Abstract
With the development and wide applications of engineered nanomaterials (ENMs), their impacts on human health have received increasing concerns. ENMs can enter human body through respiratory pathway, digestive tract, skin penetration, intravenous injection, and implantation, and then they are carried to distal organs via bloodstream and lymphatic functions to perturb physiological systems. It is very important to investigate the interactions between ENMs and biomolecules (the basic building blocks of the human body) such as phospholipid, protein, DNA, and some other small biological molecules. The chapter intends to discuss the chemical basis of interactions between ENMs and biomolecules, and the effects of the differences in surface morphology, composition, and modified groups of ENMs. The in-depth understanding of interactions between ENMs and biomolecules could lay foundations for further elucidating the effects of ENMs on human cells, organs, and physiological systems, which paves the way for human and environmental friendliness in the production and usage of ENMs.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
5.1 Brief Introduction of Interactions Between Engineered Nanomaterials and Biomolecules
5.1.1 Engineered Nanomaterials (ENMs)
Nanomaterial is defined as a material with at least one dimension in the size range between 1 and 100 nm (the usual definition of nanoscale) in principle, and it is an aggregate or agglomerate based on nanoscale units. Engineered nanomaterials (ENMs) refer to nanomaterials which are intentionally manufactured in contrast to natural ones. ENMs possess many unique optical, electronic, chemical, and biological features compared with bulk materials. In addition, all features could be influenced by surface, small size, quantum size, macroscopic quantum tunnel, and dielectric confinement effect [1,2,3]. For example, gold nanoparticles (Au NPs) exhibit a characteristic absorption peak in UV–Vis absorption spectrum [4], quantum dots (QDs) display strong fluorescence emission [5], and graphene has excellent mechanical property [6]. All these distinctive and advanced characteristics endow ENMs with fast-growing applications in medicine, pharmacy, chemical/biological sensing, manufacturing, optics, and national defense [7,8,9,10].
Most compositions of ENMs include carbon, silica, polymer, metal, and metal oxide (with shapes of sphere, rod, wire, tube, prism, sheets, and so on). The size of ENMs is often designed to be coordinate with major cellular machines and their components, so ENMs can interact with biological molecules to exert biochemical functions. Currently, the detailed chemical compositions of ENMs could be acquired by elemental analysis [11], X-ray photoelectron spectroscopy (XPS) [12], auger electron spectroscopy (AES) [13], and time-of-flight mass spectrometry (TOF-MS) [14]. Transmission electron microscopy (TEM), scanning electron microscopy (SEM), and atomic force microscopy (AFM) could characterize the morphology of ENMs [15]. And dynamic light scattering (DLS) could offer information on the hydrodynamic radii of nanoparticles in solution [16].
The surface properties of ENMs mainly refer to the surface charge and hydrophobicity, depending on different chemical modification groups. Generally, the chemical groups on the surface of nanoparticles are determined by the synthetic process. Furthermore, the group can be altered via ligand exchange or other chemical bonding methods [17]. Multiple methods have been reported to characterize the physical and chemical properties of ENMs. Ζeta-potential measurements could provide information of the surface charge properties of nanoparticles [18]. Ligands or adsorbed molecules on the surfaces could be identified by Fourier-transform infrared spectroscopy (FTIR) [19], liquid chromatography–mass spectroscopy (LC–MS) [20], and magic angle spinning nuclear magnetic resonance (MAS NMR) [21]. In addition, surface-enhanced Raman scattering (SERS) spectroscopy is another frequently used method for fingerprint identification of chemical groups [22].
5.1.2 Interaction Between ENMs and Biological Molecules
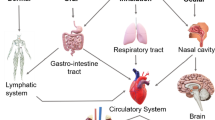
With the expanding production volume and applications of ENMs, the health and environmental effects of ENMs have attracted wide attention. The major portals of human body exposed to ENMs are skin, gastrointestinal tract, lung, nasal cavity, and eyes [23]. ENMs could then be translocated to other human organs which are distal to the site of uptake by absorbing into the bloodstream [24, 25]. In biological fluids or systems, ENMs are surrounded by excessive amounts of biological molecules, and inevitably interact with various biomolecules.
Figure 5.1 schematically illustrates the comparison of sizes and shapes between common ENMs and more familiar chemical/biological materials [24]. Materials shown for comparison are below, within or above the nanoscale range, aiming to put ENMs’ size in perspective. As seen, the sizes of biomolecules, such as lipids, proteins, and DNA, are also in nanoscale range. ENMs have super-large specific surface area and high surface energy. As a result, pristine nanoparticles are nearly nonexistent since they prefer to adsorb various molecules in order to reduce their surface energy. The driving forces for the adsorption of biomolecules by ENMs involve hydrophobicity, hydrogen bonding capability, π bonds, and stereo chemical interactions, which are related to the composition, size, shape, and surface properties of ENMs [26].
The sizes and shapes of some ENMs compared to more familiar materials. Materials shown for comparison are below, within or above the nanoscale range, aiming to put ENMs’ size in perspective. Reproduced with permission from Ref. [24]. Copyright © 2011 Yokel and MacPhail; licensee Bio Med Central Ltd. All rights reserved
Besides, the interactions between ENMs and biomolecules are also driven by the characteristics of biological settings. For instance, phospholipid, which is the main components of cell membranes, consists of a hydrophobic carbon chain (tail) and a hydrophilic phosphate group (head). The surface hydrophobicity of ENMs indicates the interaction with the hydrophilic head or the hydrophobic tail [26]. At physiological pH, proteins fold into different sizes, shapes, and net charges, and their hydrophobicity are largely dependent on the exposed amino acid residues. When ENMs encounter proteins in biological fluids, the spontaneous adsorption could occur and the stoichiometry and orientation of the combination are affected by the properties of proteins [27]. Possessing phosphate groups and base π systems, DNA molecules can bind nanoparticles through electrostatic, π–π stacking, and hydrophobic interactions [28]. Moreover, ENMs could also adsorb other small biological molecules such as amino acids [30, 35, 36], nucleobases [37, 38], and vitamins [39, 40].
Generally speaking, the morphology and surface properties of ENMs are crucial in determining the nature or the interactions between ENMs and biological molecules [31, 32]. Therefore, the primary objective of the chapter is to perform systematic summary on the interactions between ENMs and biomolecules with the diversity in morphology, composition, and modified groups of ENMs. These observations not only provide powerful evidence to understand the fundamental chemical interactions of nanoparticles and biological systems, but also provide practical approaches to produce “safe-by-design” ENMs for various industrial or medicinal applications, which would pose minimal hazard potential to human health and the environment.
5.2 The Interactions Between Engineered Nanomaterials and Small Biological Molecules
A great variety of small biological molecules play tremendous roles in various metabolic processes, and the significant reduction of their contents has been shown to elicit adverse effects in cellular functions, as evidenced of incurring injuries and illnesses [33, 34]. Due to the extremely large surface area of ENMs, small biological molecules can be adsorbed onto nanoparticles with the help of hydrophobic interactions, π–π stacking, and electrostatic interactions. For the moment, the most studied molecules include amino acids [30, 35, 36], nucleobases [37, 38], and vitamins [39, 40].
5.2.1 Amino Acids
The combination of ENMs with amino acids, the basic unit of proteins, can lead to the impairment in amino acid structure, the dysfunction of amino acids, and even the abnormal metabolism, which will perturb the intrinsic biological behaviors of biomolecules. Based on this, related researches on the interaction between ENMs and amino acids are particularly important.
For example, in Roswell Park Memorial Institute (RPMI) cell culture medium, single-walled carbon nanotubes (SWCNTs) could adsorb a variety of amino acids, vitamins, and phenol red [35]. The driving forces for adsorption were ascribed to π-π interactions and electrostatic interactions. Using the depleted media to culture HepG2 cells, their viability was reduced obviously and could be largely restored by replenishment of folate. Moreover, Casey et al. utilized spectrographic technique to demonstrate the interactions of SWCNTs and components in cell culture medium [41]. Afterward, this research group also investigated indirect cytotoxicity accounted for micronutrient depletion incurred by SWCNTs incubating A549 lung cells [36]. As seen, SWCNTs have enough large specific surface area to alter micronutrient in culture medium by adsorbing small-molecule solutes. Meanwhile, the combined data demonstrated that a broad range of small-molecule solutes, such as amino acids, vitamins, and indicator/probe dyes, is frequently influenced by nonspecific binding of physical adsorption.
Due to the excellent electrical conductivity, structure and mechanical properties of carbon nanotubes (CNTs) [42,43,44], numerous studies have focused on their applications in biological and biomedical fields. As is well known, computational chemistry has provided comprehensive information for the interaction between amino acids and nanomaterials. The binding of collagen amino acids (glycine, proline, and hydroxyproline) to graphene and Ca-doped graphene has been studied by density functional theory calculations and Ab initio molecular dynamics (AIMD) simulations [30]. The obtained data revealed that the doping calcium atoms on carbon surface dramatically enhanced the collagen amino acids binding to graphene, which was due to the electronic charge transfer from Ca to graphene and the carboxyl group of the amino acid. Rajesh et al. investigated the effect of curvature on the non-covalent interaction by comparing the interaction of amino acids with graphene and SWCNTs [45]. Figure 5.2 schematically shows a comparison of interactions between the graphene (planar) or CNT (curved) and four aromatic amino acids [45]. The binding energy followed the same trend between the planar graphene and rolled nanotube structure, but differed in the absolute magnitude. As a result, the binding strength between amino acids and CNT was weaker than that of amino acids and graphene sheet, due to the deviations of the π–π stacking resulting in the case of planar substrates. This observation indicated that the surface curvature of nanomaterials could be a key influence factor on the interactions between nanomaterials and amino acids. Besides, Mukhopadhyay and coworkers found molecular polarity could also affect the interaction of a boron nitride nanotube (BNNT) with amino acids [46]. Governed by electrostatic interactions, polar amino acid molecules exhibited easier binding with the tubular surface of BNNT. Related research results have provided fundamental insights into the interactions between ENMs and amino acids and contributed greatly to the applications of ENMs to biological and biomedical fields.
Scheme for a comparison of interactions between the graphene (planar) or CNT (curved) and four aromatic amino acids: Equilibrium geometry of the rings of the aromatic amino acids on top of graphene (a) and on top of (5, 5) CNT (b), including (i) histidine, (ii) phenylalanine, (iii) tyrosine, and (iv) tryptophan. Reproduced with permission from Ref. [45]. Copyright © 2009 American Institute of Physics. All rights reserved
5.2.2 Nucleobases
Nucleic acid bases, also named genetic molecules, are key components of deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), since they carry the information storage component of every cell in every plant or animal, and play a crucial role in life system, and has become a hot research focus over the past 5 years. Recently, there has been a profound insight in understanding the interaction between nucleobases and ENMs. This interaction not only depends on the individual susceptibility of nucleotide bases but also on the intrinsic physicochemical properties of ENMs.
Zhong et al. explored the adsorption of nucleic acid bases [adenine (A), guanine (G), cytosine (C), thymine (T), and uracil (U)] on hydrogen-passivated silicon nanowire (SiNW) by density functional theory [37]. Figure 5.3 depicts the equilibrium configurations of different nucleobases interacting with a hydrogen-passivated SiNW [37]. As shown, the calculated binding energy of the bases with SiNW was nearly the same, except that of G was higher than others. What’s more, further study indicated that the nature of bonding between a nucleobase and proposed passivated SiNW was dominated by electrostatic interactions. By using the same calculations, Akdim and coworkers calculated the adsorption energy of nucleobases on chiral C (6, 5), C (9, 1), and C (8, 3) SWCNTs [47]. And the trend of calculation was consistent with related computations and experimental work on graphitic surfaces. The interactions between nucleic acid bases with graphene [48, 49] and CNTs [50] were determined by van der Waals force, and the binding energy increased with the polarizability of nucleobases. For single-walled BNNT, the order of calculated binding energy was G > A ≈ C ≈ T ≈ U, indicating the similar interaction strength of nucleobases except for G [51]. Related structural analysis of the adsorbed nucleic acid bases has suggested that the dispersion forces with a marginal contribution from electrostatic forces could provide the stability of the bioconjugated complexes.
Scheme for interactions between ENMs and different nucleases: top and side views of the equilibrium configurations of different nucleobases interacting with a hydrogen-passivated SiNW: a A, b C, c G, d T and e U. Silicon gray, hydrogen light blue, nitrogen dark blue, carbon green, oxygen red. Reproduced with permission from Ref. [37]. Copyright 2012 Elsevier B. V. All rights reserved
Furthermore, Saha et al. theoretically investigated the interactions of different nucleotide bases (A, G, C, T, and U) and four ZnO nanomaterials (ZnO nanowires, ZnO nanotubes, ZnO quantum dots, and ZnO surface) by the self-consistent charge density functional tight binding (SCC-DFTB) method to optimize the complex systems [38]. The site-specific binding nature and the adsorption strength of these nucleobases with different ZnO nanoparticles were calculated in detail. The results manifested the binding energy order and the interaction strength of nucleobases was much dependent on the surface properties of the nanoparticles.
5.2.3 Vitamins
Vitamins are one of the organic materials required to sustain life, which plays a pivotal role in maintaining cell interstitial normal structure, enhancing blood circulation, promoting wound healing, and so on, and they are also important active substrates to keep system alive and healthy. The vitamins adsorbed by ENMs are very important in food industry and biomedical fields. For example, some ENMs with mesoporous structures, such as CNTs, are widely used for vitamin adsorption. The improvement of ENMs in uniformity and specific mesoporous properties has been seen in recent years.
Lu and coworkers added different amounts of CNTs into phenolic resin to synthesize composite spheres by suspension polymerization [40]. Then porous CNTs/activated carbon composite spheres were obtained by carbonizing above composite sphere at 600 °C and steam activating at 850 °C for more than 90 min. After being characterized by nitrogen adsorption–desorption isotherms, the pore size distribution of the obtained porous spheres was “multi-peak”, especially within 20–100 nm pores due to the aggregated pores of CNT bundles. Moreover, the amount of vitamin B12 adsorbed on porous CNTs/activated carbon composite spheres (45 wt% CNT) could be as high as 32.38 mg/g, indicating the application potential of the materials as adsorbents for middle-molecular-weight toxins or large molecules in hemoperfusion.
Shih et al. synthesized mesoporous titania (TiO2) nanocrystallite powders by means of titanium chloride and tri-block nonionic surfactant as starting materials [52]. After investigation, the distribution of pore size was in the range of 2–50 nm, and the surface area of the mesoporous material was 301 m2/g. The adsorption isotherms of vitamin E on TiO2 mesoporous nanocrystals revealed a high affinity between the vitamin E molecule and the adsorbent surface, and the results of XRD and nitrogen adsorption proved the tight packing of the vitamin E molecule inside the mesopores of TiO2 nanocrystals.
Due to the importance of vitamin to metabolism, growth, development, and health, the adsorption characteristics of nanomaterials to vitamin can be utilized in slow-release formulation. In 2015, Golubeva and coworkers investigated adsorption and in vitro release of vitamin B1 by synthetic nanoclays with montmorillonite (Mt) structure [53]. The synthetic Mt structures, with varying compositions Na2x(Al2(1–x),Mg2x )Si4O10(OH)2·nH2O (where 0 < x < 1), were prepared by hydrothermal synthesis. Modeling release of vitamin B1 was performed in simulated gastric fluid (SGF) and simulated intestinal fluid (SIF). It revealed that the adsorption of vitamin B1 depended mainly on the composition and cation-exchange capacity of Mt, and on the pH of the solution to a lesser extent. For the release of vitamin B1, the maximum amounts could reach 54 and 19 wt% in SGF and SIF, respectively, which were higher than that in natural Mt K10. Meanwhile, the data provided information on the optimal Mt compositions for further development of drug delivery systems.
5.2.4 Other Small Biological Molecules
Besides the abovementioned amino acids, nucleobases, and vitamins, other small biological molecules have also been investigated on their interactions with ENMs, such as glucose, and hormones related small biological molecules.
Ganji and coworkers studied the adsorption mechanism of glucose on intrinsic and Pt-decorated SWCNTs by using first-principles van der Waals density functional (vdW-DF) calculations [54]. Due to the higher binding energy, higher net charge transfer values, and shorter connecting distances, Pt-decorated (8, 0) SWCNT could strongly adsorb glucose molecule at the most stable state. Furthermore, the density of states demonstrated the orbital hybridization between the glucose and Pt-decorated nanotube. Therefore, Pt-decorated SWCNT is expected to be a good candidate for the design of glucose biosensors.
Because of the porous, layered structure and large surface area, soluble CNTs can be used to adsorb many environmental pollutants. Among these, the adsorption ability of CNTs for the hormones of bisphenol A (BPA) altered the properties of both BPA and CNTs, leading to different toxicities to human and living systems when BPA and CNTs were used alone. Wang et al. utilized the interactions between BPA and CNTs to investigate the endocrine disrupting effect in mice male offspring [55]. In comparison with oral exposure of BPA alone during gestation and lactation period, the male offspring suffered decreased reproductive toxicity when the mice were exposed to BPA/MWCNT–COOH (carboxylated multi-walled carbon nanotubes). BPA/MWCNT–COOH could effectively decrease malondialdehyde (MDA) level in testis and follicle-stimulating hormone (FSH) in serum, and increase the level of serum testosterone in male offspring. Their results have broadened the knowledge in nanotoxicity and provided important information on the safe application of CNTs.
In summary, ENMs have good application prospects in biomedical and pharmaceutical industry because of their small volume, high surface-activity, and easier interaction with various small biomolecules. However, the potential toxicity of ENMs has become the major constraint to their applications. Thus, it is particularly important to get insights into the impacts of nanoparticles on organisms, elaborate the mechanisms of interaction, and explore ways to minimize toxic effect. Understanding the interactions between ENMs and biomolecules would ensure us to apply the ENMs in more fields safely and effectively taking full advantages of their unique physical and chemical properties.
5.3 The Interactions Between Engineered Nanomaterials and Phospholipids
5.3.1 Current Understanding of the Interactions Between ENMs and Phospholipids
The cell membrane is mainly composed of phospholipids and proteins, and ENMs are capable to perform their functions on organisms only by entering cells through the membrane. Therefore, investigating the interactions between ENMs and phospholipids plays important roles in further studying drug delivery and reducing hazard potential of ENMs. Generally, nanoparticles can pass through cell membranes in two ways: one is direct piercing, and the other is endocytosis.
Shi et al. theoretically studied the interaction mechanisms between CNTs and lipid bilayer by coarse-grained molecular dynamics (CGMD) [56]. An analysis of the structures revealed that the van der Waals force and hydrophobic effect controlled the CNT–cell membrane interaction. As shown in Fig. 5.4, because of small diffusion distance and large curvature energy, small tubes preferred directly piercing through the membrane; on the contrary, larger tubes tended to wrapping into the cell membrane due to larger diffusion distance and lower curvature energy [56]. Wallace and coworkers also employed CGMD to simulate the phospholipid–CNT interaction [57]. When SWCNTs penetrated the dipalmitoylphosphatidylcholine (DPPC) bilayers, lipids were detected to extract and reside on the outer and the inner tube surfaces. In addition, lipids binding on the CNT interior wall could block the tube by spreading out. And the tube penetration velocity was a key influence factor for the degree of lipid lining of the inner surface.
Time-sequenced snapshots of a MWCNT (a) and SWCNT b passing through membrane. a a–f Time sequence of six snapshots of a MWCNT entering a membrane via a wrapping process driven by the van der Waals force; (g, h) fusion of the membrane neck; (i) separation of the lipid covered tube from the membrane. b (a–f) Time sequence of six snapshots of a SWCNT piercing through a membrane driven by the van der Waal force. The red particles indicate the head group of lipid molecules. Reproduced with permission from Ref. [56]. Copyright © Springer-Verlag 2008. All rights reserved
The interactions between ENMs and phospholipids can change the structures and properties of cell membrane. Jing and coworkers found that the binding of semihydrophobic nanoparticles could disrupt supported lipid bilayers (SLBs) [58]. When semihydrophobic nanoparticles reached a critical concentration, the disruption of SLBs, as well as the formation and rapid growth of lipid-poor regions which could be controlled by the concentration, size, and surface hydrophobicity of nanoparticles, was observed. Bothun investigated the effects of aqueous lipid/nanoparticle assemblies on bilayer phase and fluidity [59]. In this research, hydrophobic decanethiol-modified silver nanoparticles were embedded in DPPC bilayers as a model. It was observed that increasing nanoparticle concentration in bilayer-embedded nanoparticles could suppress the lipid pretransition temperature, reduce the melting temperature, and disrupt gel phase bilayers. And the characteristic surface plasmon resonance (SPR) peak of the embedded nanoparticles was independent of the bilayer phase; however, the SPR absorbance was related to vesicle aggregation.
5.3.2 Factors of ENMs Influencing the ENM–Phospholipid Interactions
The interaction of ENMs and phospholipids is mainly affected by the physicochemical properties of nanomaterials, such as size, shape, surface charge, hydrophobic/hydrophilic effect, crystallinity, and concentration. In a real biological system, either one or multiple above mentioned factors play dominant roles in ENM–phospholipid interactions. Moreover, to uncover the mechanisms of these influence factors, many efforts have been devoted to studying their behaviors and integrated forms in vitro and in vivo, which can facilitate the development of ENMs with high biocompatibility for safer biological applications. Here, we center on discussing a series of factors which are recognized to be major contributors that could affect the interactions of ENMs and phospholipids.
Using computer simulations, Yang and coworkers investigated the effect of ENMs with a series of shapes (spheres, ellipsoids, rods, discs, and pushpin-like particles) and volumes on the physical translocation processes [60]. It was revealed that the interaction between the nanoparticle and lipid bilayer was related to the shape anisotropy and initial orientation of ENMs, and the contact area of the particle and lipid bilayer, and the local curvature of the particle at the contact point determined the penetrating capability of a nanoparticle across a lipid bilayer. Moreover, the volume of nanoparticles could influence the translocation indirectly, and the complication of penetration process was determined by particle rotation. l-cysteine and l-glutathione capped Au NPs and gold nanorods (Au NRs) have been synthesized to discuss particle size effects on ENM–phospholipid interaction by determining surface pressure of a preformed model membrane [61]. For Au NPs of 10 and 15 nm average diameter, incorporation rate of smaller Au NPs was higher than those larger counterparts. Meanwhile, the size effect of Au NRs elicited internalization mainly but not exclusively due to longer wrapping time.
To further explore the effect of surface charge, CGMD simulations were employed to observe the interaction between charge-neutral phospholipid membranes and three kinds of nanoparticles with different surface charge densities (the uncharged one, the positively charged one, and the negatively charged one) [62]. The obtained data proved that the adhesion of a charged nanoparticle to the membrane was enhanced by the electrostatic attraction, and a full wrapping to a charged ENM occurred accompanied with the increase of electrostatic energy. At the same time, the structural transitions of fluid bilayers were induced by the adhesion of a charged nanoparticle. Furthermore, the gain in electrostatic energy drove the wrap at the cost of the elastic energy of biomembranes. Kettiger et al. compared the interaction of anionic silica nanoparticles (SNPs) and amine-modified cationic SNPs with phospholipid membranes [63]. The results from dye leakage assay and isothermal titration calorimetry (ITC) illustrated that negatively charged SNPs could react with phospholipids and made phosphatidylcholine (POPC)-based phospholipid bilayers unstable, which was driven by van der Waals forces at the level of the hydration layer on the vesicles surface.
The hydrophilic/hydrophobic property of nanoparticles is another key factor that affects the ENM–phospholipid interactions [64]. Two kinds of nanoparticles (hydrophobic and semihydrophilic) and DPPC bilayer were used as an example to simulate the interactions. Due to the difference in system-free energy, the hydrophobic nanoparticles were included into the bilayer; on the contrary, semihydrophilic nanoparticles were only adsorbed onto the membrane, which indicated that endocytosis-like mechanism was an energy-mediated process. Qiao et al. compared the translocation of fullerene C60 and its derivative C60(OH)20 across a model cell membrane [65]. Based on the molecular dynamics (MD) study, pristine C60 could translocate the membrane within a few milliseconds driven by the hydrophobic interactions. The surface functionalization rendered the hydrophilicity of fullerene, and thereby C60(OH)20 preferred to be adsorbed onto the membrane rather than the bilayer.
In short, investigating the interactions between ENMs and phospholipids, as well as considering the structure and properties of membrane and nanoscale materials, will uncover the mechanisms and elaborate the regulation strategies. It is beneficial to the amelioration of drug delivery system, decryption of its toxicological mechanisms, and development of safer nanomaterials.
5.4 The Interactions Between Engineered Nanomaterials and Proteins
Due to the active surface chemistry of ENMs, proteins in biofluids can be bound on the surfaces of nanoparticles to form bionano interfaces when ENMs are introduced into the biological settings [66]. Investigating the interactions between ENMs and proteins would expand our understanding of molecular mechanisms responsible for nanoparticles and nanoparticle–protein complexes, and would provide information for tailoring physicochemical properties to prepare “safety by design” ENMs.
5.4.1 Experimental Study of ENM–Protein Interactions
5.4.1.1 Thermodynamic and Kinetic Aspects of ENM–Protein Interactions
The thermodynamics and kinetics of the formation of ENM–protein complex are of great importance to investigate the adsorption process. De et al. used ITC to quantify the binding thermodynamics of l-amino acid-terminated monolayers functionalized Au NPs with R-chymotrypsin (ChT), histone, and cytochrome c (Cyt c) [67]. It was revealed that the changes of enthalpy and entropy for the complex formation were dependent on both physiochemical properties of nanoparticles and the intrinsic characteristics of the protein.
The effects of physiochemical properties of the specific nanoparticles on the protein adsorption can also be estimated by investigating the thermodynamics and kinetics. By using ITC, adsorption of human serum albumin (HSA) to N-iso-propylacrylamide/N-tert-butylacrylamide copolymer nanoparticles with varying size and hydrophobicity was studied [68]. It was found that a higher surface coverage occurred for the more hydrophobic particles, and lower degree of surface coverage was observed for smaller nanoparticles (70 nm) than larger counterparts (200–400 nm). Boulos and coworkers explored the adsorption of bovine serum albumin (BSA) on nanoparticles with different shapes, sizes, and surface charges using steady-state fluorescence quenching titration and affinity capillary electrophoresis (ACE) [69]. The obtained results indicated that similar binding constants were observed, which were independent of the shape and surface charge of gold nanomaterials. The limitations of these two present methods mean we are in urgent need of establishing new methods to investigate the ENM–protein interactions.
5.4.1.2 Conformational Change of Proteins Bound on ENMs
The binding of proteins on the surfaces of nanomaterials is usually accompanied by the conformational change of proteins. A significant conformational alteration at both secondary and tertiary structures of BSA upon interaction with Au NPs had been identified by various spectroscopic techniques [70]. Further fluorescence and circular dichroism studies had demonstrated that a higher pH was more inclined to elicit conformational change.
Besides the pH of the system, the size and shape of nanoparticle can affect the conformational change of the bound proteins. As shown in Fig. 5.5, the loss in α-helices content was elevated with the increase of nanoparticle size, and the activity of the fixed lysozyme was lower than that of the free one [71]. Gagner et al. investigated the effect of gold nanomaterials morphology on the structure and function of adsorbed proteins (lysozyme and α-chymotrypsin) [72]. Under saturating conditions, a higher surface density of adsorbed proteins was found on Au NRs than that of Au NPs. The adsorption of lysozyme on Au NPs and Au NRs led to a 10 and 15% loss of secondary structure, respectively. Besides, a significant diminished enzymatic activity was also observed. At low surface coverage, α-chymotrypsin could maintain most of its secondary structure and activity when binding the two kinds of gold nanomaterials; however, the losses of secondary structure and activity could reach to 40 and 86% in an approached monolayer condition, respectively.
Schematic illustration of lysozyme adsorption on silica nanoparticles with different diameters. As the size of the nanoparticle decreases, the curvature of the silica surface increases leading to a greater distance between the approaching protein molecule and the silica surface, and, thus, less interaction between the protein and smaller nanoparticles would be expected. Reproduced with permission from Ref. [71]. Copyright © 2008 American Chemical Society. All rights reserved
In addition, the surface properties of nanoparticles also influence ENM–protein interactions. By altering the ratio of the hydrophobic to hydrophilic ligands, a series of monolayer-protected metal nanoparticles (MPMN) were used to examine their interactions with Cyt c [73]. With the exaggeration of polar ligand content, adsorption of Cyt c was generally increased, indicating hydrophilic interactions between Cyt c and MPMN played a dominant role. Due to the amphipathic character of the lysine side chain, no significant structural disruption happened to Cyt c when bound to various MPMN. Moreover, the results from computational MD simulations were in qualitative agreement with experimental assay. Furthermore, the secondary and tertiary conformational changes of lysozyme relied on a higher or lower surface concentration as well [74].
5.4.2 Simulation of ENM–Protein Interactions
Though experimental methods can offer information on dynamics, thermodynamics, and conformational changes of proteins binding to ENMs, the detailed investigation of individual nanoparticle–protein conjugate is still limited. As an alternative, computer simulation could solve this problem in spite of its shortcoming that could not fully represent the situation in complex biofluids. According to the reported literatures, the nanomaterials used in simulations could be divided into two kinds: one is solid nanoparticles such as metals and their oxides, expressed by plane or solid sphere; the other is carbon-based nanomaterials with the structures of 5- and 6-member rings composed of carbon atoms.
By using discontinuous MD simulations with coarse-grained protein models, three proteins (Trp, WW, and GB3) at concentrations from 0.5 to 5 mM were adsorbed on nanoparticles with diameters ranging from 5 to 20 nm [75]. The simulation results delineated that Langmuir, Freundlich, Temkin, and Kiselev adsorption models performed well with the adsorption of Trp and WW on 10 nm nanoparticles, and provided two positive signals for developing a generalized adsorption model for a series of ENM–protein systems. Voicescu et al. employed the conformation of HSA on functionalized silver nanoparticles (Ag NPs) by the Monte Carlo simulations [76]. When interacted with Ag NPs, the α-helices of HSA diminished, and Trp residue would prefer to locate toward the proteins boundary rather than binding onto the surface of nanoparticles. Tavanti and coworkers simulated the competitive binding of insulin and fibrinogen, two of the most abundant proteins in the plasma, on citrate-coated Au NPs with a diameter of 5 nm [77]. When binding a layer of proteins, a maximum of 20 insulins could be bound and only 3 fibrinogens were able to make contemporaneous interactions with the Au NPs. The binding site of insulin was specific and mainly consisted of the C-terminal residues of the two dimer chains, whereas that of fibrinogen was less specific and generally located at the boundary between the α-nodule and the β-nodule. Once the two proteins were added simultaneously, a competitive binding process for Au NPs could be observed.
Similar to solid nanoparticles, carbon-based nanomaterials could also cause conformational changes of adsorbed proteins. Due to the structure particularity of carbon-based nanomaterials, diverse impacts could occur depending on the structure and adsorption aspects of proteins. Using the sub-domain of HSA as an example, its adsorption behaviors and features on the surfaces of CNTs were explored by MD simulation [78]. It was found that the stepwise conformation and orientation of the model protein was decided by the properties and the texture of surfaces. During its adsorption process, the secondary structures of α-helices and the random coils connecting them were slightly and strongly affected, respectively. Adopting the same simulation method, Noon and coworkers investigated the buckyball (C60)-antibody complex in detail [79]. The simulation results indicated that the complementary shape and extensive side chain interactions led to the high binding affinity and specificity between C60 and antibody molecule, and π–π stacking interaction was the regular mode for π–electron-rich carbon nanoparticles to combine biomolecules. After the tight binding of antibody, there was still 17% of the surface area of buckyball exposed to the solvent, leaving enough room for further manipulation. The adsorption of insulin on graphene with different sizes was also conducted by MD simulation [80]. Polar and charged residues and phenyl rings in the proteins could bind graphene surfaces via the van der Waals interaction and π–π stacking interaction, respectively. In addition, it is also demonstrated that the final conformation of protein was affected by the sizes of graphene and whether the graphene was fixed. Some α-helices in insulin could be protected in non-fixed graphene but not in fixed system. And when the size of graphene was smaller than protein, the interaction energy and the number of adsorption residues would increase as a function of the width of graphene.
When preparing nanomaterials, a basic and strict requirement is to obtain a good dispersion of nanoparticles. However, the adsorption of the peptide chains on the surfaces of nanoparticles can prevent nanoparticles from agglomeration; this finding offers new ways for the synthesis of nanomaterials. Containing hydrophobic valine, aromatic phenylalanine residues, hydrophilic glutamic acid, and lysine residues, the designed amphiphilic helical peptide nano-1 by Chiu et al. [81] was used to investigate its characteristics in different water/hydrophobic interfaces (water/oil, water/graphite, and water/SWCNT) by atomistic MD simulations. Compared with the other two interfaces, nano-1 curved on the SWCNT surface was in α-helical conformation, which maximized its hydrophobicity to contact with the SWCNTs and its hydrogen to bind with water. Also, Chiu et al. studied the properties of the pentamer/(6,6) SWCNT complex and the hexamer/(8, 8) SWCNT complex [82]. As observed in Fig. 5.6, the adsorbed peptides still kept α-helical conformation and formed inter-peptide H-bonds through their Lys and Glu residues [82]. Just like the single peptide system, the peptides in the multipeptide/SWCNT complexes also had as much contact with the SWCNTs, indicating nano-1 was an excellent dispersal agent for SWCNTs.
Snapshots from MD simulations for pentamer/(6, 6) SWCNT (a, b) and hexamer/(8, 8) SWCNT (c, d) systems. a and c illustrate the initial conformations, whereas b and d illustrate the final conformations after 33 ns of simulation. The upper panels display views perpendicular to the SWCNT long axis, and the lower panels display perspectives to the SWCNT long axis. SWCNTs are represented using the vdW model, and peptide backbones are visualized using ribbons. Side chains are shown with a stick model, where Phe and Val residues are emphasized using thicker lines colored in purple and tan, respectively. The peptides are marked as P0 (blue), P1 (red), P2 (gray), P3 (orange), and P4 (yellow) for both systems, and in addition P5 (tan) for the hexamer/(8,8) SWCNT system. The P1 peptide in (d) has slightly unfolded at its C-terminus and uses its His residue to interact with the SWCNT sidewall. Reproduced with permission from Ref. [82]. Copyright © 2009 Wiley Periodicals, Inc. All rights reserved
In a word, there are various factors influencing the interactions between ENMs and proteins, including physicochemical properties of nanoparticles (size, shape, and surface modification) and environmental factors (the concentrations of proteins and nanoparticles) [66, 83, 84]. Confirming these factors contributes to the understanding of the relationships between synthetic chemistry and biological sciences. Modification of ENMs could prevent the unexpected interactions or enhance the desirable interactions, which will facilitate rapid development of nanotechnology-based drug delivery and rational nanotoxicity reduction, and safer ENMs design for biomedical applications.
5.5 The Interactions Between Engineered Nanomaterials and DNA
As one of the basic macromolecules in organisms, DNA with different sequences of base pairs stores a large amount of genetic information. DNA is the polymer of deoxynucleotides which is consisted of a pentose, a phosphate, and a base. All the deoxynucleotides are linked by phosphodiester bond, and two single-stranded DNA (ssDNA) with specific base pairing could form double helix structure [double-stranded DNA (dsDNA)] by hydrogen bonding. Due to the particularity of molecular structure, DNA could strongly interact with ENMs. The interaction between DNA and nanomaterials is related to the properties of both the DNA molecules and the nanoparticles. According to the reported literatures, gold nanoparticles (Au NPs), carbon-based nanomaterials, and quantum dots (QDs) are the hot spots in this research area.
5.5.1 Gold Nanoparticles (Au NPs)
The binding of DNA on the surfaces of Au NPs is attributed to electrostatic interactions. A kinetic study of the interaction of dsDNA with N-(2-mercaptopropionyl) glycine capped Au NPs was investigated by Prado-Gotor and coworkers [85]. The obtained kinetic curves revealed that the interaction involved a simple three-step series mechanism reaction scheme: the first step is fast and involves diffusion-controlled formation of a precursor complex; the second step is the formation of compound (DNA/Au NPs) I dependent on the binding affinity; and the third step is the conformational change from the compound (DNA/Au NPs) I to a more complex form (DNA/Au NPs) II. Komarov et al. employed computer simulation to study the metallization of DNA fragments by the assembly of Au NPs [86]. Due to electrostatic attraction and the short-range attraction between the metallic nanoparticles, Au NPs could aggregate on template DNA. As illustrated in Fig. 5.7, at D = 1σ, the thickness of Au NPs aggregated monolayer covering was close to the diameter of the nanoparticles, and the distribution of the nanoparticles on the template surface was similar to the charge distribution of the template (Fig. 5.7a); however, at D = 2σ, the number of aggregated nanometers was the same as the smaller nanoparticles, indicating that nanoparticles at D = 2σ could aggregate until the negative charge of the DNA fragment was almost compensated (Fig. 5.7b); at D = 3σ, most of the aggregated nanoparticles were in direct contact with the surface of the template (Fig. 5.7c); and, the final aggregation was formed by polydisperse nanoparticles, and the distribution of the nanoparticles was less even in comparison with the case of monodisperse nanoparticles of 2σ size. It was manifested that the size of Au NPs and the charge of the DNA fragment influenced the structure of metallic coat on template DNA. What’s more, the adsorption constants and the number of binding sites on the surfaces of Au NPs were obtained by analyzing adsorption isotherms via model-based Scatchard and Langmuir methods [87].
Typical snapshots of two different projections of the final aggregate formed by monodisperse nanoparticles. Green represents DNA and yellow represents nanoparticles. a D = 1σ, the nanoparticles do form the monolayer covering of the thickness not much larger than the diameter of a nanoparticle, the distribution of the nanoparticles on the template surface roughly duplicates the charge distribution of the template. b D = 2σ, the thickness of metallic covering is about two times larger than the diameter of a nanoparticle, a considerable number of nanoparticles do not contact with the template surface immediately. c D = 3σ, the metallic coat in this case is still more uneven than at D = 2σ, the aggregation number does not depend on the charge of the nanoparticles because the diameter of the nanoparticles becomes larger than the electrostatic screening length. d A typical snapshot of the final aggregate formed by polydisperse nanoparticles, the distribution of the nanoparticles is less even in comparison with the case of monodisperse nanoparticles at D = 2σ. σ = 10 Å is the unit of length. Copyright 2012 Elsevier B.V. All rights reserved Reproduced with permission from Ref. [86]. Copyright © 2008 American Institute of Physics. All rights reserved
Au NP–DNA interactions are mainly affected by the characters of DNA, nanoparticles, and the microenvironment. The binding ability of unmodified DNA to colloidal and surface-confined Au NPs has been investigated by colorimetric and electrochemical technologies [88]. Only ssDNA rather than dsDNA can be adsorbed by colloidal and naked surface-confined Au NPs, and the binding mode was electrostatic interaction and covalent interaction, respectively. To explore the variables which influenced the coverage of DNA on Au NPs, a series of factors, including nanoparticle size, salt concentration, spacer composition, and degree of sonication, had been discussed in detail [89]. Compared to Au NPs with small particle size (13–30 nm), the amount of DNA loading for larger nanoparticles (250 nm) was two orders of magnitude higher. For a particular Au NPs with a diameter of 15 nm, DNA loading can be increased by using 0.7 M NaCl for salt aging and PEG as spacer instead of common nucleobase (A or T) spacers. Lazarus and coworkers found Au NPs functionalized with cationic polyelectrolytes (poly-ethyleneimine and poly-l-lysine) presented a higher electrophoretic mobility when bound to linear DNA than the supercoiled and nicked configuration [90]. In addition, the binding affinities of DNA to Au NPs varied with the change of pH, which could be used to distinguish human p53 gene from sequences with single-base mismatch [91].
The interactions of DNA and Au NPs also elicit alterations in DNA molecules. For example, the relaxation of supercoiled DNA (scDNA) was enhanced when bound to the surfaces of trimethylammonium (TMA) C12 capped Au NPs, which promoted the potency of Au NPs in the treatment of diseases [92]. Yang et al. found small Au NPs inhibited the hybridization of ssDNA with complementary DNA sequences [93]. The nonspecific binding was strong enough to break pre-existing hydrogen bonds in short dsDNA and weakened with the increase of the particle size. The assembly of small metal nanoparticles by DNA hybridization also has provided new ideas for synthesizing larger nanoparticles via core–shell assisted growth method. Octanethiol and 11-trimethylammonium-undecanethiol functionalized Au NPs (2 nm) displayed high binding ability toward DNA driven by electrostatic attraction, and the Au NPs–DNA complex possessed sufficient affinity to inhibit recognition and transcription of T7 RNA polymerase from producing RNA products, indicating the useful application of ENMs to biological researches [94].
5.5.2 Carbon-Based Nanomaterials
In recent years, molecular interactions between carbon-based nanomaterials and DNA have attracted special attention owing to the excellent properties and promising applications carbon-based nanomaterials. The driving forces for the binding of DNA on carbon-based nanomaterials mainly include hydrophobic effect, π–π stacking, and electrostatic force [95]. The adsorption of DNA could alter not only the properties of carbon-based nanomaterials but also the conformations changes of DNA molecules.
5.5.2.1 Carbon Nanotubes (CNTs)
In order to explore the self-assembly mechanisms, structure, and energetic properties of SWCNTs–ssDNA, classical all-atom MD simulations have been adopted [96]. As shown in Fig. 5.8, a random 14-base DNA was initially in a helical-stacked conformation (Fig. 5.8a), and a conformational change of nucleobases within the first 500 ps enabled individual nucleobases when bound on the sidewall of SWCNTs in radial direction via the π–π stacking interaction; Afterward, the additional nucleobases were bound to the sidewall within 5.5 ns accompanied with the approach of entire ssDNA backbone to SWCNTs (Fig. 5.8b). Over the next 16 ns, ssDNA spontaneously wrapped around SWCNTs into a compact right- or left-handed helices, which was driven by electrostatic and torsional interactions (Fig. 5.8c). Using the method of replica exchange molecular dynamics (REMD), Roxbury et al. found that the ordered structures of ssDNA on SWCNTs were dependent on DNA sequence and SWCNT size [97]. Experimental measurements manifested that DNA sequence (TAT)4 on smaller diameter (6,5)-SWCNT and larger diameter (8,7)-SWCNT formed an ordered right-handed helical strand and a small loop configuration, respectively, indicating the selectivity of SWCNT size. In addition, homopolymer (T)12 formed a left-handed wrap on the (6,5)-SWCNT via intrastrand hydrogen bonding, showing the effect of DNA sequence. Albertorio and coworkers quantified the base-dependent ssDNA–SWCNT interactions by probing the specific base dissociation temperatures of homo-oligonucleotide/SWCNT hybrids [98]. Qiu et al. proved that the interaction of DNA–CNT was also governed by the monovalent salt concentration in aqueous solutions [99]. Besides the interaction of ssDNA–SWCNT, the binding of fragmented double-stranded (fds-) DNA (100–500 base pairs, containing both double- and single-stranded regions) to SWCNT should be considered as well [100]. Compared with ssDNA, the interaction of fds-DNA–SWCNT was less efficient. And the formation of hybrids started from the binding of untwisted ss-regions of DNA, and then the whole polymer could wrap on the wall of the tube.
The configuration changes of ssDNA in self-assemble DNA–carbon nanotube hybrid in aqueous solution. a Initial configuration. A simulation was performed on a random 14-base oligonucleotide initially separated from a (11, 0) SWCNT by about 1.5 nm. ssDNA was initialized in a helical-stacked conformation. b Configuration after 5.5 ns. Within 5.5 ns, the entire ssDNA backbone is drawn close to SWCNT, which permits additional nucleobases to bind to the side wall. c Final configuration after 21 ns. Over the next 16 ns, many of the remaining unbound nucleobases adsorb and ssDNA spontaneously warps around SWCNT into a left-handed helix. Reproduced with permission from Ref. [96]. Copyright © 2008 American Chemical Society. All rights reserved
A number of studies have documented the genotoxicity of CNTs in vivo and vitro. There is also a possible interruption of genetic integrity, because DNA could readily insert into CNTs. The MD simulations have proved that the insertion is a spontaneous process, and the van der Waals and hydrophobic forces play dominant roles in the interaction [101]. MD simulations have verified that external electric field could regulate and control the translocation of ssDNA through polarized CNTs [102]. When the electric field strength is inferior to the critical field strength, the translocation event could be inhibited. The translocation process is related not only to the electric field strength, but also to the tube size and nonbonded interactions. Similar to electric field, gravitational acceleration field also has a significant influence on the DNA translocation process [103]. Figure 5.9 shows that DNA could translocate through (10, 10)–(14, 14) CNTs under the gravitational field, and an existing energy barrier indicated that DNA translocation inside a CNT channel was different from DNA translocation into a CNT from outside. Alshehri et al. explored interaction of ssDNA inside SWCNTs, and obtained that the minimum and optimum radiuses of SWCNTs to enclose ssDNA were 12.30 and 12.8 Å [104].
Schematic diagrams for effect of the gravitational field: Snapshots of DNA translocation through (10, 10)–(14, 14) CNTs under the gravitational field of ag = 2.6 × 1013 m/s2 at a time = 0 ps, b time = 330 ps, and c time = 600 ps. Reproduced with permission from Ref. [103]. Copyright © 2008 American Institute of Physics. All rights reserved
5.5.2.2 Graphene Oxide (GO)
The complex DNA–GO has been widely used in biosensing and biomedicine. However, the cognition of GO-mediated genotoxicity is still scarce; thus far, exploring the interaction between DNA and GO is very meaningful. Xu et al. adopted SPR technique to explore DNA–GO binding [105]. In comparison with dsDNA, the binding of ssDNA toward GO was much stronger, which was driven by hydrogen bonding. As a result, an ultra-sensitive sensor for the detection of ssDNA could be established based on the ssDNA/dsDNA discrimination ability of GO. And a homogenous fluorescence polarization assay for the measurement of the DNA of HIV A T7 was also exploited by using the binding difference between dsDNA and ssDNA [106]. Though ssDNA can be bound to GO more easily, the binding force is also connected with the length of DNA. In order to explain the difference caused by DNA length, he and coworkers deeply investigated the binding mechanism of ssDNA with GO by using fluorescence spectroscopy [107]. After calculation, the binding constant of short ssDNA with GO was much lower than that of long ssDNA. According to this conclusion, a novel sensing strategy for determination of S1 nuclease and its inhibitor has been developed. Similarly, Zhang et al. reported a turn-on fluorescence-sensing technique for glucose determination [108]. Additionally, the adsorption kinetics of nano-sized GO (NGO) to shorter DNA was proved faster than that of micro-sized large GO by Lee and coworkers [109].
5.5.3 Quantum Dots (QDs)
Possessing unique properties of emitting narrow and symmetric fluorescence peak, high fluorescence intensity, and excellent anti-photobleaching ability, QDs have been widely used in contaminant and biomolecule sensing, bio-labeling, and immunoassay. Investigating the interactions between QDs and DNA is of great importance for the further application in biology, and a series of methods have been used to study it. Wang and coworkers adopted UV–vis adsorption spectroscopy and electrochemical method to explore the interaction of CdSe/CdS QDs with herring sperm DNA (hs-DNA) [110]. Deduced from the changes of UV–vis adsorption spectroscopy, the apparent association constants of dsDNA–QDs and ssDNA–QDs were 4.94 × 103 and 2.39 × 102 M−1, respectively. The high affinity of hs-DNA toward QDs is attributed to electrostatic force, hydrogen bonds, and van der Waals interactions. Xu et al. investigated the interaction between dsDNA and CdTe QDs by an indirect electrochemical method with the help of Co(phen)33+/2+ (phen = 1,10-phenanthroline) [111]. It was found that the presence of CdTe QDs accelerated the dissociation of Co(phen)33+/2+ from dsDNA modified gold electrode. The results indicated that the major groove of dsDNA was probably the binding site of CdTe QDs, and the same conclusion was also obtained by capillary electrophoresis with laser-induced fluorescence detection and gel electrophoresis [112]. Besides experimental methods, a prototypical model consisting of a capped CdSe QDs and a DNA molecule has also been chosen to compute the interaction by Ab initio electronic structure (based on density functional theory) calculations [113].
The nature of QD surface cations can exert an influence on the interaction with DNA [114]. For instance, the fluorescence of surface-activated CdS QDs by Cd2+, Mg2+, and Zn2+ could be remarkably quenched after binding DNA with straight, bent, and linked structures. And CdS QDs activated with harder Mg2+ and Zn2+ showed higher affinity to different DNAs than those activated with softer Cd2+ ions; however, the ability to distinguish different oligonucleotide shapes was lower because Mg2+ and Zn2+ could cause DNA bending. The complex QDs–DNA could also be used to evaluate the interaction and conformational change of DNA. Employing electrochemiluminescence resonance energy transfer (ECRET), Li et al. [115] and Hu et al. [116] successfully determined them by using luminol-DNA-CdSe/ZnS QDs and CdSe/ZnS QDs-DNA-Cy5 systems, respectively.
Due to the distinctive structure of the DNA molecule that can be connected through the strict principle of base pairing, DNA molecules can combine with metal nanoparticles or graphene to synthesize self-assembly nanomaterials. In addition, exploring the interactions between ENMs and DNA has important practical significance on disease treatment and genetic detection.
5.6 Conclusions and Outlook
Possessing outstanding properties of diverse morphologies, large specific surface area and strong surface reactivity, ENMs have good application prospect in biomedical field. Due to the high affinities to biomolecules, the toxic and side effects of ENMs on organisms impose restrictions on their wide usages. In recent years, remarkable progress has been witnessed in the investigation between nanomaterials and biomolecules by a fair amount of theoretical and experimental research. Current results have revealed that the binding ability of ENMs is related to their size, shape, and surface properties when they react with small biological molecules, phospholipid, protein, and DNA. Meanwhile, the nature of biomolecules is also affected, such as the change of protein structure and DNA conformation.
However, the interaction mechanisms between ENMs and biomolecules are still far from clear understanding, which need further investigations. In-depth understanding of the ENM–biomolecule interactions can do help to explore the interaction mechanisms of ENMs–organelle and ENMs–cell, which contribute to proposing new methods to effectively reduce the toxicity of nanoparticles. In addition, investigating the binding of biomolecules on the surfaces of ENMs also makes people realize the characteristics of the materials more clearly. As a result, we can make more safe and effective use of ENMs with excellent physical and chemical properties in more fields.
References
Liang WZ, Wang XJ, Yokojima S, Chen GH (2000) Electronic structures and optical properties of open and capped carbon nanotubes. J Am Chem Soc 122:11129–11137
Eychmuller A (2000) Structure and photophysics of semiconductor nanocrystals. J Phys Chem B 104:6514–6528
Leroueil PR, Hong SY, Mecke A, Baker JR, Orr BG, Holl MMB (2007) Nanoparticle interaction with biological membranes: does nanotechnology present a janus face? Acc Chem Res 40:335–342
Ray PC (2010) Size and shape dependent second order nonlinear optical properties of nanomaterials and their application in biological and chemical sensing. Chem Rev 110:5332–5365
Jin T, Yoshioka Y, Fujii F, Komai Y, Seki J, Seiyama A (2008) Gd3+-functionalized near-infrared quantum dots for in vivo dual modal (fluorescence/magnetic resonance) imaging. Chem Commun 44:5764–5766
Zhang YY, Wang CM, Cheng Y, Xiang Y (2011) Mechanical properties of bilayer graphene sheets coupled by sp3 bonding. Carbon 49:4511–4517
Bobo D, Robinson KJ, Islam J, Thurecht KJ, Corrie SR (2016) Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm Res 33:2373–2387
He H, Pham-Huy LA, Dramou P, Xiao DL, Zuo PL, Pham-Huy C (2013) Carbon nanotubes: applications in pharmacy and medicine. Biomed Res Int 2013:578290
Islam MS, Deng Y, Tong LY, Faisal SN, Roy AK, Minett AI, Gomes VG (2016) Grafting carbon nanotubes directly onto carbon fibers for superior mechanical stability: towards next generation aerospace composites and energy storage applications. Carbon 96:701–710
Zheng TY, Bott S, Huo Q (2016) Techniques for accurate sizing of gold nanoparticles using dynamic light scattering with particular application to chemical and biological sensing based on aggregate formation. ACS Appl Mater Interfaces 8:21585–21594
Nagao A, Higashimine K, Huaman JLC, Iwamoto T, Matsumoto T, Inoue Y, Maenosono S, Miyamura H, Jeyadevan B (2015) Formation of Pt decorated Ni-Pt nanocubes through low temperature atomic diffusion—time-resolved elemental analysis of nanoparticle formation. Nanoscale 7:9927–9934
Powell CJ, Werner WSM, Shard AG, Castner DG (2016) Evaluation of two methods for determining shell thicknesses of core-shell nanoparticles by X-ray photoelectron spectroscopy. J Phys Chem C Nanomater Interfaces 120:22730–22738
Chao LC, Yang SH (2017) Growth and Auger electron spectroscopy characterization of donut-shaped ZnO nanostructures. Appl Surf Sci 253:7162–7165
Chhoden T, Clausen PA, Larsen ST, Norgaard AW, Lauritsen FR (2015) Interactions between nanoparticles and lung surfactant investigated by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 29:1080–1086
Rao CNR, Biswas K (2009) Characterization of nanomaterials by physical methods. Annu Rev Anal Chem 2:435–462
Zimbone M, Calcagno L, Messina G, Baeri P, Compagnini G (2011) Dynamic light scattering and UV–vis spectroscopy of gold nanoparticles solution. Mater Lett 65:2906–2909
Ding Y, Wang S, Li J, Chen L (2016) Nanomaterial-based optical sensors for mercury ions. TrAC Trends Anal Chem 82:175–190
Chen LX, Fu XL, Li JH (2013) Ultrasensitive surface-enhanced Raman scattering detection of trypsin based on anti-aggregation of 4-mercaptopyridine-functionalized silver nanoparticles: an optical sensing platform toward proteases. Nanoscale 5:5905–5911
Yao H, Dai QL, You ZP (2015) Fourier transform infrared spectroscopy characterization of aging-related properties of original and nano-modified asphalt binders. Constr Build Mater 101:1078–1087
Zhao PY, Huang BY, Gu KJ, Zou N, Pan CP (2015) Analysis of triallate residue and degradation rate in wheat and soil by liquid chromatography coupled to tandem mass spectroscopy detection with multi-walled carbon nanotubes. Int J Environ Anal Chem 95:1413–1423
Kim HJ, Lee HC, Lee JS (2007) Al-27 triple-quantum magic-angle spinning nuclear magnetic resonance characterization of nanostructured alumina materials. J Phys Chem C 111:1579–1583
Lang XF, You TT, Yin PG, Tan EZ, Zhang Y, Huang YF, Zhu HP, Ren B, Guo L (2013) In situ identification of crystal facet-mediated chemical reactions on tetrahexahedral gold nanocrystals using surface-enhanced Raman spectroscopy. Phys Chem Chem Phys 15:19337–19342
Schwerha JJ (2010) Fantastic voyage and opportunities of engineered nanomaterials: what are the potential risks of occupational exposures. J Occup Environ Med 52:943–946
Yokel RA, MacPhail RC (2011) Engineered nanomaterials: exposures, hazards, and risk prevention. J Occup Med Toxicol 6:7
Kapralov AA, Feng WH, Amoscato AA, Yanamala N, Balasubramanian K, Winnica DE, Kisin ER, Kotchey GP, Gou PP, Sparvero LJ, Ray P, Mallampalli RK, Klein-Seetharaman J, Fadeel B, Star A, Shvedova AA, Kagan VE (2012) Adsorption of surfactant lipids by single-walled carbon nanotubes in mouse lung upon pharyngeal aspiration. ACS Nano 6:4147–4156
Mu QX, Jiang GB, Chen LX, Zhou HY, Fourches D, Tropsha A, Yan B (2014) Chemical basis of interactions between engineered nanoparticles and biological systems. Chem Rev 114:7740–7781
Ambike A, Rosilio V, Stella B, Lepêtre-Mouelhi S, Couvreur P (2011) Interaction of self-assembled squalenoyl gemcitabine nanoparticles with phospholipid–cholesterol monolayers mimicking a biomembrane. Langmuir 27:4891–4899
Xu JW, Yang LL, Han YY, Wang YM, Zhou XM, Gao ZD, Song YY, Schmuki P (2016) Carbon-decorated TiO2 nanotube membranes: a renewable nanofilter for charge-selective enrichment of proteins. ACS Appl Mater Interfaces 8:21997–22004
Munk M, Ladeira LO, Carvalho BC, Camargo LSA, Raposo NRB, Serapiao RV, Quintao CCR, Silva SR, Soares JS, Jorio A, Brandao HM (2016) Efficient delivery of DNA into bovine preimplantation embryos by multiwall carbon nanotubes. Sci Rep 6:33588
Cazorla C (2010) Ab initio study of the binding of collagen amino acids to graphene and A-doped (A = H, Ca) graphene. Thin Solid Films 518:6951–6961
Albanese A, Tang PS, Chan WC (2012) The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng 14:1–16
Baer DR, Gaspar DJ, Nachimuthu P, Techane SD, Castner DG (2010) Application of surface chemical analysis tools for characterization of nanoparticles. Anal Bioanal Chem 396:983–1002
Shan CS, Yang HF, Song JF, Han DX, Ivaska A, Niu L (2009) Direct electrochemistry of glucose oxidase and biosensing for glucose based on graphene. Anal Chem 81:2378–2382
McCallum EA, Hyung H, Do TA, Huang CH, Kim JH (2009) Adsorption, desorption, and steady-state removal of 17 beta-estradiol by nanofiltration membranes. J Membr Sci 319:38–43
Guo L, Von Dem Bussche A, Buechner M, Yan AH, Kane AB, Hurt RH (2008) Adsorption of essential micronutrients by carbon nanotubes and the implications for nanotoxicity testing. Small 4:721–727
Casey A, Herzog E, Lyng FM, Byrne HJ, Chambers G, Davoren M (2008) Single walled carbon nanotubes induce indirect cytotoxicity by medium depletion in A549 lung cells. Toxicol Lett 179:78–84
Zhong XL, Slough WJ, Pandey R, Friedrich C (2012) Interaction of nucleobases with silicon nanowires: a first-principles study. Chem Phys Lett 553:55–58
Saha S, Sarkar P (2014) Understanding the interaction of DNA-RNA nucleobases with different ZnO nanomaterials. Phys Chem Chem Phys 16:15355–15366
Shen WZ, Wang H, Guan RG, Li ZJ (2008) Surface modification of activated carbon fiber and its adsorption for vitamin B1 and folic acid. Colloid Surface A 331:263–267
Lu YM, Gong QM, Lu FP, Liang J (2014) Synthesis of porous carbon nanotubes/activated carbon composite spheres and their application for vitamin B12 adsorption. Sci Eng Compos Mater 21:165–171
Casey A, Davoren M, Herzog E, Lyng FM, Byrne HJ, Chambers G (2007) Probing the interaction of single walled carbon nanotubes within cell culture medium as a precursor to toxicity testing. Carbon 45:34–40
Geim AK, Novoselov KS (2007) The rise of graphene. Nat Mater 6:183–191
Zhang Y, Tan YW, Stormer HL, Kim P (2005) Experimental observation of the quantum Hall effect and Berry’s phase in graphene. Nature 438:201–204
Abanin DA, Lee PA, Levitov LS (2006) Spin-filtered edge states and quantum Hall effect in graphene. Phys Rev Lett 96:176803
Rajesh C, Majumder C, Mizuseki H, Kawazoe YA (2009) Theoretical study on the interaction of aromatic amino acids with graphene and single walled carbon nanotube. J Chem Phys 130:124911
Mukhopadhyay S, Scheicher RH, Pandey R, Karna SP (2011) Sensitivity of boron nitride nanotubes toward biomolecules of different polarities. J Phys Chem Lett 2:2442–2447
Akdim B, Pachter R, Day PN, Kim SS, Naik RR (2012) On modeling biomolecular-surface nonbonded interactions: application to nucleobase adsorption on single-wall carbon nanotube surfaces. Nanotechnology 23:165703
Ortmann F, Schmidt WG, Bechstedt F (2005) Attracted by long-range electron correlation: adenine on graphite. Phys Rev Lett 95:186101
Le D, Kara A, Schröder E, Hyldgaard P, Rahman TS (2012) Physisorption of nucleobases on graphene: a comparative van der Waals study. J Phys Condens Mater 24:424210
Gowtham S, Scheicher RH, Pandey R, Karna SP, Ahuja R (2008) First-principles study of physisorption of nucleic acid bases on small-diametercarbon nanotubes. Nanotechnology 19:125701
Mukhopadhyay S, Gowtham S, Scheicher RH, Pandey R, Karna SP (2010) Theoretical study of physisorption of nucleobases on boron nitride nanotubes: a new class of hybrid nano-biomaterials. Nanotechnology 21:165703
Shjh CJ, Lin CT, Wu SM (2010) Adsorption of vitamin E on mesoporous titania nanocrystals. Mater Res Bull 45:863–869
Golubeva OY, Pavlova SV, Yakovlev AV (2015) Adsorption and in vitro release of vitamin B-1 by synthetic nanoclays with montmorillonite structure. Appl Clay Sci 112:10–16
Ganji MD, Skardi FSE (2014) Adsorption of glucose molecule onto platinum-decorated single-walled carbon nanotubes: a dispersion-corrected DFT simulation. Fuller Nanotub Carbon Nanostruct 23:273–282
Wang WW, Jiang CJ, Zhu LD, Liang NN, Liu XJ, Jia JB, Zhang CK, Zhai SM, Zhang B (2014) Adsorption of bisphenol A to a carbon nanotube reduced its endocrine disrupting effect in mice male offspring. Int J Mol Sci 15:15981–15993
Shi XH, Kong Y, Gao HJ (2008) Coarse grained molecular dynamics and theoretical studies of carbon nanotubes entering cell membrane. Acta Mech Sin 24:161–169
Wallace EJ, Sansom MSP (2008) Blocking of carbon nanotube based nanoinjectors by lipids: a simulation study. Nano Lett 8:2751–2756
Jing BX, Zhu YX (2011) Disruption of supported lipid bilayers by semihydrophobic nanoparticles. J Am Chem Soc 133:10983–10989
Bothun GD (2008) Hydrophobic silver nanoparticles trapped in lipid bilayers: size distribution, bilayer phase behavior, and optical properties. J Nanobiotechnol 6:13
Yang K, Ma YQ (2010) Computer simulation of the translocation of nanoparticles with different shapes across a lipid bilayer. Nat Nanotechnol 5:579–583
Ábrahám N, Csapó E, Bohus G, Dékány I (2014) Interaction of biofunctionalized gold nanoparticles with model phospholipid membranes. Colloid Polym Sci 292:2715–2725
Li Y, Gu N (2010) Thermodynamics of charged nanoparticle adsorption on charge-neutral membranes: a simulation study. J Phys Chem B 114:2749–2754
Kettiger H, Québatte G, Perrone B, Huwyler J (2016) Interactions between silica nanoparticles and phospholipid membranes. BBA Biomembr 1858:2163–2170
Li Y, Chen X, Gu N (2008) Computational investigation of interaction between nanoparticles and membranes: hydrophobic/hydrophilic effect. J Phys Chem B 112:6647–16653
Qiao R, Roberts AP, Mount AS, Klaine SJ, Ke PC (2007) Translocation of C60 and its derivatives across a lipid bilayer. Nano Lett 7:614–619
Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S (2011) Protein-nanoparticle interactions: opportunities and challenges. Chem Rev 111:5610–5637
De M, You CC, Srivastava S, Rotello VM (2007) Biomimetic interactions of proteins with functionalized nanoparticles: a thermodynamic study. J Am Chem Soc 129:10747–10753
Lindman S, Lynch I, Thulin E, Nilsson H, Dawson KA, Linse S (2007) Systematic investigation of the thermodynamics of HSA adsorption to N-iso-propylacrylamide/N-tert-butylacrylamide copolymer nanoparticles. Effects of particle size and hydrophobicity. Nano Lett 7:914–920
Boulos SP, Davis TA, Yang JA, Lohse SE, Alkilany AM, Holland LA, Murphy CJ (2013) Article-protein interactions: a thermodynamic and kinetic study of the adsorption of bovine serum albumin to gold nanoparticle surfaces. Langmuir 29:14984–14996
Shang L, Wang Y, Jiang J, Dong S (2007) pH-dependent protein conformational changes in albumin: gold nanoparticle bioconjugates: a spectroscopic study. Langmuir 23:2714–2721
Vertegel AA, Siegel RW, Dordick JS (2004) Silica nanoparticle size influences the structure and enzymatic activity of adsorbed lysozyme. Langmuir 20:6800–6807
Gagner JE, Lopez MD, Dordick JS, Siegel RW (2011) Effect of gold nanoparticle morphology on adsorbed protein structure and function. Biomaterials 32:7241–7252
Hung A, Mwenifumbo S, Mager M, Kuna JJ, Stellacci F, Yarovsky I, Stevens MM (2011) Ordering surfaces on the nanoscale: implications for protein adsorption. J Am Chem Soc 133:1438–1450
Wu X, Narsimhan G (2008) Effect of surface concentration on secondary and tertiary conformational changes of lysozyme adsorbed on silica nanoparticles. BBA Proteomics 1784:1694–1701
Shao Q, Hall CK (2016) Protein adsorption on nanoparticles: model development using computer simulation. J Phys Condens Mater 28:414019
Voicescu M, Ionescu S, Angelescu DG (2012) Spectroscopic and coarse-grained simulation studies of the BSA and HSA protein adsorption on silver nanoparticles. J Nanopart Res 14:1–13
Tavanti F, Pedone A, Menziani MC (2015) Competitive binding of proteins to gold nanoparticles disclosed by molecular dynamics simulations. J Phys Chem C 119:22172–22180
Shen JW, Wu T, Wang Q, Kang Y (2008) Induced stepwise conformational change of human serum albumin on carbon nanotube surfaces. Biomaterials 29:3847–3855
Noon WH, Kong YF, Ma JP (2002) Molecular dynamics analysis of a buckyball-antibody complex. Proc Natl Acad Sci USA 99:6466–6470
Lj Liang, Wang Q, Wu T, Shen JW, Kang Y (2009) Molecular dynamics simulation on stability of insulin on graphene. Chin J Chem Phys 22:627–634
Chiu CC, Dieckmann GR, Nielsen SO (2008) Molecular dynamics study of a nanotube-binding amphiphilic helical peptide at different water/hydrophobic interfaces. J Phys Chem B 112:16326–16333
Chiu CC, Dieckmann GR, Nielsen SO (2009) Role of peptide-peptide interactions in stabilizing peptide-wrapped single-walled carbon nanotubes: a molecular dynamics study. Biopolymers 92:156–163
Shvedova AA, Kagan VE, Fadeel B (2010) Close encounters of the small kind: adverse effects of man-made materials interfacing with the nano-cosmos of biological systems. Annu Rev Pharmacol Toxicol 50:63–88
Moghimi SM, Hunter AC, Andresen TL (2012) Factors controlling nanoparticle pharmacokinetics: an integrated analysis and perspective. Annu Rev Pharmacol Toxicol 52:481–503
Prado-Gotor R, Grueso E (2011) A kinetic study of the interaction of DNA with gold nanoparticles: mechanistic aspects of the interaction. Phys Chem Chem Phys 13:1479–1489
Komarov PV, Zherenkova LV, Khalatur PG (2008) Computer simulation of the assembly of gold nanoparticles on DNA fragments via electrostatic interaction. J Chem Phys 128:124909
Akhlaghi Y, Kompany-Zareh M, Ebrahimi S (2015) Model-based approaches to investigate the interactions between unmodified gold nanoparticles and DNA strands. Sensor Actuators B Chem 221:45–54
Wu Y, Liu LK, Liang ZQ, Shen ZM, Zhu XL (2011) Colorimetric and electrochemical study on the interaction between gold nanoparticles and unmodified DNA. Curr Nanosci 7:359–365
Hurst SJ, Lytton-Jean AKR, Mirkin CA (2006) Maximizing DNA loading on a range of gold nanoparticle sizes. Anal Chem 78:8313–8318
Lazarus GG, Revaprasadu N, Lopez-Viota J, Singh M (2014) The electrokinetic characterization of gold nanoparticles, functionalized with cationic functional groups, and its’ interaction with DNA. Colloids Surf B 121:425–431
Sun LP, Zhang ZW, Wang S, Zhang JF, Li H, Ren L, Weng J, Zhang QQ (2009) Effect of pH on the interaction of gold nanoparticles with DNA and application in the detection of human p53 gene mutation. Nanoscale Res Lett 4:216–220
Foley EA, Carter JD, Shan F, Guo T (2005) Enhanced relaxation of nanoparticl-bound supercoiled DNA in X-ray radiation. Chem Commun 3192–3194
Yang J, Lee JY, Too HP, Chow GM (2006) Inhibition of DNA hybridization by small metal nanoparticles. Biophys Chem 120:87–95
McIntosh CM, Esposito EA, Boal AK, Simard JM, Martin CT, Rotello VM (2001) Inhibition of DNA transcription using cationic mixed monolayer protected gold clusters. J Am Chem Soc 123:7626–7629
Wang Z, Fang H, Wang S, Zhang F, Wang DG (2015) Simulating molecular interactions of carbon nanoparticles with a double-stranded DNA fragment. J Chem 2015:1–6
Johnson RR, Johnson ATC, Klein ML (2008) Probing the structure of DNA-carbon nanotube hybrids with molecular dynamics. Nano Lett 8:69–75
Roxbury D, Mittal J, Jagota A (2012) Molecular-basis of single-walled carbon nanotube recognition by single-stranded DNA. Nano Lett 12:1464–1469
Albertorio F, Hughes ME, Golovchenko JA, Branton D (2009) Base dependent DNA-carbon nanotube interactions: activation enthalpies and assembly-disassembly control. Nanotechnology 20:395101
Qiu XY, Ke FY, Timsina R, Khripin CY, Zheng M (2016) Attractive interactions between DNA-carbon nanotube hybrids in monovalent salts. J Phys Chem C 120:13831–13835
Gladchenko GO, Karachevtsev MV, Leontiev VS, Valeev VA, Glamazda AY, Plokhotnichenko AM, Stepanian SG (2006) Interaction of fragmented double-stranded DNA with carbon nanotubes in aqueous solution. Mol Phys 104:3193–3201
Gao HJ, Kong Y, Cui DX, Ozkan CS (2003) Spontaneous insertion of DNA oligonucleotides into carbon nanotubes. Nano Lett 3:471–473
Xie YH, Kong Y, Soh AK, Gao HJ (2007) Electric field-induced translocation of single-stranded DNA through a polarized carbon nanotube membrane. J Chem Phys 127:225101
Pei QX, Lim CG, Cheng Y, Gao HJ (2008) Molecular dynamics study on DNA oligonucleotide translocation through carbon nanotubes. J Chem Phys 129:125101
Alshehri MH, Cox BJ, Hill JM (2012) Interaction of double-stranded DNA inside single-walled carbon nanotubes. J Math Chem 50:2512–2526
Xue TY, Cui XQ, Guan WM, Wang QY, Liu C, Wang HT, Qi K, Singh DJ, Zheng WT (2014) Surface plasmon resonance technique for directly probing the interaction of DNA and graphene oxide and ultra-sensitive biosensing. Biosens Bioelectron 58:374–379
Wang LJ, Tian JN, Huang Y, Lin XW, Yang W, Zhao YC, Zhao SL (2016) Homogenous fluorescence polarization assay for the DNA of HIV A T7 by exploiting exonuclease-assisted quadratic recycling amplification and the strong interaction between graphene oxide and ssDNA. Microchim Acta 183:2147–2153
He Y, Jiao BN, Tang HW (2014) Interaction of single-stranded DNA with graphene oxide: fluorescence study and its application for S1 nuclease detection. RSC Adv 4:18294–18300
Zhang H, Huang H, Lin ZH, Su XG (2014) A turn-on fluorescence-sensing technique for glucose determination based on graphene oxide-DNA interaction. Anal Bioanal Chem 406:6925–6932
Lee J, Yim Y, Kim S, Choi MH, Choi BS, Lee Y, Min DH (2016) In-depth investigation of the interaction between DNA and nano-sized graphene oxide. Carbon 97:92–98
Wang QS, Yang L, Fang TT, Wu S, Liu P, Min XM, Li X (2011) Interactions between CdSe/CdS quantum dots and DNA through spectroscopic and electrochemical methods. Appl Surf Sci 257:9747–9751
Xu Q, Wang JH, Wang Z, Yin ZH, Yang Q, Zhao YD (2008) Interaction of CdTe quantum dots with DNA. Electrochem Commun 10:1337–1339
Stanisavljevic M, Chomoucka J, Dostalova S, Krizkova S, Vaculovicova M, Adam V, Kizek R (2014) Interactions between CdTe quantum dots and DNA revealed by capillary electrophoresis with laser-induced fluorescence detection. Electrophoresis 35:2587–2592
Anandampillai S, Zhang X, Sharma P, Lynch GC, Franchek MA, Larin KV (2008) Quantum dot-DNA interaction: computational issues and preliminary insights on use of quantum dots as biosensors. Comput Methods Appl Mech Eng 197:3378–3385
Mahtab R, Sealey SM, Hunyadi SE, Kinard B, Ray T, Murphy CJ (2007) Influence of the nature of quantum dot surface cations on interactions with DNA. J Inorg Biochem 101:559–564
Li MY, Li J, Sun L, Zhang XL, Jin WR (2012) Measuring interactions and conformational changes of DNA molecules using electrochemiluminescence resonance energy transfer in the conjugates consisting of luminol, DNA and quantum dot. Electrochim Acta 80:171–179
Hu XF, Zhang XL, Jin WR (2013) Applications of electrochemiluminescence resonance energy transfer between CdSe/ZnS quantum dots and cyanine dye (Cy5) molecules in evaluating interactions and conformational changes of DNA molecules. Electrochim Acta 94:367–373
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Wang, S., Ji, Y., Yin, K., Lv, M., Chen, L. (2017). The Interactions Between Engineered Nanomaterials and Biomolecules. In: Yan, B., Zhou, H., Gardea-Torresdey, J. (eds) Bioactivity of Engineered Nanoparticles. Nanomedicine and Nanotoxicology. Springer, Singapore. https://doi.org/10.1007/978-981-10-5864-6_5
Download citation
DOI: https://doi.org/10.1007/978-981-10-5864-6_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-5863-9
Online ISBN: 978-981-10-5864-6
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)