Abstract
The genus, Babuvirus of the family Nanoviridae contains three virus species, Abaca bunchy top virus (ABTV), Banana bunchy top virus (BBTV) and Cardamom bushy dwarf virus (CBDV). In India, only two babuviruses, BBTV and CBDV are known to affect banana and large cardamom, respectively. BBTV, which causes bunchy top disease in banana, is a nationally important virus as it is widely prevalent in all the banana growing states including North-East region. Whereas, CBDV, which causes foorkey disease of large cardamom, is of regional importance as it is restricted only in the North-Eastern sub-Himalayan mountains. Early infection of these babuviruses cause 100% yield loss. Bunchy top of banana and foorkey disease of large cardamom are known in India for a long time and considerable information has been generated. This chapter summarises the biological and molecular properties of both the babuviruses occurring in India.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Multi-component ssDNA containing small isometric plant viruses were initially classified under the genus Nanovirus. The legume infecting nanoviruses being substantially different from banana bunchy top virus (BBTV), the nanoviruses were reclassified in the eighth report of International Committee on Taxonomy of Viruses (ICTV) where a family, Nanoviridae containing two genera, Nanovirus and Babuvirus were created (Vetten et al. 2005). The genus Nanovirus, included all the legume infecting nanoviruses and the genus Babuvirus included only, BBTV. Later, two more babuviruses were discovered, Abaca bunchy top virus (ABTV) from Philippines (Sharman et al. 2008) and Cardamom bushy dwarf virus (CBDV) from India (Mandal et al. 2013).
Foorkey disease of large cardamom was the first babuvirus associated disease known in India, which was recorded in 1935 in Darjeeling hills. Bunchy top disease of banana, although was known in Oceania region in early twentieth century, it was recorded in India during 1940s. In India, only two babuviruses so far has been identified, BBTV causing banana bunchy top disease and CBDV causing foorkey disease of large cardamom. Banana is one of the most important fruit crop cultivated throughout India. BBTV has been characterised from at least from three geographic regions of India (Vishnoi et al. 2009; Banerjee et al. 2014; Selvarajan et al. 2010). The wide spread occurrence and serious yield limiting ability of BBTV made it to be one of the top most important phytopathogens in India. The other babuvirus, CBDV that caused foorkey disease of large cardamom is prevalent only in the North-East sub-Himalayan mountains. Large cardamom is an important spice plantation crop grown under the special organic environmental conditions prevailing in the North-East sub-Himalayan mountains. CBDV infection results into complete loss of large cardamom clump. As CBDV is prevalent in a particular agro-ecological area of the country, it is of regional significance. Babuviruses were known to naturally infect the plant species of the family Musaceae. The current research work on CBDV provided the evidence of a new babuvirus species that infects plant species of the family Zingiberaceae. CBDV is the only member of the family Nanoviridae that was discovered in India. A few review papers are available for the nanoviruses and babuviruses (Mandal 2010; Selvarajan 2015). As both the babuviruses, BBTV and CBDV are economically significant viral pathogens in India, a large body of literature has been generated. This chapter presents the work on the biological and molecular characterization of these babuviruses occurring in India.
2 Banana Bunchy Top Virus
2.1 Occurrence and Significance
Banana bunchy top disease (BBTD) caused by BBTV was first recorded in Fiji in 1889. It is believed that BBTD spread to Australia and Sri Lanka in 1913 through infected suckers from Fiji and later during 1940’s it was presumed to be introduced to Kerala state of India (Jones 2000). Upon gaining entry in the southern tip of India, the virus has spread to all the banana growing states of the country. BBTD remains a major problem in Kerala, Andhra Pradesh, Tamil Nadu, Orissa, Maharashtra, Madhya Pradesh, Gujarat, Bihar, Karnataka, West Bengal, Assam and Uttar Pradesh (Singh 2003). It has also been recorded in wild and cultivated bananas in Nagaland, Meghalaya, Arunachal Pradesh, Mizoram, Sikkim and Tripura.
Of all the viruses known to infect banana in India, BBTV is the most serious and destructive virus. In lower Pulney hills of Tamil Nadu, a very famous elite dessert banana cultivar, Virupakshi (Pome group, AAB) having unique flavor and distinct aroma, has been near extinct due to BBTV since 1970’s and the area under this banana has been reduced from 18,000 ha to 2000 ha (Kesavamoorthy 1980). A survey conducted during May 2009 in Lower Pulney hills (Kodaikanal) recorded 15.26–83.88% incidence in Hill banana across the plantations (Selvarajan et al. 2011). Metha et al. (1964) reported a loss of about ₹ 40 million annually reported in Kerala. Emergence of BBTV in tissue culture plantations during 2007–2011 in Jalgaon, Maharashtra and Kodur, Andhra Pradesh caused an annual loss of production worth of US$50 million (Selvarajan and Balasubramanian 2014). An outbreak of BBTD in 2011 in Theni district of Tamil Nadu recorded an infection in 0.3 million plants of both tissue culture and conventional sucker grown plants. As BBTD causes significant yield reduction and reduces the productivity in India and other parts of the world, it is considered as one of the most destructive diseases of banana in the world.
3 Biology of BBTV
3.1 Disease Symptom and Dissemination
BBTV infected plants express discontinuous dark green flecks and streaks of variable length on leaf sheath, midrib, leaf veins and petioles of infected plants (Fig. 3.1). Leaves that are produced after the infection are progressively shorter both in width and length with limited elongation of petioles and remain abnormally erect (Sharma 1988). Infected leaves are narrow and brittle in texture, display marginal yellowing or chlorosis and leaves bunches at the top, hence the name is “bunchy top” disease (Fig. 3.1). BBTV occurs in the phloem tissues of banana and incites symptoms such as leaf chlorosis, vein clearing, dwarfing and leaf atrophy (Wu and Su 1990; Su et al. 2007). Mostly, BBTV infected plants fail to produce bunch, however, in late infections the plant may produce bunch but the fingers are malformed and not fit for sale. Occasionally, bracts of male flower bud turn to leafy green structures and exhibit dark green dots and streaks (Thomas et al. 1994). Any daughter suckers emerging from infected plant exhibit severe symptoms. Late infection of BBTV in cultivar Grand Naine banana leads to throw bunch with extremely long or very short peduncle. Sometimes affected Grand Naine banana fingers appear like a non Cavendish type.
Disease symptom of BBTV. (a) Field view of hill banana cultivation in lower pulney hills. (Insert) Severe symptom of BBTD; (b) Vein flecking on leaf lamina; (c) Dark green dots and streaks on petiole; (d) Chlorosis; (e) Greenish leafy tips in male bud of BBTV infected Cavendish banana plant; (f) Aphids vector – Pentalonia nigronervosa
In BBTV infection, the virus resides in the plant without exhibiting any visible symptom which is termed as latency. BBTV has been found to express visual symptoms only 23–25 days after inoculation but the virus could be detected early from young roots or cortex tissue even before the symptom expression (Hafner et al. 1995). The shortest time for the diagnosis of BBTV using polymerase chain reaction (PCR) is reported as 15 days after infection (Hooks et al. 2008). Sometimes, the infected plants (PCR positive) do not show any symptoms even up to 560 days under pot culture experiment (R. Selvarajan, unpublished). Samraj et al. (1970) have reported that a minimum time of 5 days and maximum 10–15 days is required for the down ward movement of the virus after inoculation with the aphid and this might change depending upon the vigour of the plant. A short, but significant, latent period of 20–28 h is required for vector transmission (Anhalt and Almeida 2008) and it has been found that the optimum temperature range for acquisition of virus by the vector was 25–27°C.
The only confirmed hosts of BBTV are species within the genus Musa (M. balbisiana) (Espino et al. 1993; Magee 1927), M. ornate (Thomas and Dietzgen 1991), M. acuminate ssp zebrina, M. velutina, M. coccinia (Thomas and Iskra-Caruana 2000), M. sinensis, M. paradisica and their hybrids and Fei’ bananas and Ensete ventricosum (Selvarajan and Balasubramanian 2013). This disease has been observed on a wild species, Musa itenerans that occurs as a feral in hills of Arunachal Pradesh (Selvarajan et al. 2010). Colocasia esculenta has been reported to be a host for BBTV from Pune (Ram and Summanwar 1984) but it was disproved later (Geering and Thomas 1997; Hu et al. 1996).
BBTV is primarily spread through the use of infected vegetative propagules, including the suckers, corms, (Magee 1948) and tissue-cultured plants (Drew et al. 1989). Drew et al. (1989) have demonstrated that BBTD is transmitted through micro propagated plantlets established from infected plants. When these tissue culture plantlets were established in the greenhouse, only 73% of plant developed characteristic symptoms and the remaining 27% of plants appeared healthy without typical symptoms of the BBTD. BBTV is secondarily, naturally transmitted by the banana black aphid vector, Pentalonia nigronervosa (Hemiptera, Aphididae) (Fig. 3.1) in a persistent circulative manner (Anhalt and Almeida 2008; Selvarajan et al. 2006). Recently, another closely related species, P. caladii, has been shown to transmit BBTV under experimental conditions with a lower level of efficiency than P. nigronervosa (Watanabe et al. 2013).
The aphids are usually found clustered around the unfurled heart-leaf and the sheathing leaf base of petioles which are ideal locations for feeding and protection. They are also found on the base of the pseudostem and on very young suckers. Menon and Christudas (1967) reported the life history and population dynamics of Pentalonia nigronervosa in Kerala, India and stated that climatic conditions existing during the summer months and rainy weather are unfavourable for the banana aphid in Kerala.
4 Genomic Properties of BBTV in India
BBTV genome consists of six circular single stranded DNA individually packed in six particles (Table 3.1). The genome of each component is about 1.1 kbp in size and each encodes for a single open reading frame (ORF) for different functional proteins from the virion sense strand such as DNA-R (replication initiation protein), -S (coat protein), -M (movement protein), -C (cell cycle link protein), -N (nuclear shuttle protein), and -U3 (a protein of unknown function) (Vetten et al. 2012).
In India, complete genome of three BBTV isolates, one each from Lucknow (UP), Bhagalpur (Bihar) and Lower Pulney Hills (TN) have been sequenced and characterised (Fig. 3.2) and found that they belong to Pacific-Indian Oceans (PIO) group (Islam et al. 2010; Selvarajan et al. 2010; Vishnoi et al. 2009). Complete genome sequences of these Indian isolates showed high degree of similarity with the corresponding sequences of BBTV isolates originating from Fiji, Egypt, Pakistan, and Australia. An analysis of the coat protein sequences of 16 Indian isolates with distinct geographical origins revealed that they belong to the PIO group, except the isolates from Shevroy and Kodaikanal hills of Tamil Nadu (Selvarajan et al. 2010). Recently a novel BBTV isolate from Umiam (Meghalaya) which has a deletion of about 20 nucleotides in the DNA-R component has been characterized (Banerjee et al. 2014).
Organization of the genomic components of the BBTV-Hill banana isolate. The DNA component- R, -U, -S, -M, -C and -N are illustrated diagrammatically. The positions and orientations of genes are indicated with shaded arrows. Also the positions of the stem loop common region (CR-SL), consensus TATA box and polyadenylation signal sequences are shown
5 Sequence Diversity
Earlier, BBTV was grouped into the South Pacific group comprising isolates from Australia, Burundi, Egypt, Fiji, India, Tonga and Western Samoa and the Asian group, comprising isolates of Vietnam, Philippines and Taiwan based on sequence analysis of BBTV DNA-R, -S and -N (Karan et al. 1994, 1997; Wanitchakorn et al. 2000). Karan et al. (1994) determined the maximum variability of South Pacific isolates to be 3.8% with a mean of 1.9%, whereas the value for Asian isolates was found to be 4.2% with a mean of 3% for DNA-R components. The two groups of BBTV differ by an average of 90.6% (DNA-R), 11.86% (DNA-S) and 14.5% (DNA-N) over the entire nucleotide sequence with an average difference of 32% (DNA-R), 38.6% (DNA-S) and 27% (DNA-N) within CR-M (Karan et al. 1994, 1997; Wanitchakorn et al. 2000). A more recent analysis, based on a much larger sample set, determined the mean variation to be approximately 1.6% and 2.9% for South Pacific and Asian isolates respectively (Hu et al. 2007). The genetic diversity of BBTV isolates among countries is very low, viz., India (except isolates from north eastern region) (Vishnoi et al. 2009; Islam et al. 2010; Selvarajan et al. 2010), Pakistan (Amin et al. 2008), Africa (Niyongere et al. 2013, 2015), and Oceania (Stainton et al. 2012). However, in India, relatively a greater diversity for BBTV was observed in the North-eastern region (Banerjee et al. 2014). Recently, based on the phylogenetic relationships among the DNA-R component sequences, various BBTV isolates were grouped into two different lineages: (i) the Pacific-Indian Oceans (PIO) group (formerly South Pacific group) comprising isolates in Africa, Australia, Hawaii, south Asia, Myanmar, and Tonga; and (ii) the South-East Asian (SEA) group (formerly Asian group) comprising isolates from China, Indonesia, Japan, the Philippines, Taiwan, and Vietnam (Fig. 3.3) (Stainton et al. 2012; Yu et al. 2012; Banerjee et al. 2014; Kumar et al. 2015). Motif-based analysis revealed that several unusual recombination events occurred and those events have contributed to the evolution of BBTV genome components (Wang et al. 2013).
6 BBTD in North-East (NE) India
The North-eastern region of India, comprising eight states viz., Assam, Arunachal Pradesh, Meghalaya, Manipur, Mizoram, Nagaland, Tripura and Sikkim, represents a distinct agro-climatic zone of the country. The NE India is a bio-diversity hotspot and possesses diverse germplasm of banana of both wild and cultivated. However, little was known about BBTV occurring in the NE India, except the preliminary information on BBTV coat protein (Selvarajan et al. 2010). Recently, a new isolate of the virus (BBTV-Umiam) was identified and characterized from banana growing in mid-hills of Meghalaya in NE India (Fig. 3.3) (Banerjee et al. 2014). The overall genome organization of BBTV-Umaim was mostly identical with previously reported isolates from India except having some distinct features viz., deletion of 20 nucleotides in the intergenic region of DNA R, absence of predicted ORF in DNA U3 and probability for a small ORF in DNA U3. The BBTV-Umiam is supposed to be a PIO group member due to its geographical origin (Umiam, Meghalaya, India). The earlier reported Indian BBTV isolates always grouped within PIO cluster instead of SEA cluster indicating the possible introduction of BBTV into southern India from Australia through Fiji and Srilanka by infected planting materials (Wardlaw 1972). Similarly, in phylogenetic grouping the BBTV isolate from Meghalaya (BBTV-Umiam) grouped within PIO cluster sharing 95.5% nucleotide sequence identity (Fig. 3.4). However, all the BBTV isolates from plains of India clustered together suggesting their separate and independent evolution, but BBTV-Umiam was found to be the most distinct member among the PIO isolates identified so far (Fig. 3.4). Although, overall sequence analysis of genomic components, as well as, ORFs clearly indicated strong similarity of BBTV-Umiam with PIO group, but BBTV-Umiam shared relatively less nucleotide identity with PIO group for each genomic component (85.0–95.4%) and corresponding ORFs (93.8–97.5%) than that of earlier PIO isolates (91.5–99.6% and 96.0–99.3%, respectively) (Banerjee et al. 2014). Recombination analysis revealed two intra-component (in DNA U3 and DNA N) and five inter-component recombination events [DNA U3 (1), DNA S (2), DNA M (1) and DNA N (1)] around CR-M and CR-SL region of BBTV-Umiam, but none of them was unique (Banerjee et al. 2014). The BBTV DNA U3 being the most recombined one (Hyder et al. 2011; Stainton et al. 2012) showed similar trend in BBTV-Umiam, while an event involving the transfer of a DNA N fragment from PIO group virus resembling those found in India, Pakistan and Australia into the progenitor of a group of viruses from Tonga was reported earlier (Stainton et al. 2012). Earlier workers indicated CR-M region as a major recombination hotspot and CR-SL region as a minor hotspot (Stainton et al. 2012). Though in case of BBTV-Umiam, maximum inter-component recombination events were identified around CR-SL (Banerjee et al. 2014). Moreover, recombination had no role in the deletion of 20 nucleotides in the intergenic region of BBTV-Umiam DNA R. Generally, in plant pararetroviruses (Caulimovirus), this kind of natural deletion is thought to be the outcome of recombination (Howarth et al. 1981). Thus, the genetic distinctness of BBTV-Umiam was not the outcome of genetic cross-over. On the other hand, all the PIO DNA R components were reported to have descended from a common recombinant ancestor deriving a proportion of its Rep encoding sequence from an unknown babuvirus (Stainton et al. 2012). Banerjee et al. (2014) provided strong evidence for considering BBTV-Umiam (KC119098) and BBTV-HaiKou-4 (HQ378190) as the hypothetical major and minor parental sequences, respectively for intra-component recombination of DNA R of all Tonga isolates of BBTV (JF957625-JF957636) (Fig. 3.5).
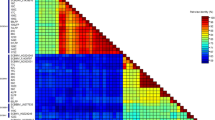
Phylogenetic relationships based on nucleotide sequences of DNA-R of BBTV-Umiam with previously reported PIO and SEA isolates of BBTV. The evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (shown only when > 50%). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the ‘Maximum Composite Likelihood’ method and are in the units of the number of base substitutions per site. Each sequence is labelled with the GenBank accession number followed by origin and isolate name
Intra-component recombination analysis of BBTV DNA-R of Tonga isolates. (a) Phylogenetic tree of nucleotide sequence of the recombined region showing clustering of recombinant isolates JF957625-JF957636 (Tonga isolates) with HQ378190 (BBTV-HaiKou-4); (b) Bootstrap support plot showing a graphical overview of the recombination event in JF957625 (the representative Tonga isolate) at position 150–373 nucleotide. The recombinant isolate shows similarity to KC119098 (BBTV-Umiam, major parent) throughout the entire genome but shares very high identity with HQ378190 (BBTV-HaiKou-4, minor parent) only in the recombined region; (c) Phylogenetic tree of the nucleotide sequence of the non-recombined region showing clustering of Tonga isolates with KC119098 (BBTV-Umiam).
Further studies has confirmed occurrence of BBTV in Assam, Arunachal Pradesh, Nagaland, Manipur, Mizoram, Sikkim and Tripura including samples from commercial orchards, road side banana mats and even in tissue culture raised plant materials (Banerjee et al. 2015). Altogether, ten BBTV isolates distributed throughout the surveyed areas were characterized based on DNA R segment. The full DNA R sequences of each isolate except the isolate from Mizoram shared >97.0% similarity with BBTV isolates reported from plains of India. However, these isolates showed relatively less similarity (~95.0%) with BBTV-Umiam. Interestingly, the Mizoram isolate shared only 91.0–92.0% similarity with both PIO and SEA group members. While, during phylogenetic analysis the Mizoram isolate including other isolates from NE India clustered within PIO group, but the clustering pattern indicated the distinctiveness of Mizoram isolate as of previously reported BBTV-Umiam from Meghalaya (Banerjee et al. 2015). Recent survey in seven districts of Tripura viz., North Tripura, Dhalai, Khowai, West Tripura, Shipahijala, Gomati, and South Tripura showed prevalence of BBTV in all parts of Tripura. Phylogenetic analysis based on complete nucleotide sequence of BBTV DNA R confirmed Tripura isolates of BBTV as PIO group members. However, Tripura isolates formed two different clades within the PIO group. Tripura isolates showed on an average 10.19% variation with SEA group and 2.79% variation with PIO groups. Moreover, Tripura isolates were more identical with the isolates reported from plains of India rather than the distinct isolate reported from Meghalaya (BBTV-Umiam). Thus, the source of planting material from plains of India, as well as, from neighboring countries could have contributed to the geographical expansion of PIO isolates of BBTV within NE India. However, the existence of distinct PIO isolates in naturally growing banana mats of Meghalaya and Mizoram further strengthened the possibility of differential evolution of BBTV in this isolated region. The natural occurrence of hybrids of Musa balbasiana and M. acuminate in this geographically isolated region could be the contributing factor in accumulating genetic distinctiveness in BBTV-Umiam which needs further characterization.
7 Large Cardamom Bushy Dwarf Virus
Large cardamom (Amomum subulatum), a perennial herb belongs to the family Zingiberaceae, order Scitaminae, is an important spice crop of India. Large cardamom is cultivated in the eastern sub-Himalayan mountains at altitudes ranging between 800–3000 m above mean sea level under of the forest cover conditions. Sikkim and Darjeeling hills of West Bengal have the major areas (30,000 ha) under large cardamom cultivation. Usually, the large cardamom plantations are established with seedlings or suckers (tiller with roots separated from rhizome) or tissue culture raised plants and the plantations are maintained for many years. The plant produces large red or black coloured aromatic capsules at the base near rhizome. The dried capsules containing seeds are used for flavoring food and preparing medicine. The capsule size, colour and aroma are distinct from small cardamom. Viral diseases known as ‘chirke’ and ‘foorkey’ are the major limiting factors in cultivation of large cardamom (Varma and Capoor 1964). The virus associated with the foorkey disease has been identified as cardamom bushy dwarf virus (CBDV), a new babuvirus so far recorded only in India.
7.1 The Foorkey Disease
Foorkey is one of the earliest known viral diseases in India, which was first documented in 1936 from Darjeeling hills (Mitra 1936). In Nepalese language, foorkey means bushy dwarf. The typical symptoms appear in the newly developed tillers from the infected rhizome. Initially, the affected tillers grow to the height up to 6–12 in. with slight bunchy top appearance and sometimes leaves are not unfurled (Fig. 3.6). Subsequently, numerous stunted plantlets of about 2–3 in. with small pale green leave proliferate from the rhizome giving a bushy dwarf appearance at the base. The CBDV infected clumps survive for a few years but the clumps become sterile and unproductive. The recognition of initial disease symptoms of foorkey under field conditions is difficult, however, when the symptoms are fully expressed, it is easy to spot in the plantation. Most of the commonly grown cultivars such as Golsey, Ramsey, Sawaney and Varlangey are susceptible. The disease incidence varies from place to place and higher incidence is generally observed in lower altitudes ranging from 300 to 1380 m above MSL. The high incidence of foorkey up to 39.3% was observed in Kooldhara and Khaptali Gaon in Darjeeling hills (Mandal et al. 2013).
Symptoms of foorkey disease of large cardamom cv. Varlangey. (a) A healthy clump bearing flower; (b) Large cardamom spike and capsules from healthy and diseased plants; (c) A clump showing initial symptoms of shorter lateral pseudostems containing pale yellow leaves; (d) The emerging leaves from infected plants are unfurled and twisted; (e) Excessive stunted shoots developing in a clump; Proliferation of stunted shoots giving a bushy appearance of a clump
The virus associated can be readily detected by PCR or dot-blot hybridization using the primers or probe from the replication associated protein gene (Rep) of DNA-R. PCR is most effective in detecting the CBDV. The primer pair AV5F: tggcgcgatatgtggtatgc and AV6R: tcagcaagaaaccaactttattc derived from the Rep gene amplify a ~ 0.84 kb DNA fragment. The PCR successfully detected the virus in the different plant parts of large cardamom viz., leaf, leaf sheath, meristem, stem, root and mother rhizome. Another primer pair from stem and loop (S&L) region of the Rep genome component of the virus (AV32F: ggggcttattattacccccagcg and AV33R: agcgcttacgtggcgcactaact) amplified 1.1 kb fragment. The Rep or S&L primers are useful tools for the monitoring of the occurrence of the virus by PCR.
7.2 Resolving the Etiology of Foorkey
The existence of foorkey disease was known in large cardamom for a long time, however, the virus associated with the disease was not known. Foorkey was considered as a viral disease based on the fact that the disease could be transmitted through an aphid vector. The virus identity was elusive because no virus particle could be readily observed in transmission electron microscopy. However, a few isometric virus particles of 17–20 nm were observed in the partially purified preparation as well as in some field samples collected from Kalimpong during 2003–2004. During this time, BBTV was considered as a member of the genus Nanovirus, containing the similar virion morphology to that observed in the foorkey samples. Therefore, the observation of the virion morphology was an important clue that directed to further examination of foorkey samples by ELISA and PCR with the antiserum and primer of BBTV, respectively. The ELISA showed weak serological relationship. The sequence (859 nucleotides) generated from the PCR product showed 82% sequence identity with the DNA-R of BBTV and 47.6–48.5% identity with the other nanoviruses. The previous studies showed that the virus associated with the foorkey disease was different from BBTV by the fact that it did not infect banana and was vectored by a different aphid species (Basu and Ganguly 1968; Varma and Capoor 1964). The serological and partial genome sequence information further provided evidence of existence of a new nanovirus, which was named as CBDV (Mandal et al. 2004).
7.3 Mode of Spread
7.3.1 Through Planting Materials
CBDV is not known to spread through seed or contact of the infected plant materials. In the experimental conditions, the virus is also not transmitted through sap inoculation. The most important mode of spread of the virus is through vegetative propagative materials derived through the infected clump. The suckers, which are commonly used for raising new plantation, are the initial source of CBDV as it is difficult to judge the initial infection by the visual observation of clump. Tissue culture planting materials also can potentially circulate the virus if the plantlets are derived from the infected clump.
7.3.2 Through the Aphid Vector
The banana aphid, Pentalonia nigronervosa was initially reported as vector of CBDV (Varma and Capoor 1964). Later, Basu and Ganguly (1968) published a brief note that the aphid, Mycromyzus kalimpongensis transmitted CBDV but not P. nigronervosa. As both the studies reported contradictory findings, it was necessary to confirm the findings. Further, there was no studies on the natural occurrence of the aphid species on large cardamom as well as the role of aphid vector in natural dissemination of CBDV. The study was initiated during 2012–2015 in the Regional Station of IARI, Kalimpong to understand the temporal occurrence of aphid species on large cardamom in Darjeeling and Sikkim hills (Ghosh et al. 2016a). This study for the first time documented the natural occurrence and seasonal dynamics of three aphid species, M. kalimpongensis, P. nigronervosa and Aulacorthum solani on large cardamom. The abundance and colonization habits of these aphids on large cardamom are different and it was demonstrated that only M. kalimpongensis, which persists in the plantation throughout the year and colonises mainly in the roots and other underground parts of large cardamom, could transmit CBDV. This study (Ghosh et al. 2016a) further confirmed M. kalimpongensis but not P. nigronervosa is the vector of CBDV. In 2012, while surveying a large cardamom plantation in the Darjeeling hill, an interesting observation was encountered that the foorkey affected clumps contained the higher number of M. kalimpongensis in the underground plant parts compared to that in the healthy clumps. Further surveys confirmed the consistence association of M. kalimpongensis with the foorkey affected plants irrespective of seasons, altitudes and large cardamom cultivars. This specific behavior of M. kalimpongensis was studied in the laboratory and contained field experiments, which revealed that the infection of large cardamom by CBDV influences the aphid to migrate the infected plants (Ghosh et al. 2016b). In the contained field experiment, it was observed that the aphids colonizing on the infected plants had reduced nymphal period and increased longevity and fecundity compared to those grown on the healthy plants. Therefore, CBDV infected plants facilitates emergence of more number of M. kalimpongensis. When, foorkey affected plants gradually dries, the viruliferous aphids migrate to the nearby clumps and eventually spread CBDV. This study suggests a general pattern of dissemination of CBDV by M. kalimpongensis in the plantation, where CBDV infected plants attract and stimulate emergence of more viruliferous aphids. The most of the members of the family Nanoviridae are vectored by aphids, but, the alteration of behavior of aphid vector by the virus infected plants that favors its dissemination of the virus was first demonstrated through the interaction of CBDV, large cardamom and M. kalimpongensis (Ghosh et al. 2016b).
CBDV appears to be naturally transmitted through aphid from large cardamom to large cardamom as there is no alternate host of CBDV is known so far. CBDV could not be transmitted from large cardamom to Musa sapientum, Gladiolus sp, Canna indica, Zingiber officinale, Triticum aestivum, Sorghum vulgare and Zea mays (Varma and Capoor 1964). The only experimental host of CBDV known is small cardamom (Elettaria cardamom), which although is not grown in the area where large cardamom is cultivated.
7.4 Molecular Properties of CBDV
7.4.1 Cloning of the Genome Components
CBDV was considered as a new member of the genus Babuvirus based on the distinct sequence of Rep gene of DNA-R genome component. In order to establish it as a distinct virus species, Mandal et al. (2013) cloned and analysed the complete set of genome components that established CBDV as the third distinct species of the genus Babuvirus, where the other two species were BBTV and ABTV. DNA-R was the first genome component that was cloned by designing a pair of abutting primers based on the sequence of the Rep gene of CBDV. Babuviruses are known to contain six DNA components eg., DNA-R, -S, -M, -N, -C and U3. Therefore, CBDV was also expected to contain similar set of genomic components. The each genome components of the members of the family Nanoviridae have common features containing a coding region of a single protein and a non-coding region containing a stem and loop (SL) structure and major common region (CR-M), TATA box and poly-A signal. The SL structure contains the nonanucleotides is highly conserved among the genome components. A pair of abutting primer (AV32F&AV33R) designed from the SL region of the DNA-R, was expected to amplify the other genome components of CBDV. However, this approach resulted in cloning only DNA-S, DNA-M and an unknown component (DNA-U1). To obtain the DNA-N and -C components, partial genome fragment was amplified based on the primers designed based on the respective components of BBTV and ABTV and the specific abutting primers from this partial sequence resulted in amplification and cloning of the full-length DNA. Further the rolling circle amplification (RCA) followed by restriction digestion resulted in several putative clones with unit-length inserts. Sequencing of these RCA clones led to identify the satellite Rep component (Sat-Rep), DNA-U2 and DNA-U3.
7.4.2 Major Genome Components
The sequence analysis of all the clones obtained through PCR and RCA revealed existence of six DNA components (DNA-R, -S, -M, -N, -C and U3), which in the analogy of the BBTV and ABTV, were considered as the integral components of CBDV (Fig. 3.7, Table 3.1). In addition, three more DNA components (U1, U2 and Sat-Rep) that were associated with the foorkey affected samples were also identified. Association of these novel nine DNA components with the foorkey disease established CBDV as a new virus species of the genus Babuvirus family Nanoviridae (Mandal et al. 2013).
Genome organization of Cardamom bushy dwarf virus. DNA-R encoding replication initiation protein, DNA-S encoding coat protein, DNA-M encoding movement protein, DNA-N encoding nuclear shuttle protein, DNA-C encoding cell cycle link protein. ORF open reading frame, SL stem and loop structure, CR-SL stem-loop common region, CR-M major common region. Nucleotide count was shown from the beginning of 5′ region of SL (Adopted from Mandal et al. 2013)
The CBDV DNA components were composed of 1079–1134 nucleotides. The sequence comparison between the DNA components showed the CR-SL was 49–70 nucleotides, which contained the nonanucleotide, TATTATTAC in the loop region. The sequence in the loop of SL region was slightly different among the DNA components. DNA-R, -S and -M were different from DNA-N, -C, -U3 and -U2 by AC in place of CT. The CR-SL of CBDV genome components was highly different sharing only 46.3–97.1% sequence identity with BBTV and ABTV. The iteron sequences, R (GTCCC), F1 (GGGAC) and F2 (GGAAC) adjacent to the stem region were present in all the components of CBDV. The CR-M was 73–80 nucleotides long sharing 55.9–86.0% similarities among the components and contained two highly conserved regions, the 17 nt GC rich identical sequence (AAGGGCCGAAGGCCCGT) and the near identical 17 nt sequence (CGC/AAA/CTTAT/A/CGACCTGTC). The CR-M of CBDV is also highly divergent from BBTV and ABTV with only 18.8–56.0% sequence identity.
The DNA-R contained 1102 nucleotides that encoded a major protein of 33.6 kDa, potentially associated with replication (Rep). In addition, a small ORF encoding a 5.1 kDa protein of unknown function was present within the Rep ORF. The Rep protein of CBDV shared the closest sequence identity of 85.3–87.7% with the master-Rep (m-Rep) of BBTV and ABTV, whereas 53.8–56.9% with the other species of the member of the family Nanoviridae. The DNA-S contains 570–571 nucleotides intergenic region and 513 nucleotides region that encodes a 19.5 kDa capsid protein. CBDV shares a closer sequence identity (80.5%) with ABTV than BBTV (75.8%) in the amino acids sequence of capsid protein. The DNA-M component encodes 13.49 kDa movement protein, which is highly different from that of BBTV and CBDV (59.8–66.6%). The DNA-N genome encodes 18.2 kDa nuclear shuttle protein (NSP). Interestingly, a TATA box is located 57 nt after the initiation codon of the NSP ORF. The NSP of CBDV is distantly related to that of both ABTV and BBTV with 68.3–69.0% amino acid sequence identity.
The DNA-C component encodes a 18.5 kDa cell cycle linking protein (Clink) that contains retinoblastoma like-binding motif LFCDE (LXCXE). The DNA-U3 contained CR-SL and CR-M structures similar to that in the other major components but do not contain any significant ORF. The U3 of CBDV shared low sequence similarities (43.8–45.1%) with BBTV and ABTV.
7.4.3 Satellite Component
A Sat-Rep DNA component was isolated through RCA from the foorkey affected sample. Sat-Rep is composed of 1134 nucleotides, which is 6 nucleotides shorter than DNA-R. The DNA contains a major ORF of 855 nucleotides encoding 33kDa protein. The dNTP binding motif, GNEGKS, which is present in the Sat-Rep protein, is different from that in DNA-R of CBDV. The nucleotide sequence length of loop and stem and the nonanucleotides of Sat-Rep are different from that of DNA-R of CBDV. The nucleotide sequence of Sat-Rep is significantly different (42.1–43.5%) from DNA-R of babuvirus including CBDV.
7.4.4 The Other Unknown Components
In addition to U3 genome component, two other unknown components, U1 (1080 nucleotides) and U2 (1078 nucleotides) were isolated from foorkey affected plant. None of these DNA contains any major ORF. These unknown components, however, contained CR-SL and CR-M as found in the major genome components of CBDV. The SL structure of these components is almost identical to the DNA-R, -S and -M components. The BLAST search showed no sequence similarity in the region other than CR-SL and CR-M with the members of Nanoviridae in the database.
7.4.5 Phylogenetic Relationships and Diversity in CBDV
The mean genetic distance of CBDV of six major genome components is 38.0–40.2% from the other two babuviruses, BBTV and ABTV. Phylogenetically, CBDV shows distinct divergence from the other two babuviruses. There is a variation in phylogenetic relationships with reference to the DNA components of CBDV. DNA-R, -N and -U3 showed closer phylogenetic relatedness with BBTV, DNA-M and -S with ABTV, whereas DNA-C is highly different from both the babuviruses. The Sat-Rep component of CBDV shares a closer phylogenetic relations with S1 satellite DNA of BBTV. The unknown components, U1 and U2 of CBDV are distinctly related from U3 of ABTV, BBTV and CBDV.
CBDV having multipartite genomes may lead to reassortment of whereby entire genome components of different strains. Savory and Ramakrishnan (2014) have extensively analyzed the reassortment among 163 CBDV isolates collected in North-East India. They have showed the evidence of recombination, which might have played a role in the evolutionary dynamics of populations. By sequencing six discrete genome components for each isolate, they demonstrated that over 40% of the isolates displayed evidence of at least one reassortment event during their evolutionary histories. They also observed that DNA-M and DNA-N components are the most predisposed to reassortment. The comparisons of the common regions of different genome components revealed signatures of concerted evolution mediated by frequent inter-component homologous recombinations.
8 Concluding Remarks
BBTV is internationally and nationally a serious threat to banana and plantains and has been characterized at molecular level in India. CBDV is restricted to large cardamom growing regions of Sikkim and West Bengal. The genomic properties of BBTV and CBDV have been studied well and so far a total of 1325 accession numbers (324 for BBTV and 1001 for CBDV) have been contributed to the GenBank database from India (Fig. 3.8). However, the infectivity of the cloned DNA components is required to confirm that these genome components are necessary to cause bunchy top and foorkey disease, respectively in these crop species. So far, infectivity of the cloned genome components has been demonstrated only for Faba bean necrotic yellows virus (FBNYV), a, nanovirus, but not for any babuviruses (Mandal 2010). The genome of babuviruses (BBTV, ABTV and CBDV) is composed of six DNA components, DNA-R, -S, -M, -N, -C and U3 (Mandal et al. 2004; Vetten et al. 2012). The DNA-U3 is considered as an integral part of the babuvirus genome, but CBDV U3 did not contain any major ORF as in case of the other two babuviruses. However, some of the BBTV isolates were shown to contain a small ORF in U3 genome encoding 9–10 kDa protein (Beetham et al. 1999; Vishnoi et al. 2009). Two additional unknown components (U1 and U2) were found in CBDV affected large cardamom, which did not contain any major protein coding sequence. The virus associated with Cardamom bushy dwarf disease (CBDD) is biologically distinct from BBTV that its vector is M. kalimpongensis (Basu and Ganguly 1968) whereas banana black aphid, Pentalonia nigronervosa transmits BBTV and banana is a non-host for CBDV (Varma and Capoor 1964). Serologically, CBDV is different from BBTV (Mandal et al. 2004). Nine novel genomic components of CBDV are associated with the CBDD of large cardamom whereas only eight components including two satellite DNA’s are associated with BBTV. The overall genome sequence identity and phylogeny showed evolutionary divergence of CBDV from the existing members within the genus Babuvirus. The genus, Babuvirus was created with the sole species, BBTV (Vetten et al. 2005). In 2008, another babuvirus species, ABTV infecting abaca was reported from Philippines (Sharman et al. 2008) and so far it has not been known in India. The study of Mandal et al. (2013) showed the evidence of the third member under genus Babuvirus and thrown light on the further diversity in babuvirus. Further, eradication of these viruses in India need integrated approach combining surveillance, monitoring, destruction of infected material and replanting with virus free plants and controlling vector using systemic insecticides would bring down the disease incidence in the area where plantations are perennial for both of these two crops. In case of banana, the implementation of National certification system for tissue culture raised plants (NCS-TCP) by DBT is a major step in containing the incidences of BBTV in tissue culture raised plants. At present, 270 million tissue culture banana plants are supplied by the 95 tissue culture recognized units across India. Among 270 million plants, only 35 million plants are certified during 2015–16. In 2009–11, a severe outbreak of BBTV in Jalgaon, Maharastra and Kodur, AP should be taken as lesion by the TC banana producers and all the TCPUs must enter into the NCS-TCP system and must get virus free certification before dispatching the plants to the farmers. The impact of the certification system in managing BBTV in banana needs to be studied.
References
Amin I, Qazi J, Mansoor S, Ilyas M, Briddon RW (2008) Molecular characterisation of banana bunchy top virus (BBTV) from Pakistan. Virus Genes 36:191–198
Anhalt MD, Almeida RPP (2008) Effect of temperature, vector life stage, and plant access period on transmission of banana bunchy top virus to banana. Phytopathology 98:743–748
Banerjee A, Roy S, Beherea GT, Roy SS, Dutta SK, Ngachana SV (2014) Identification and characterization of a distinct banana bunchy top virus isolate of Pacific-Indian Oceans group from North-East India. Virus Res 183:41–49
Banerjee A, Singh R, Roy S, Behere GT, Roy SS, Dutta SK, Ranebennur H, Ngachan SV (2015) Genetic diversity of Banana bunchy top virus from Northeast India showed existence of distinct PIO isolates in naturally growing banana mats. In: Proceedings of IPS zonal symposium on Holistic Plant Health Management in Organic Agriculture.10–11 February. ICAR Research Complex for NEH Region, Umiam, Meghalaya, p 54
Basu AN, Ganguly B (1968) A note on the transmission of foorkey disease of large cardamom by the aphid, Micromyzus kalimpongensis Basu. Ind Phytopathol 21:127
Beetham PR, Harding RM, Dale JL (1999) Banana bunchy top virus DNA-2 to -6 are monocistronic. Arch Virol 144:89–105
Drew RA, Moisander JA, Smith MK (1989) The transmission of banana bunchy top virus in micropropagated bananas. Plant Cell Tissue Org Cult 16:187–193
Espino RC, Magnaye LV, Johns AP, Juanillo C (1993) Evaluation of Philippine banana cultivars for resistance to bunchy-top and fusarium wilt. In: Valmayor RV, Hwang SC, Ploetz R, Lee SC, Roa NV (eds) Proceedings: International symposium on recent developments in banana cultivation technology, 14–18 December 1992, Chiuju, Pingtung, Taiwan, Philippines: Taiwan Banana Research Institute. INIBAP/ASPNET, Los Banos, pp 89–102
Geering ADW, Thomas JE (1997) Search for alternative hosts of banana bunchy top virus in Australia. Aus Plant Pathol 26:250–254
Ghosh A, Das A, Lepcha R, Mandal B (2016a) Identification, distribution and temporal occurrence of aphids infesting large cardamom and their efficiency in transmitting large cardamom viruses in northeastern sub-Himalayan region. Aus Plant Pathol. doi:10.1007/s13313-016-0437-0
Ghosh A, Das A, Vijayanandra S, Mandal B (2016b) Cardamom bushy dwarf virus infection in large cardamom alters plant selection preference, life stages and fecundity of aphid vector, Micromyzus kalimpongensis (Aphididae: Hemiptera). Environ Entomol 45:178–184
Hafner GJ, Harding RM, Dale JL (1995) Movement and transmission of banana bunchy top virus DNA component one in bananas. J Gen Virol 76:2279–2285
Hooks CRR, Wright MG, Kabasawa DS, Manandhar R, Almeida RPP (2008) Effect of banana bunchy top virus infection on morphology and growth characteristics of banana. Ann Appl Biol 153:1–9
Howarth AJ, Gardner RC, Messing J, Shepherd R (1981) Nucleotide sequence of naturally occurring deletion mutants of cauliflower mosaic virus. Virology 112:678–685
Hu JS, Wang M, Sether D, Xie W, Leonhardt KW (1996) Use of polymerase chain reaction (PCR) to study transmission of banana bunchy top virus by the banana aphid (Pentalonia nigronervosa). Ann Appl Biol 128:55–64
Hu JM, Fu HC, Lin CH, Su HJ, Yeh HH (2007) Reassortment and Concerted Evolution in Banana Bunchy Top Virus Genomes. J Virol 81(4):1746–1761
Hyder MZ, Shah SH, Hameed S, Naqvi SMS (2011) Evidence of recombination in the banana bunchy top virus genome. Infect Genet Evol 11:1293–1300
Islam MN, Naqvi AR, Jan AT, Mohd Q, Haq R (2010) Genetic diversity and possible evidence of recombination among Banana Bunchy Top Virus (BBTV) Isolates. Int Res J Microbiol 1(1):001–012
Jones DR (2000) Diseases of Banana. Abaca and Enset, CAB International, Wallingford
Karan M, Harding RM, Dale JL (1994) Evidence for two groups of banana bunchy top virus isolates. J Gen Virol 75:3541–3546
Karan M, Harding RM, Dale JL (1997) Association of banana bunchy top virus DNA components 2 to 6 with banana bunchy top disease. Mol Plant Pathol:1–16
Kesavamoorthy RC (1980) Radical changes in ecosystem in the Pulney hills. In: Muthukrishnan CR, Abdul Chaser, J B. M. Md. Proc. 13th national seminar on banana production technology, TNAU, Coimbatore. pp 23–28
Kumar PL, Selvarajan R, Iskra-Caruana ML, Chabannes M, Hanna R (2015) Biology, Etiology, and Control of Virus Diseases of Banana and Plantain. Adv Virus Res 91:229–269
Magee CJP (1927) Investigations of the bunchy top disease of bananas. Bulletin of the Council for Scientific and Industrial Research (Australia). 30: 64
Magee CJP (1948) Transmission of banana bunchy top to banana varieties. J Aus Inst Agri Sci 14:18–24
Mandal B, Mandal S, Pun KB, Varma A (2004) First report of the association of a Nanovirus with ‘Foorkey’ disease of large cardamom in India. Plant Dis 88:428
Mandal B (2010) Advances in small isometric multicomponent ssDNA viruses infecting plants. Ind J Virol 21:18–30
Mandal B, Shilpi S, Roy-Barman A, Mandal S, Varma A (2013) Nine novel DNA components associated with the foorkey disease of large cardamom: Evidence of a distinct babuvirus species in Nanoviridae. Virus Res 178:297–305
Menon MR, Christudas SP (1967) Studies on the population of aphid Pentalonia nigronervosa Coq. On banana plants in Kerala. Agri Res J Kerala 5:84–86
Metha PR, Joshi NC, Rao MH, Renjhen PL (1964) Bunchy top serious disease of banana in India. Sci Cult 30:259–263
Mitra M (1936) Report of the imperial mycologist. Scientific Report of Agricultural Research Institute, Pusa. 1933–1934
Niyongere C, Lepoint P, LosengeT BG, Ateka EM (2015) Towards understanding the diversity of banana bunchy top virus in the Great Lakes region of Africa. Afr J Agri Res 10(7):702–709
Niyongere C, Losenge T, Ateka EM, Ntukamazina N, Ndayiragije P, Simbare A et al (2013) Understanding banana bunchy top disease epidemiology in Burundi for an enhanced and integrated management approach. Plant Pathol 62:562–570
Ram RD, Summanwar AS (1984) Colocasia esculenta (L) Schott. A reservoir of bunchy top disease of banana. Curr Sci 53:145–146
Samraj J, Menon MR, Christudas SP (1970) The movement of banana bunchy top virus in plant. Agri Res J Kerala 8:106–108
Savory FR, Ramakrishnan U (2014) Asymmetric patterns of reassortment and concerted evolution in Cardamom bushy dwarf virus. Infect Genet Evol 24:15–24
Selvarajan R, Balasubramanian V (2013) Natural occurrence of banana bunchy top virus in Ensete superbum in India. Ind J Virol 24:97–98
Selvarajan R, Balasubramanian V (2014) Host interaction host–virus interactions in banana-infecting viruses. In: Gaur RK, Hohn T, Sharma P (eds) Plant virus–host interaction molecular approaches and viral evolution. Elsevier Academic Press, Waltham, pp 57–78
Selvarajan R, Balasubramanian V, Sathiamoorthy S (2006) Vector transmission of banana bract mosaic and banana streak viruses in India. In: Abstracts of XVI annual convention and international symposium on management of vector-borne viruses, ICRISAT, 7–10 February 2006, p 110
Selvarajan R, Mary Sheeba M, Balasubramanian V, Rajmohan R, Lakshmi Dhevi N, Sasireka T (2010) Molecular characterization of geographically different banana bunchy top virus isolates in India. Ind J Virol 21:110–116
Selvarajan R, Balasubramanian V, Sheeba MM, Raj Mohan R, Mustaffa MM (2011) Virus-indexing technology for production of quality banana planting material: a boon to the tissue-culture industry and banana growers in India. In: Van den Bergh I et al. (eds) Acta Horticulturae: 897. Proceedings of international ISHS-ProMusa symposium on global perspectives on Asian challenges, pp 463–469
Selvarajan R (2015) Viral diseases of banana. CAB Rev 10. No. 050
Sharma SR (1988) Banana bunchy top virus. Int J Trop Plant Dis 6:19–41
Sharman M, Thomas JE, Skabo S, Holton TA (2008) Abaca’ bunchy top virus, a new member of the genus Babuvirus (family Nanoviridae). Arch Virol 153:135–147
Singh SJ (2003) Viral diseases of Banana, 1st edn. Kalyani Publishers, Ludhiana, pp 10–40
Stainton D, Kraberger S, Walters M, Wiltshire EJ, Rosario K, Halafihi M et al (2012) Evidence of inter-component recombination, intra-component recombination and reassortment in banana bunchy top virus. J Gen Virol 93:1103–1119
Su HJ, Hwang AS, Lee SY, Chao CP (2007) Conservation, disease indexing and utilization of pathogen free citrus and banana genetic resources in Taiwan. International training workshop on the conservation and utilization of Tropical/ Subtropical plant genetic resources, In, pp 1–24
Thomas JE, Dietzgen RG (1991) Purification, characterization and serological detection of virus-like particles associated with banana bunchy top virus in Australia. J Gen Virol 72:217–224
Thomas JE, Iskra-Caruana ML (2000) Bunchy top. In: Jones DR (ed) Diseases of banana, abacá and enset. CABI Publishing, Wallingford, pp 241–253
Thomas JE, Iskra-Caruana ML, Jones DR (1994) Banana bunchy top disease, Musa disease fact sheet No.4, INIBAP, Montpellier, p 2
Varma PM, Capoor SP (1964) ‘Foorkey’ disease of large cardamom. Ind J Agri Sci 34:56–62
Vetten HJ, Chu PWG, Dale JL, Harding R, Hu J, Katul L, Kojima M, Randles JW, Sano Y, Thomas JE (2005) Family Nanoviridae. In: Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier, London, pp 343–352
Vetten HJ, Dale JL, Grigoras I, Gronenborn B, Harding R, Randles JW., et al (2012) Family Nanoviridae. In: Virus taxonomy: ninth report of the international committee on taxonomy of viruses, pp 395–404
Vishnoi R, Raj SK, Prasad V (2009) Molecular characterization of an Indian isolate of banana bunchy top virus based on six genomic DNA components. Virus Genes 38:334–344
Wang HI, Chang CH, Lin PH, Fu HC, Tang C, Yeh HH (2013) Application of Motif-Based Tools on Evolutionary Analysis of Multipartite Single-Stranded DNA Viruses. PLoS One 8(8):e71565. doi:10.1371/journal.pone.0071565
Wanitchakorn R, Harding RM, Dale JL (2000) Sequence variability in the coat protein gene of two groups of banana bunchy top isolates. Arch Virol 145:593–602
Wardlaw CN (1972) Banana Diseases: Including plantains and abaca. William Clowes and Sons Ltd., London, pp 68–115
Watanabe S, Greenwell AM, Bressan A (2013) Localization, concentration, and transmission efficiency of banana bunchy top virus in four asexual lineages of Pentalonia aphids. Viruses 5:758–775
Wu RY, Su HJ (1990) Purification and characterization of banana bunchy top virus. J Phytopathol 128:153–160
Yu NT, Zhang YL, Feng TC, Wang JH, Kulye M, Yang WJ et al (2012) Cloning and sequence analysis of two banana bunchy top virus genomes in Hainan. Virus Genes 44:488–494
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Selvarajan, R., Mandal, B., Balasubramanian, V., Banerjee, A., Vijayanandraj, S., Ghosh, A. (2017). Biology and Molecular Biology of Babuviruses Occurring in India. In: Mandal, B., Rao, G., Baranwal, V., Jain, R. (eds) A Century of Plant Virology in India. Springer, Singapore. https://doi.org/10.1007/978-981-10-5672-7_3
Download citation
DOI: https://doi.org/10.1007/978-981-10-5672-7_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-5671-0
Online ISBN: 978-981-10-5672-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)