perform a series of short centrifugation/concentration steps, with careful mixing of the sample using a pipette, as opposed to one long centrifugation step that may result in an unwanted concentration gradient at the sample-membrane interface which may cause the sample to aggregate. Therefore, it may be necessary to monitor the sample e.g., using DLS, to ensure that aggregates do not form during the concentration steps.
Buffer exchange/adjustment can also be performed with a desalting column that is a similar process that occurs during SEC. By choosing the correct column length and sample load, the macromolecules of a sample will be too large to enter the pores of the desalting resin and will quickly pass through the column. Buffer salts and other small molecules will, on the other hand, enter the pores of the resin. After equilibrating the desalting column in a desired buffer for the SAXS measurements and passing the sample over it, the original buffer components will remain trapped by the resin, while the macromolecule of interest will flow through and be recovered in the SAXS buffer. The disadvantage of using a desalting column is that the sample undergoes significant dilution. In addition, our experience shows that with this method as well as during SEC, the buffer does not always undergo complete exchange.
2.2.4 Organization of the Experiment
Once the sample is obtained in sufficient quantities, final arrangements for shipping/transporting the samples can be made. Figure 2.4 demonstrates some scenarios on how to organize the experiment depending on the nature of the sample.
Practical aspects for shipping samples can include taking care of required papers/documentations for customs and/or the transport of dried ice, etc. For synchrotron SAXS, it is always advisable to contact the local beam line responsible in advance and ask for the specific shipping address (to avoid delays in delivery). Furthermore, it is always important to include clear instructions on the storage conditions for the person receiving the sample package. If further sample preparation is required, e.g., overnight dialysis, access to the samples and laboratories for after working hours should be arranged in advance. In this respect, consumables (dialysis accessories, syringes and needles, concentrators, etc.) should be shipped with the samples. For some samples it might become necessary to perform the final purification steps on-site. Many beam lines offer access to laboratory facilities, which can be booked prior to arrival (for an example see Boivin et al. 2016). In such instances, it is best to discuss the experimental procedures in detail with the local responsible to ensure that all equipment and consumables that are needed, are indeed available.
Finally, and very importantly, it can be very easy under the stresses of sample preparation to focus entirely on the sample for a SAXS experiment and forget about the buffer. Remember to include sufficient buffer for your standard SAXS measurements (for example 50 ml). For SEC-SAXS up to 1 liter may be required, or alternatively, pack the dried ingredients to reconstitute the desired buffer on-site, immediately prior to the experiment.
2.3 Trouble-Shooting at the Beam Line
Once at the beam line make sure all your equipment has arrived and take time to orient yourself around the facility and laboratories. If applicable, thaw sample and buffer keeping in mind that this can take some time. Before continuing, it is advised to give the samples a quick spin in a centrifuge (e.g., 10,000×g for 10 min) or pass the samples through a 0.22 μm filter to remove any large aggregates or insoluble particulate matter (this is particularly important for SEC-SAXS samples). UV spectroscopy is a good method to determine if any sample was lost during transport and storage. Synchrotron SAXS experiments can be hectic affairs as there is often pressure to measure as many samples as possible within an allocated beam time period. Consequently, there may be little time to conduct a thorough analysis of the data. However, thanks to automated pipelines at most SAXS beam lines, data is analyzed on-the-fly in near real-time and as a consequence if problems arise regarding the sample (e.g., the presence of aggregates) decisions can be made on-the-spot to ameliorate the sample conditions to improve data quality (Blanchet et al. 2015). Basic trouble-shooting approaches include
-
Detecting and handling radiation damage.
-
Dealing with aggregation.
-
Dealing with miss-matched buffer
-
Dealing with concentration dependent effects.
2.3.1 Dealing with Radiation Damage
Unavoidably, some samples will be susceptible to radiation damage that manifest as a continual increase in scattering intensity during the measurement. To detect such an effect, a number of frames are typically collected during one exposure, e.g., instead of recording a single continuous one second exposure, twenty 50 ms frames are recorded over one second. The individual frames are then compared to each other and those frames showing traces of radiation damage are discarded (Franke et al. 2015). In some cases, the onset of radiation damage may occur late during the measurement so that a sufficient number of frames can be averaged to produce a SAXS profile with an acceptable signal-to-noise ratio. However, if a sample is overly susceptible to radiation damage, it maybe that only one frame can be collected, that may or may not, be influenced by damaged species. In this scenario, there are several adaptions that can be made to reduce the effects of radiation damage that include altering the data collection strategy or changing the sample environment (Jeffries et al. 2015). If the amount of sample is sufficient, the flow-rate of the sample through the X-ray beam can be increased; the beam can be attenuated to reduce the X-ray flux, or the beam can be defocused. Alternatively, the chemical composition of the sample can be changed via the addition of equal quantities of small-molecules to the sample and matched buffer. These small molecules are referred to as radical scavengers and may include–but are not limited to–dithiothreitol (DTT) and ascorbic acid. Although not scavengers per se, polyols such as glycerol, ethylene glycol as well as sucrose influence long-range protein-protein interactions and are effective in reducing radiation-induced aggregation. In Fig. 2.5a the effect of adding DTT and glycerol to the sample are shown.
All of these options to curb the effects of radiation damage come with an associated cost. Changes in data collection strategy may require the repeated measurement of more sample; addition of polyols result in a reduced contrast; the addition of classical scavengers can potentially cause deleterious chemical alterations (e.g. DTT may reduce disulfide bonds).
2.3.2 Dealing with Aggregation
Another common observation during the measurements is the presence of aggregates. Aggregates are often detected by non-linear or ‘upturn’ features in the Guinier plot of the SAXS data (ln(I(q)) vs q 2) at very low-angles (qR g <1.3; where R g is radius of gyration) or an greater than expected MW estimate of the sample determined from the SAXS data (Guinier 1939). Figure 2.5b shows an example in which the presence of aggregates is detectable in one of two storage conditions. In these cases, a dilution series of the sample becomes important; it may be that aggregates are concentration-dependent and simple dilution can remove their influence. However, if the aggregates remain persistent, a few options can be explored to remove them from solution. Giving the sample a strong spin with a (ultra-) centrifuge can sometimes be useful (e.g., 20.000×g for 15 min); the insoluble aggregates collect at the bottom of the tube and the soluble protein remains in the supernatant. A faster option is to try filtering the sample; either using a syringe filter tip or small centrifugal device (keeping in mind that some proteins may interact with membranes and be adsorbed, altering their structure). Another option is the use of online SEC-SAXS. Here, aggregates are separated from the sample during the size-exclusion step. This option of course requires more time, sample volume and concentration and often has to be organized in advance, but can be well worth the investment (especially for samples not prone to radiation damage).
2.3.3 Dealing with Miss-Matched Buffer
As mentioned above, incorrectly-matched sample buffers required for the accurate subtraction of background scattering contributions is often reason why SAXS experiments are unsuccessful. The detection of miss-matched buffer is unfortunately not always that straight-forward and not always unambiguous. There are, however, a few indications when looking at the distinct scattering plots (often automatically generated by integrated pipelines) especially for over-subtracted buffer contributions that produce negative scattering intensities at high-angle (i.e., at high-q) in the subtracted SAXS profiles. Figure 2.5c, shows the effect of under- as well as over-subtracting the background (blue and red curves, respectively). Once buffer miss-match is detected and time permits then a new dialysis can be set-up. Alternatively, a diafiltration device as described above can be used if time is scarce.
2.3.4 Dealing with Concentration Dependent Effects
As mentioned above it may be important to measure the sample at different concentrations in order to detect and consequently ameliorate concentration dependent effects in the sample that impact the scattering data. For example, as depicted in Fig. 2.5d, a significant decrease in R g as the sample concentration increases may indicate that repulsive interactions are present between the macromolecules of the sample (i.e., coulombic repulsion). Conversely, if the Rg significantly increases as the sample concentration goes up, attractive interactions between the sample macromolecules are likely present that may result in sample oligomerization, sample polydispersity, or in the worst case scenario, sample aggregation. As a result, samples may require additional dilutions to nullify concentration effects or, more radically, alterations to the supporting solvent such as the addition of salts and changes in pH to alter protein surface potentials responsible for concentration-dependent effects.
2.3.5 Other Considerations
Finally, it is advised to always check the structural parameters derived from experimental scattering data with the results obtained from complementary methods. This is especially important for validating the molecular weight of the sample. For one thing, errors such as accidental swapping of tubes, loading of air bubbles, questionable dilution series etc. can be best identified at the beam line allowing for ‘on-the-spot’ corrections.
In summary, SAXS experiments are straightforward and the demands for sample preparation are feasible for most systems. There are, however, a few essential steps to keep in mind when planning, preparing and performing the SAXS experiments. In understanding how these sample preparative steps directly influence the scattering process, enables the SAXS user to successfully produce high quality samples and ultimately high quality SAXS data.
References
Ahrer K, Buchacher A, Iberer G, Josic D, Jungbauer A (2003) Analysis of aggregates of human immunoglobulin G using size-exclusion chromatography, static and dynamic light scattering. J Chromatogr A 1009(1–2):89–96
Blanchet CE, Spilotros A, Schwemmer F, Graewert MA, Kikhney A, Jeffries CM, Franke D, Mark D, Zengerle R, Cipriani F, Fiedler S, Roessle M, Svergun DI (2015) Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J Appl Crystallogr 48:431–443
Boivin S, Kozak S, Meijers R (2013) Optimization of protein purification and characterization using thermofluor screens. Protein Expr Purif 91(2):192–206
Boivin S, Kozak S, Rasmussen G, Nemtanu IM, Vieira V, Meijers R (2016) An integrated pipeline for sample preparation and characterization at the EMBL@PETRA3 synchrotron facilities. Methods 95:70–77
David G, Perez J (2009) Combined sampler robot and high-performance liquid chromatography: a fully automated system for biological small-angle X-ray scattering experiments at the synchrotron SOLEIL SWING beamline. J Appl Crystallogr 42:892–900
Franke D, Jeffries CM, Svergun DI (2015) Correlation map, a goodness-of-fit test for one-dimensional X-ray scattering spectra. Nat Methods 12(5):419–422
Graewert MA, Svergun DI (2013) Impact and progress in small and wide angle X-ray scattering (SAXS and WAXS). Curr Opin Struct Biol 23:748–754
Graewert MA, Franke D, Jeffries CM, Blanchet CE, Ruskule D, Kuhle K, Flieger A, Schäfer B, Tartsch B, Meijers R, Svergun DI (2015) Automated pipeline for purification, biophysical and x-ray analysis of biomacromolecular solutions. Sci Rep 5:10734
Guinier A (1939) La diffraction des rayons X aux tr‘es petits angles: application ‘a l’étude de phénomenes ultramicroscopiques ed. Univ. de Paris, Paris, pp 3, p l, 80, 2, p 1 l
Hong P, Koza S, Bouvier ES (2012) Size-exclusion chromatography for the analysis of protein biotherapeutics and their aggregates. J Liq Chromatogr Relat Technol 35(20):2923–2950
Jacques DA, Trewhella J (2010) Small-angle scattering for structural biology–expanding the frontier while avoiding the pitfalls. Protein Sci 19:642–657
Jacques DA, Guss JM, Svergun DI, Trewhella J (2012a) Publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution. Acta Crystallogr Sect D Biol Crystallogr 68:620–626
Jacques DA, Guss JM, Trewhella J (2012b) Reliable structural interpretation of small-angle scattering data from bio-molecules in solution—the importance of quality control and a standard reporting framework. BMC Struct Biol 12:9
Jeffries CM, Graewert MA, Svergun DI, Blanchet CE (2015) Limiting radiation damage for high-brilliance biological solution scattering: practical experience at the EMBL P12 beamline PETRAIII. J Synchrotron Radiat 22:273–279
Kikhney AG, Svergun DI (2015) A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett 589(19 Pt A):2570–2577. doi:10.1016/j.febslet.2015.08.027. Epub 2015 Aug 29
Lebowitz J, Lewis MS, Schuck P (2002) Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci., 2002 11(9):2067–2079
Li X, Wu Y, Zhang L, Cao Y, Li Y, Li J, Zhu L, Wu G (2014) Comparison of three common DNA concentration measurement methods. Anal Biochem 451:18–24
Mathew E, Mirza A, Menhart N (2004) Liquid-chromatography-coupled SAXS for accurate sizing of aggregating proteins. J Synchrotron Radiat 11:314–318
Petoukhov MV, Billas IM, Takacs M, Graewert MA, Moras D, Svergun DI (2013) Reconstruction of quaternary structure from X-ray scattering by equilibrium mixtures of biological macromolecules. Biochemistry 52(39):6844–6855
Round A, Brown E, Marcellin R, Kapp U, Westfall CS, Jez JM, Zubieta C (2013) Determination of the GH3.12 protein conformation through HPLC-integrated SAXS measurements combined with X-ray crystallography. Acta Crystallogr D 69:2072–2080
Round A, Felisaz F, Fodinger L, Gobbo A, Huet J, Villard C, Blanchet CE, Pernot P, McSweeney S, Roessle M, Svergun DI, Cipriani F (2015) BioSAXS sample changer: a robotic sample changer for rapid and reliable high-throughput X-ray solution scattering experiments. Acta Crystallogr Sect D Biol Crystallogr 71:67–75
Sharon M, Horovitz A (2015) Probing allosteric mechanisms using native mass spectrometry. Curr Opin Struct Biol 34:7–16
Trewhella J (2016) Small-angle scattering and 3D structure interpretation. Curr Opin Struct Biol 40:1–7
Vestergaard B (2016) Analysis of biostructural changes, dynamics, and interactions – small -angle X-ray scattering to the rescue. Arch Biochem Biophys 602:69–79
Whitten AE, Cai S, Trewhella J (2008) MULCh: modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J Appl Crystallogr 41:222–226
Acknowledgments
This work was supported by the Bundesministerium fur Bildung und Forschung (BMBF) project BIOSCAT, grant 05K12YE1 by iNEXT, grant number 653706, funded by the Horizon 2020 programme of the European Union and by a Human Frontier Science Program research grant, HFSP Ref: RGP0017/2012. M.A.G. was supported by an EMBL Interdisciplinary Postdoc Programme (EIPOD) and Marie Curie COFUND actions.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Graewert, M.A., Jeffries, C.M. (2017). Sample and Buffer Preparation for SAXS. In: Chaudhuri, B., Muñoz, I., Qian, S., Urban, V. (eds) Biological Small Angle Scattering: Techniques, Strategies and Tips. Advances in Experimental Medicine and Biology, vol 1009. Springer, Singapore. https://doi.org/10.1007/978-981-10-6038-0_2
Download citation
DOI: https://doi.org/10.1007/978-981-10-6038-0_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-6037-3
Online ISBN: 978-981-10-6038-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)