Abstract
Post-transcriptional gene silencing (PTGS) is a successful technology for the investigation of functions of gene in plants. In general, this phrase refers to the capability of a cell to avert the expression of a definite gene. PTGS can be achieved either by RNA interference (RNAi) or virus-induced gene silencing (VIGS). Tobacco Streak Virus (genus Ilarvirus and family Bromoviridae) consists of a tripartite genome and infects plants by causing symptoms like necrosis and leaf puckering. Ilarvirus are the most imperative viruses, thus causing enormous economic losses worldwide by plummeting crop production by its quantity and quality. Virus infection in plants is known to activate the silencing pathway in which siRNAs are produced. There are numerous reports for the genus Ilarvirus, which have confirmed that RNAi is engineered to target viral RNA in plants. RNA silencing is a high-throughput tool for restraining gene expression carried out by sequence-specific manner, chiefly via transcriptional repression or RNA degradation. As a retort to this defence mechanism, many ilarviruses programme gene silencing suppressor proteins performing at diverse stages in the silencing pathway.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
24.1 Introduction
A number of plant species are vulnerable to a variety of plant viruses (Marwal et al. 2013a). One of such kind is the genus Ilarvirus, belonging to Bromoviridae family (Bujarski et al. 2012). The genus Ilarvirus comprises of 19 virus species. Tobacco Streak Virus (Johnson 1936) is the main species of this genus prevailing in the world. The rest of the species of Ilarvirus are divided into six subgroups: subgroup 1, subgroup 2, subgroup 3, subgroup 4, subgroup 5 and subgroup 6. Tobacco Streak Virus and Parietaria Mottle Virus belong to subgroup 1. Citrus Leaf Rugose Virus, Asparagus Virus 2, Elm Mottle Virus, Citrus Variegation Virus, Tulare Apple Mosaic Virus and Spinach Latent Virus fall in subgroup 2. Whereas subgroup 3 comprises of Apple Mosaic Virus (Fenner 1976), Humulus japonicus Latent Virus (Francki et al. 1991) and Prunus Necrotic Ringspot Virus (Candresse et al. 1998), Fragaria chiloensis Latent Virus (van Regenmortel et al. 2000) and Prune Dwarf Virus (Boari et al. 1998) belong to subgroup 4. Only one species, i.e. American Plum Line Pattern Virus (Matthews 1982; Alayasa et al. 2003), is in the subgroup 5. Finally subgroup 6 contains Lilac Leaf Chlorosis Virus (James et al. 2010). The four species, viz. Blackberry Chlorotic Ringspot Virus (Tzanetakis et al. 2004), Blueberry Shock Virus (Jones et al. 2006), Lilac Ring Mottle Virus (Matthews 1979; Scott and Zimmerman 2008) and Strawberry Necrotic Shock Virus (Tzanetakis et al. 2010), are unassigned ilarviruses. Tobacco Streak Virus causes serious crop production losses and decreases in the product quality as well (Walter et al. 1995).
These viruses are isometric in shape and acquire a single-stranded, mainly tripartite RNA genome. Tobacco Streak Virus has been studied the most, and similarly a good deal of knowledge has been attained for Prunus Necrotic Ringspot Virus (Sharman et al. 2009). Developing new plant varieties with resistance to Tobacco Streak Virus (Ladhalakshmi et al. 2009) and other viral pathogens is considered highly necessary for farmers. Even if procreation for virus-resistant varieties has been followed for a long time by simple breeding techniques, the advancement remains sluggish because of the inherent genetic complexity of resistance (Barba et al. 1992; Waterhouse and Helliwell 2003). Wherein molecular biology makes it promising to manoeuvre and improve plant resistance to Tobacco Streak Virus, such molecular biology skill highlights post-transcriptional gene silencing (PTGS) (Baulcomb 2004). The 2b protein of Tobacco Streak Virus and other ilarviruses are responsible for RNA silencing and the viral movement for long distances in the plant (Guo and Ding 2002). Here, we are presenting a mini review on silencing of Tobacco Streak Virus and other ilarviruses mediated via RNAi (RNA interference) (Watson et al. 2005) and understanding the effect of various genes through VIGS (virus-induced gene silencing) (Burch-Smith et al. 2004; Constantin et al. 2004) from the agronomical and horticultural point of view.
24.2 Optimization Through RNAi-Mediated Silencing
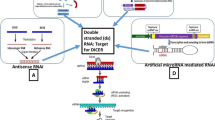
RNA interference (RNAi) takes place in a broad range of living beings; this includes plants, fungi and animals (Bass 2000; Saunders et al. 2004). RNA degradation progression is a sequence-specific RNA silencing mechanism that is activated either by the formation of dsRNA or otherwise by unusual RNAs associated with transgenes viruses and transposons (Vaucheret 2006; Marwal et al. 2013b). Double-stranded RNA (dsRNA) is generally cleaved in plants by the cellular machinery into short interfering RNAs (siRNAs), which are efficient inducers of gene silencing (Fusaro et al. 2006; Kerschen et al. 2004). RNAs with hairpin with a loop structures are particularly actual inducers of PTGS in plants (Ikegami et al. 2011; Yoshikawa et al. 2013). The dsRNAs trigger an RNA-mediated defence system resulting in their cleavage into small-interfering RNAs (siRNAs) by Dicer-like enzymes. In RNA-induced silencing complex, the siRNAs further act upon the degradation of RNAs, which has identical sequences to those of the inserted fragment and viral genome (Baulcomb 2004; Lecellier and Voinnet 2004; Marwal et al. 2013c; Meister and Tuschi 2004).
Prunus species are harmfully pretentious by a major pollen scattered Ilarvirus, i.e. Prunus Necrotic Ringspot (PNRSV) (Amari et al. 2007). RNA interference (RNAi) vector pART27–PNRSV was created, enclosed with an inverted repeat (IR) region consisting of PNRSV. This construct was then inoculated into two hybrid cherry rootstocks [‘Gisela 6’ (GI 148–1) and ‘Gisela 7’ (GI 148–8)] which were tolerant and sensitive, respectively, to PNRSV infection (Lacomme et al. 2003). After 1 year of inoculation with PNRSV plus Prune Dwarf Virus, nontransgenic ‘Gisela 6’ doesn’t exhibit any indication of virus disease but does possessed a noteworthy PNRSV titre. The transgenic ‘Gisela 6’ was devoid of symptoms and encountered with negligible PNRSV titre. In the course of this experiment, the nontransgenic ‘Gisela 7’ trees don’t survive, while the transgenic ones, i.e. ‘Gisela 7’ trees, continue to exist (Song et al. 2013).
A number of leading viruses critically impinge on Prunus L. fruit production (Aparicio et al. 2010). It is exceedingly required by growers and breeders that the expansions of new varieties resistant to these viruses are quite exigent. For engineering multivirus resistance in plants, a post-transcriptional gene silencing foundation was accounted. For this approach, a solo chimeric transgene, i.e. PTRAP6, was fashioned by the amalgam of around 400–500-base pair (bp) gene fragments from six major Prunus fruit viruses, consisting of Peach Mosaic Virus, American Plum Line Pattern Virus, Prunus Necrotic Ringspot Virus, Prune Dwarf Virus (PDV), Plum Pox Virus (PPV) and Tomato Ringspot Virus (ToRSV). Devoid of any splicing intrusion, it was found that the two strands of PTRAP6 created a 2.5 kb transcript in plant when being transcribed.
PTRAP6i was shaped by insertion of two copies of PTRAP6 in an inverted repeat under the command of the Cauliflower Mosaic Virus 35S promoter and divides by an intron spacer fragment for inducing gene silencing/virus resistance. Out of 28 R0 PTRAP6i transgenic lines, only 12 were resistant to ToRSV which were earlier inoculated in Nicotiana benthamiana plants. The symptoms range from mild visualization to phenotypes which were devoid of any symptoms. Detailed analysis of two of the three highly resistant homozygous R3 generation lines demonstrated that they were resistant to PPV, PDV and ToRSV. The rest of the three viruses targeted by PTRAP6i were either unavailable for this study or were unable to systemically infect N. benthamiana (Lui et al. 2007).
In another incident, Prune Dwarf Virus (PDV) was found causing systemic infection in some almond trees and other Prunus sp. which were spread by means of pollen grains (Abou-Jawdah et al. 2004). An approach that was focused on the coat protein (cp) gene subjected to restrict PDV replication in host plant cells has been studied. To construct the cDNA of the cp gene, a Portuguese isolate of PDV was acquired from infected almond leaves. To seek for the transgenic expression of the new or customized Prune Dwarf Virus coat protein (cpPDVSense and cpPDVMutated), a range of constructs was organized based on this sequence. Similar aspects were made in case of cpPDV RNA (cpPDVAntisense and cpPDV without start codon) for its expression. Widespread molecular characterization and controlled infections were achieved on transformants and their offspring, where all constructs were tested in a PDV host model, Nicotiana benthamiana.
As evaluated by DAS-ELISA on newly developed leaves, transgenic plants exhibiting cp RNA were capable of blocking the propagation of Prune Dwarf Virus isolate, thus contributing nearly 91% homology with the isolate used for cpPDV cloning. With cp expression, the obstruction of PDV propagation in lately formed leaves was only accomplished due to the mutated construct of cpPDV, where arginine was replaced by alanine due to substitution in the coat protein at the 14th aa residue position. The experiment emphasizes the possible responsibility of the mutated amino acid in the virus capability to replicate and proliferate. The following study expressed the likelihood for accomplishing defence against Prune Dwarf Virus via mutated cp sequence or by coat protein RNA (Raquel et al. 2008).
Prunus domestica L were transformed with the Plum Pox Virus coat protein gene (PPV-CP). Transgenic plums were extremely challenging to PPV infection since it exhibits post-transcriptional gene silencing (PTGS). In order to test the consequence of heterologous viruses on the usefulness and constancy of PTGS against PPV, transgenic C5 trees were graft inoculated with diverse amalgamation of Prunus Necrotic Ringspot Virus (PNRSV), Apple Chlorotic Leaf Spot Virus (ACLSV), Prune Dwarf Virus (PDV) and PPV-D strain (Sasaki et al. 2011).
The possibility for suppression of the silencing system mediated by these viruses was evaluated. Confront experiments were performed under greenhouse, nursery and field conditions in Romania and Spain, including two different environments, continental and Mediterranean, respectively. Virus infections were appraised by visual supervises of symptom and by molecular and serological study. Resistance against Plum Pox Virus for C5 transgenic plums was engineered, which was firm and was not obscured by the occurrence of the challenging heterologous viruses. This study was carried over a period of 3 year in all trials (Zagrai et al. 2008).
Three experiments were undertaken in Romania, where transgenic plums of Prunus domestica L. as the subjects were inoculated with PPV-CP (coat protein gene of Plum Pox Virus). With the influence of natural infection, the transgenic clones such as C2, C3, C4, C5, C6 and PT3 were assessed for sharka resistance. The highest resistance was observed in transgenic clone C5 (named as ‘HoneySweet’). Up to 10 years, transgenic C5 trees were devoid of any visible symptoms caused by naturally infected aphids. This is due to post-transcriptional gene silencing (PTGS) exhibited by the resistant C5 lines. The second study evaluated the consequence of two heterologous viruses (i.e. Prune Dwarf Virus and Prunus Necrotic Ringspot Virus) based on the effectiveness and stability of PTGS-mediated resistance to Plum Pox Virus demonstrated by the C5 plum. This engineered resistance to Plum Pox Virus in the C5 transgenic plums was firm and doesn’t concealed by the existence of the examined heterologous viruses (Zagrai et al. 2011).
One of the most efficiently important viruses infecting several crop plants in India is the Tobacco Streak Virus (TSV). RT-PCR with TSV replicase gene-specific primers was carried on indicative samples collected from sunflower and okra fields. In order to build up tobacco transgenic plants resistance to Tobacco Streak Virus (TSV) by articulating hairpin RNA transcript (hpRNA), the replicase (Rep) genes of these isolates were sequenced. A 99% nucleotide sequence identity of replicase gene of these isolates with Tamil Nadu okra isolate was revealed. The position 3065–3405 of the TSV replicase gene was used for building of pHANNIBAL vector, i.e. a conserved nucleotide sequence having a hairpin construct.
The Rep hairpin construct was cloned into pART27 and congregate into Agrobacterium tumefaciens LBA4404 and commenced into tobacco by Agrobacterium-mediated transformation. Taking the genomic DNA from transformed tobacco plants, the T0 plants produced were subjected to PCR and Southern blot examination. Corresponding to nptII gene and Rep gene, the transformants produced ~299 bp and 340 bp amplicons, respectively. The single- and multiple-copy integration of the transgenes was confirmed by Southern blot analysis. Upon mechanical inoculation of TSV, the transgenic T0 tobacco plants illustrate resistance against TSV without showing any visible symptoms; resistance was also confirmed by DAC-ELISA (Suppaiah et al. 2015).
24.3 Engineering by VIGS: A Versatile Tool
Viruses that derived small-interfering RNAs (siRNAs) are the hallmarks of an innate immune response in plants that targets invading viruses through post-transcriptional gene silencing (PTGS). Virus-induced gene silencing (VIGS) has a great potential as a reverse genetic tool in plant genomics (Burch-Smith et al. 2004; Marwal et al. 2014; Robertson 2004). In plants, PTGS has been widely studied, and like PTGS that is distinguished by sequence-specific resistance against virus infection, viruses also induced an RNA-mediated defence system in plants. Tobacco Mosaic Virus (TMV) was the first RNA virus used as silencing vectors (Godge et al. 2008).
VIGS involves using a vector containing the piece of gene of interest that causes the silencing of specific gene expression (Gleba et al. 2007). siRNA is an important method for evaluating gene functionality and is being exploited for the development of new approaches to control plant viruses (Mourrain et al. 2000; Covey et al. 1997; Marwal et al. 2012; Ratcliff et al. 1997; Lu et al. 2003). VIGS engross the release of a recombinant virus to plants containing a portion of the plant gene that is proposed to be silenced. The plant defence mechanism system then diminishes not only the virus but also the targeted endogenous plant gene expression through post-transcriptional gene silencing (Robertson 2004).
Asparagus Virus 2 (AV-2) is another member of the genus Ilarvirus. The coat protein (CP) and the 2b protein (2b) genes of AV-2 isolates were cloned from Asparagus plants from a variety of province, and it was established that the sequence for CP and for 2b was extremely conserved among the isolates, signifying that AV-2 from around the world is almost indistinguishable (Xin et al. 1998). Later an AV-2 infectious clone was created by instantaneous inoculation with in vitro transcripts of RNAs 1–3 of AV-2 and in vitro-synthesized CP, which is obligatory for initial infection. Because 2b of cucumoviruses in Bromoviridae can hold back systemic silencing as well as confined silencing, it was analysed whether there is practical synteny of 2b protein between AV-2 and Cucumovirus. By means of the AV-2 infectious clone, the Ilarvirus 2b job as an RNA silencing suppressor is now evident; AV-2 2b has suppressor bustle against systemic silencing but not confined silencing (Shimura et al. 2013).
For molecular characterization of gene functions in plants, RNA silencing is a dominant skill. Genetic transformation is a generally used method for the introduction of RNA silencing. The best potent substitute is to use a customized viral vector for virus-induced gene silencing (VIGS) to demean RNA molecules partaking similar nucleotide sequence. Due to a long immature stage and intractable to genetic transformation, unfortunately genomic studies in many allogamous woody perennials such as peach are sternly delayed. The construction of a viral vector imitative from Prunus Necrotic Ringspot Virus (PNRSV), a prevalent fruit tree virus that is endemic in all Prunus fruit production countries and regions in the world, was reported.
It was affirmed that the modified PNRSV vector, an anchor ageing the sense-orientated objective gene sequence of 100–200 bp in length in genomic RNA 3, could impressively trigger the silencing of a transgene or an endogenous gene in the model plant Nicotiana benthamiana. It was further demonstrated that vector formed by Prunus Necrotic Ringspot Virus can be easily manoeuvre to cause silencing of endogenous genes in peach similar to translation initiation factor 4E isoform (eIF(iso)4E) of eukaryotic, a host factor of many potyviruses including Plum Pox Virus (PPV). Moreover, the eIF(iso)4E-knocked down peach plants were resistant to PPV (Cui and Wang 2016).
Functional genomics authorize knockdown of expression of individual genes or closely linked gene families through virus-based gene silencing systems which is a well thought-out influential tool. TSV shows recovery from initial symptoms and efficiently invades both meristems and developing embryos in soybean making it an excellent candidate for a virus-based silencing system for those tissues. TSV RNAs 1, 2, 3 and 4 were cloned into pHST40, a pUC-based plasmid vector, and pCASS-4RZ, an Agrobacterium tumefaciens-compatible binary vector. Both sets of clones were infectious in soybean and tobacco. 2b gene of pHST40-RNA2 was truncated, and multicloning site was introduced, and the clone was stably transmitted in soybean seed.
Obvious leaf yellowing typical for silencing of MgCh mRNA was exhibited, when magnesium chelatase (MgCh) gene parts of 105 nt and 175 nt were put into the truncated 2b vector and were stable in systemic leaves of inoculated ‘Williams82’ plants. RNA 3 of the pCASS-4RZ clone was partitioned between two RNAs, one with only the movement protein (pCASS-R3Mp) and the other expressing only the coat protein (pCASS-R3Cp). Full-length green fluorescent protein (GFP) and phytoene desaturase (PDS) coding regions were inserted into pCASS-R3Mp and pCASS-R3Cp, respectively. Tobacco plants illustrated steady expression of GFP and photo bleaching symptoms when inoculated, which is reliable with silencing of PDS mRNA (Jossey et al. 2011).
Dahlia (Dahlia variabilis) flower colour has been credited with black, due to the elevated levels of anthocyanins which are cyaniding compounds (Chen et al. 2004). This pattern transpires because of flavone synthesis, as it is reduced for the reason that of post-transcriptional gene silencing (PTGS) of flavone synthase II (DvFNS). Apart from the black colour, purple-coloured flowers are also known, which has appeared from a black cultivar ‘Kokucho’. It was found that the purple colour of flower is not the result of mutation but due to the infection of Tobacco Streak Virus, which suppresses the PTGS of DvFNS. When Tobacco Streak Virus was eradicated from the purple flowering ‘Kokucho’ by leaf primordia-free shoot apical meristem culture, the resultant flowers again restore their black colour (Deguchi et al. 2015).
It was portentous that Tobacco Streak Virus has a silencing suppressor, as due to Tobacco Streak Virus which was infecting purple flowers showed lower numbers of siRNAs than black flowers. Tobacco Streak Virus-infected dahlia distorted the flower colour severely by the graft inoculation of other black cultivars apart from ‘Fidalgo Blacky’, which is a very deep black cultivar with the highest amount of cyaniding-based anthocyanins. The flowers of all six Tobacco Streak Virus-infected Dahlia cultivars mount up augmented quantity of flavones and reduced quantity of cyaniding-based anthocyanins. There was no change in the accumulation of pigments in ‘Fidalgo Blacky’ and thus remained black whereas in Dahlia plants infected with Tobacco Streak Virus still had higher level of cyaniding-based anthocyanins than other cultivars.
24.4 Conclusion
This review makes noticeable that the RNAi tactic is beneficial for developing viral resistance in plants and such transgenics have imminent to augment production of customized varieties (nongenetically) thus evading concern regarding transgene flow. Transgene-wide and siRNA species were detected along with vanishing of transgene transcript in the resistant lines, representing that PTGS underlies the method of resistance. This review presents confirmation that RNAi is able to bestow gene silencing-based resistance to multiple ilarviruses (Hamilton and Baulcombe 1999).
Virus-induced gene silencing (VIGS) is a successful technology for the investigation of functions of gene in plants (Gronlund et al. 2008; vanKammen 1997; Zhang and Ghabrial 2006). This work opens a potential avenue for the control of virus diseases in plants via viral vector-mediated silencing of host factors, and vector may serve as a powerful molecular tool for functional genomic studies. Ultimately, the two approaches discussed above are used to produce virus-resistant cultivars. Researchers around the world are currently developing approaches to engineer multivirus resistance in plants to address the serious virus problems encountered in agricultural practice.
References
Abou-Jawdah Y, Sobh H, Cordahi N, Kawtharani H, Nemer G, Maxwell DP, Nakhla MK (2004) Immunodiagnosis of Prune Dwarf Virus using antiserum produced to its recombinant coat protein. J Virol Methods 121:31–38
Alayasa N, Al Rwahnih M, Myrta A, Herranz MC, Minafra A, Boscia D, Pallás V (2003) Identification and characterization of an American Plum Line Pattern Virus isolate from Palestine. J Plant Pathol 85:3–7
Amari K, Burgos L, Pallás V, Sánchez-Pina MA (2007) Prunus Necrotic Ringspot Virus early invasion and its effects on apricot pollen grains performance. Phytopathology 97:892–899
Aparicio F, Sánchez-Navarro JA, Pallás V (2010) Implication of the C terminus of the Prunus Necrotic Ringspot Virus movement protein in cell-to-cell transport and in its interaction with the coat protein. J Gen Virol 91:1865–1870
Barba M, Martino L, Lauretti F (1992) Comparison of different methods to produce virus free stone fruits. Acta Hortic 309:385–392
Bass BL (2000) Double-stranded RNA as a template for gene silencing. Cell 101:235–238
Baulcomb D (2004) RNA silencing in plants. Nature 431:356–363
Boari A, Boscia D, Di Terlizzi B, Savino V (1998) Study on seed transmission of Prune Dwarf Virus (PDV) in Prunus mahaleb L. Adv Hortic Sci 12:89–92
Bujarski J, Figlerowicz M, Gallitelli D, Roossinck MJ, Scott SW (2012) Family Bromoviridae. In: Virus taxonomy. Ninth report of the international committee on taxonomy of viruses. Elsevier Academic, San Diego, pp 965–976
Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39:734–746
Candresse T, Kofalvi SA, Lanneau M, Dunez J (1998) A PCRELISA for the simultaneous detection and identification of Prunus Necrotic Ringspot (PNRSV) and Apple mosaic (APMV) ilarviruses. Acta Hortic 472:219–224
Chen J-C, Jiang C-Z, Gookin TE, Hunter DA, Clark DG, Reid MS (2004) Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Mol Biol 55:521–530
Constantin GD, Krath BN, MacFarlane SA, Nicolaisen M, Johansen IE, Lund OS (2004) Virus-induced gene silencing as a tool for functional genomics in a legume species. Plant J 40:622–631
Covey SN, Al-Kaff NS, La ´ngara A, Turner DS (1997) Plants combat infection by gene silencing. Nature (London) 385:781–782
Cui H, Wang A (2016) An efficient viral vector for functional genomic studies of Prunus fruit trees and its induced resistance to Plum Pox Virus via silencing of a host factor gene. Plant Biotechnol J 15(3):344–356
Deguchi A, Tatsuzawa F, Hosokawa M, Doi M, Ohno S (2015) Tobacco Streak Virus (strain dahlia) suppresses posttranscriptional gene silencing of flavone synthase II in black dahlia cultivars and causes a drastic flower color change. Planta 242:663–675
Fenner F (1976) Classification and nomenclature of viruses. Second report of the international committee on taxonomy of viruses. Intervirology 7:1–115
Francki RIB, Fauquet CM, Knudson DL, Brown F (1991) Classification and nomenclature of viruses. Fifth report of the international committee on taxonomy of viruses. Arch Virol Suppl 2:452
Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, Waterhouse PM (2006) RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep 7:1168–1175
Gleba Y, Klimyuk V, Marillonnet S (2007) Viral vectors for the expression of proteins in plants. Curr Opin Biotechnol 18:134–141
Godge MR, Purkayastha A, Dasgupta I, Kumar PP (2008) Virus-induced gene silencing for functional analysis of selected genes. Plant Cell Rep 27:209–219
Gronlund M, Constantin G, Piednoir E, Kovacev J, Johansen IE, Lund OS (2008) Virus-induced gene silencing in Medicago truncatula and Lathyrus odorata. Virus Res 135:345–349
Guo HS, Ding SW (2002) A viral protein inhibits the long range signalling activity of the gene silencing signal. EMBO J 21:398–407
Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950–952
Ikegami M, Kon T, Sharma P (2011) RNA silencing and viral encoded silencing suppressors. In: Gaur RK, Gafni Y, Gupta VK, Sharma P (eds) RNAi technology. CRC Press, Boca Raton, pp 209–240
James D, Varga A, Leippi L, Godkin S, Masters C (2010) Sequence analysis of RNA 2 and RNA 3 of Lilac Leaf Chlorosis Virus: a putative new member of the genus Ilarvirus. Arch Virol 155:993–998
Johnson J (1936) Tobacco streak, a virus disease. Phytopathology 26:285–292
Jones AT, McGavin WJ, Gepp V, Scott SW, Zimmerman MT (2006) Purification and properties of Blackberry Chlorotic Ringspot, a new virus species in Su bgroup 1 of the genus Ilarvirus found naturally infecting blackberry in the UK. Ann Appl Biol 149:125–135
Jossey S, Singh AK, Ghabrial SA, Domier LL (2011) Development of a Tobacco Streak Virus (TSV) based gene silencing vector for soybean seed development. APS IPPC Joint Meeting, Honolulu
Kerschen A, Napoli C, Jorgensen R, Muller A (2004) Effectiveness of RNA interference in transgenic plants. FEBS Lett 566:223–228
Lacomme C, Hrubikova K, Hein I (2003) Enhancement of virus induced gene silencing through viral-based production of inverted-repeats. Plant J 34(4):543–553
Ladhalakshmi D, Ramaiah M, Ganapathy T, Krishna Reddy M, Khabbaz SE, Babu M, Kamalakannan A (2009) First report of the natural occurrence of Tobacco Streak Virus on blackgram (Vigna mungo). Plant Pathol 12:55
Lecellier CH, Voinnet O (2004) RNA silencing: no mercy for viruses? Immunol Rev 198:285–303
Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC (2003) Virus-induced gene silencing in plants. Methods 30:296–303
Lui Z, Scorza S, Hily JM, Scott SW, James D (2007) Engineering resistance to multiple Prunus Fruit Viruses through expression of chimeric hairpins. J Am Soc Hortic Sci 132(3):407–414
Marwal A, Sahu A, Prajapat R, Choudhary DK, Gaur RK (2012) First report of association of begomovirus with the leaf curl disease of a common weed, Datura inoxia. Virus Dis 23(1):83–84
Marwal A, Sahu A, Sharma P, Gaur RK (2013a) Transmission and host interaction of geminivirus in weeds. Plant virus-host interaction: molecular approaches and viral evolution. Elsevier Chapter 7:143–161
Marwal A, Sahu A, Sharma P, Gaur RK (2013b) Molecular characterizations of two begomoviruses infecting Vinca rosea and Raphanus sativus in India. Virol Sin Virol Sin 28(1):053–056
Marwal A, Sahu A, Choudhary DK, Gaur RK (2013c) Complete nucleotide sequence of a begomovirus associated with satellites molecules infecting a new host Tagetes patula in India. Virus Genes 47(1):194–198
Marwal A, Sahu A, Gaur RK (2014) First report of airborne begomovirus infection in Melia azedarach (pride of India), an ornamental tree in India. Aerobiologia 30(2):211–215
Matthews REF (1979) Classification and nomenclature of viruses. Third report of the international committee on taxonomy of viruses. Intervirology 12:129–296
Matthews REF (1982) Classification and nomenclature of viruses. Fourth report of the international committee on taxonomy of viruses. Intervirology 17:1–199
Meister G, Tuschi T (2004) Mechanisms of gene silencing by double stranded RNA. Nature 431:343–349
Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Remoue K, Sanial M, Vo TA, Vaucheret H (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533–542
Raquel H, Lourenco T¸ Moita C, Oliveira MM (2008) Expression of prune dwarf Ilarvirus coat protein sequences in Nicotiana benthamiana plants interferes with PDV systemic proliferation. Plant Biotechnol Rep 2:75–85
Ratcliff FG, Harrison BD, Baulcombe DC (1997) A similarity between viral defence and gene silencing in plants. Science 276:1558–1560
Robertson D (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55:495–519
Sasaki S, Yamagishi N, Yoshikawa N (2011) Efficient virus-induced gene silencing in apple, pear and Japanese pear using apple latent spherical virus vectors. Plant Methods 7:15
Saunders K, Norman A, Gucciardo S, Stanley J (2004) The DNA- b satellite component associated with ageratum yellow vein disease encodes an essential pathogenicity protein (bC1). Virology 324:37–47
Scott SW, Zimmerman MT (2008) Partial nucleotide sequences of the RNA 1 and RNA 2 of Lilac Ring Mottle Virus confirm that this virus should be considered a member of subgroup 2 of the genus Ilarvirus. Arch Virol 153:2169–2172
Sharman M, Persley DM, Thomas JE (2009) Distribution in Australia and seed transmission of Tobacco Streak Virus in Parthenium hysterophorus. Plant Dis 93:708–712
Shimura H, Masuta C, Yoshida N, Sueda K, Suzuki M (2013) The 2b protein of Asparagus virus 2 functions as an RNA silencing suppress or against systemic silencing to prove functional synteny with related cucumoviruses. Virology 442:180–188
Song GQ, Sink KC, Walworth AE, Cook MA, Allison RF, Lang GA (2013) Engineering cherry rootstocks with resistance to Prunus Necrotic Ring Spot Virus through RNAi-mediated silencing. Plant Biotechnol J 11(6):702–708
Suppaiah R, Muthuraj R, Gandhi K (2015) Conserved sequence of replicase gene mediated resistance in Nicotiana tabacum L. cv Abirami through RNA silencing. Eur J Plant Pathol. doi:10.1007/s10658-015-0658-z
Tzanetakis IE, Mackey IC, Martin RR (2004) Strawberry Necrotic Shock Virus is a distinct virus and not a strain of Tobacco Streak Virus. Arch Virol 149:2001–2011
Tzanetakis IE, Martin RR, Scott SW (2010) Genomic sequences of Blackberry Chlorotic Ringspot Virus and Strawberry Necrotic Shock Virus and the phylogeny of viruses in subgroup 1 of the genus Ilarvirus. Arch Virol 155:557–561
vanKammen A (1997) Virus-induced gene silencing in infected and transgenic plants. Trends Plant Sci 2:409–411
van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB (2000) Virus taxonomy, Seventh report of the International Committee on Taxonomy of Viruses. Academic, San Diego. p1162
Vaucheret H (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20:759–771
Walter MH, Wyatt SD, Kaiser WJ (1995) Comparison of the RNAs and some physiochemical properties of the seed transmitted Tobacco Streak Virus isolate Mel 40 and the infrequently seed-transmitted isolate Mel F. Phytopathology 85:1394–1399
Waterhouse PM, Helliwell CA (2003) Exploring plant genomes by RNA-induced gene silencing. Nat Rev Genet 4:29–38
Watson JM, Fusaro AF, Wang MB, Waterhouse PM (2005) RNA silencing platforms in plants. FEBS Lett 579:5982–5987
Xin HW, Ji LH, Scott SW, Symons RH, Ding SH (1998) Ilarviruses encode a Cucumovirus-like 2b gene that is absent in other genera within the Bromoviridae. J Virol 72(8):6956–6959
Yoshikawa M, Iki T, Tsutsui Y, Miyashita K, Poethig RS, Habu Y, Ishikawa M (2013) 3′ fragment of miR173 programmed RISC cleaved RNA is protected from degradation in a complex with RISC and SGS3. Proc Natl Acad Sci U S A 110:4117–4122
Zagrai I, Capote N, Ravelonandro M, Cambra M, Zagrai L, Scorza R (2008) Plum Pox Virus silencing of c5 transgenic plums is stable under challenge inoculation with heterologous viruses. PT J Plant Pathol 90(1):S1.63–S1.71
Zagrai I, Ravelonandro M, Zagrai L, Scorza R, Minoiu N (2011) Overview of the investigations of transgenic plums in Romania. Acta Horticult ISHS 899:153–158
Zhang C, Ghabrial SA (2006) Development of Bean Pod Mottle Virus based vectors for stable protein expression and sequence-specific virus-induced gene silencing in soybean. Virology 344:401–411
Acknowledgements
The authors are thankful to the Science and Engineering Research Board – Department of Science and Technology, New Delhi, India, for the financial assistance (File No. YSS/2015/000265) and also to the University Grants Commission, New Delhi, for providing financial assistantship under Research Award for Teacher (F.30-1/2014/RA-2014-16-GE-RAJ-4696 (SA-II).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Marwal, A., Gaur, R.K. (2017). Understanding Functional Genomics of PTGS Silencing Mechanisms for Tobacco Streak Virus and Other Ilarviruses Mediated by RNAi and VIGS. In: Singh, D., Singh, H., Prabha, R. (eds) Plant-Microbe Interactions in Agro-Ecological Perspectives. Springer, Singapore. https://doi.org/10.1007/978-981-10-5813-4_24
Download citation
DOI: https://doi.org/10.1007/978-981-10-5813-4_24
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-5812-7
Online ISBN: 978-981-10-5813-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)