Abstract
Retinoic acid (RA) is a vitamin A-derived morphogen controlling important developmental processes in vertebrates, and more generally in chordates, including axial patterning and tissue formation and differentiation. In the embryo, endogenous RA levels are controlled by RA synthesizing and degrading enzymes and the RA signal is transduced by two retinoid receptors: the retinoic acid receptor (RAR) and the retinoid X receptor (RXR). Both RAR and RXR are members of the nuclear receptor superfamily of ligand-activated transcription factors and mainly act as heterodimers to activate the transcription of target genes in the presence of their ligand, all-trans RA. This signaling pathway was long thought to be a chordate innovation, however, recent findings of gene homologs involved in RA signaling in the genomes of a wide variety of non-chordate animals, including ambulacrarians (sea urchins and acorn worms) and lophotrochozoans (annelids and mollusks), challenged this traditional view and suggested that the RA signaling pathway might have a more ancient evolutionary origin than previously thought. In this chapter, we discuss the evolutionary history of the RA signaling pathway, and more particularly of the RARs, which might have experienced independent gene losses and duplications in different animal lineages. In sum, the available data reveal novel insights into the origin of the RA signaling pathway as well as into the evolutionary history of the RARs.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Retinoic acid (RA) is a fat-soluble morphogen derived from vitamin A that, in vertebrates, controls the organization of the anteroposterior axis and the formation and differentiation of various tissues during development. It is well known that one of the major actors of the RA signaling pathway is the retinoic acid receptor (RAR), a member of the nuclear receptor superfamily of ligand-activated transcription factors. RARs mainly act as heterodimers with the retinoid x receptors (RXRs) to activate the transcription of target genes in the presence of their ligand, all-trans RA (reviewed in [36]) (Fig. 4.1). In most vertebrates, including human and mouse, there are three rar genes (rarα, rarβ and rarγ) and three rxr genes (rxrα, rxrβ and rxrγ) (Fig. 4.2), each encoding several isoforms (reviewed in [51]). In fact, at the origin of vertebrates two whole genome duplications took place [4, 21, 57, 82] giving rise to the multiple RAR and RXR paralogs in vertebrates. Thus, a number of different heterodimers can be formed between these receptors and it is believed that there are more than thirty different RAR-RXR heterodimer associations, taking into account the many RAR and RXR isoforms produced, that transduce signals in the presence of all-trans RA [51, 59]. It is also important to mention that RXR is believed to be a receptor for a specific RA isomer, 9-cis RA, although the in vivo relevance of this observation is still debated ([11, 47, 68] and references therein). Moreover, other ligands such as docohexaenoic acid or phytanic acid, among others, have been proposed as ligands for RXR [22, 60]. The functional role of RXR in the heterodimer is outside the scope of this chapter, but it is nonetheless important to point out that RXR is not a passive partner, and the true nature of its in vivo role remains to be described (see [32] and references therein).
History: Production, Metabolism and Signaling by Retinoids in Vertebrates
The Structure of the RAR-RXR Heterodimer
Like other members of the nuclear receptor superfamily, RARs are modular proteins, possessing several domains, which carry out specific functions required for their activities as ligand-regulated transcription factors (Fig. 4.1). The RAR-RXR heterodimer binds to DNA on specific sequences called retinoic acid response elements (RAREs), which, most frequently, consist of direct repeats (DRs) [2, 15] (Fig. 4.1).
RA signaling pathway. a Schematic representation of RA metabolism and signaling. Retinol is converted to RA in two steps, first retinol (vitamin A) is oxidized into retinal by a reversible reaction and then retinal is oxidized into RA. RA is transported into the nucleus where it binds to the retinoic acid receptor (RAR), which forms a heterodimer with the retinoid X receptor (RXR). Upon binding of RA, the RAR-RXR heterodimer will recruit co-activators and activate the transcription of target genes. The colors represent the evolutionary conservation: RXR and RALDH are in red because they are present in all metazoans and CYP26 and RAR are in orange because they are absent in ecdysozoans and cnidarians. ADH alcohol dehydrogenase; CRBP cellular retinol binding protein; CRABP cellular retinoic acid binding protein; CYP26 cytochrome P450 subfamily 26; RA retinoic acid; RALDH retinaldehyde dehydrogenase; RAR retinoic acid receptor; RARE retinoic acid response element; RXR retinoid X receptor; SDR short chain dehydrogenase/reductase. b Overview of the structure of the RAR protein: N-terminal ligand-independent transcriptional activation domain (AF-1), a centrally located DNA-binding domain (DBD) consisting of a highly conserved core region, which contains two zinc finger modules, a hinge that allows flexibility between the N- and C-terminal portion of the molecule and a C-terminal ligand binding domain (LBD), which interacts with the ligand, allowing receptor dimerization and additionally serving as a ligand-activated transcriptional activation function (AF-2) domain
In the apo form, which corresponds to the receptor without the ligand, RAR, together with RXR, interacts with transcriptional co-repressors, such as NCoR or SMRT, and represses the transcription of target genes (reviewed in [36]). Following ligand binding, the LBD undergoes a conformational change during which the most C-terminal helix (H12) forms a lid that closes the hydrophobic pocket in which the ligand is buried (reviewed in [36]). It has been shown that this conformational change significantly alters the composition of the proteins that interact with the LBD. The conformational change causes dissociation of the co-repressors and the binding of co-activators, such as the members of the p160 proteins (SRC1, 2 and 3) that induce histone acetylation and subsequently, transcriptional activation (reviewed in [66]. This is, however, an oversimplified presentation of RAR-RXR functions, as many different types of co-repressors, co-activators and chromatin remodeling complexes work together to orchestrate the remodeling of chromatin linked to transcriptional regulation [3, 34]. Recently, older models of unliganded RAR action have been challenged by new genome-wide analyses of RAR target genes (see Chaps. 9 and 10). It has become clear that ligand binding generally increases the ability of RAR-RXR to interact with the regulatory regions of at least some target genes [10, 67, 69].
Regulation of Endogenous RA Levels
Many studies have thoroughly characterized the function of RAR in vertebrates, focusing mainly on developmental and physiological aspects [35, 86]. Other studies have characterized the biochemical pathways that produce RA in vivo. These data have revealed that the RA precursor, retinol (which is vitamin A), is catabolized to retinal in a reversible manner by alcohol dehydrogenase (ADH) [8, 54, 103] or the short-chain dehydrogenase/reductase (SDR) enzymes, such as RDH10 [102]. This reaction is followed by an irreversible oxidation step of retinal to RA by retinal dehydrogenases (RALDHs), which is the rate-limiting step of this pathway [25]. There are three RALDH enzymes in most vertebrates, and RALDH2 is the most relevant RALDH during embryonic development, with its expression allowing us to infer, at least grossly, the regions of the embryo characterized by high endogenous RA levels (see [78]). Importantly, this zone of high endogenous RA levels gives rise to two RA gradients, one oriented anteriorly and one posteriorly in the embryo, as has recently been revealed in developing zebrafish using a visualization technique based on fluorescence resonance energy transfer (FRET) [91].
Within a target cell, retinol is associated with cellular retinol binding protein (CRBP), and RA is associated with the cellular retinoic acid binding protein (CRABP) [72]. Cellular and tissue concentrations of RA are generally regulated through homeostatic processes involving both RA production and degradation, with enzymes of the cytochrome P450 subfamily 26 (CYP26) principally mediating the catalysis of RA to inactive products like 4-oxo RA and 4-hydroxy RA [76]. Three genes encoding CYP26 enzymes (CYP26A1, CYP26B1 and CYP26C1) exist in vertebrates [99]. Moreover, in vitro experiments have identified further RA-degrading CYP enzymes that may also be involved in the RA catalysis process, such as CYP1A2, 2A4, 2A6, 1B1, 2B1, 2B6, 2C3, 2C7, 2C8, 2C9, 2D6, 2E1, 2E2, 2G1, 3A4/5, 3A6, 3A7, and 4A11. However, the in vivo functions of these enzymes remain unclear [37, 61, 81].
Development of the Field: Evolutionary Origins of RA Signaling
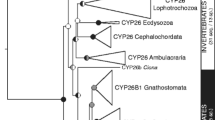
In contrast to the situation in vertebrates, where the roles and mode of action of RA are relatively well understood, very little is known about the origins of the RA signaling pathway. Only a few studies have focused on the functions of RAR in non-vertebrate animals. Among non-vertebrates, only representative RAR receptors found in other chordates, i.e. in tunicates and cephalochordates, have been functionally characterized [27, 28, 30, 31]. However, the recent identification of some components of the RA machinery in the genomes of non-chordate animals has provided some useful hints that shed new light on the action of RA and the presence of a RA signaling pathway in metazoan animals. For example, homologs of rar, cyp26 and raldh have been identified in the genomes of ambulacrarians (sea urchins and acorn worms) and lophotrochozoans (annelids and mollusks) (Fig. 4.2) [1, 12, 13, 65, 92]. Our current understanding of the origin of RAR suggests the evolutionary history of RAR was influenced by specific losses and duplications in particular animal lineages. Altogether, this analysis allows us to conclude that, evolutionarily speaking, the RA signaling pathway is more ancient than originally thought.
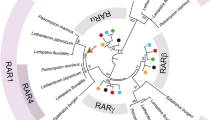
Evolution of RAR. Schematic phylogenetic distribution of RARs and RXRs in bilaterian animals: RARs are generally present in chordates (vertebrates, tunicates and cephalochordates) and have recently been identified in the genomes of ambulacrarians (hemichordates and echinoderms) and lophotrochozoans (annelids and mollusks). In general, RARs seem to be absent from ecdysozoan genomes. RAR might thus have already been present in the last common ancestor of deuterostomes and protostomes, called Urbilateria [12]. Given the absence from ecdysozoan genomes, RAR might have secondarily been lost in this lineage. Moreover, RAR has probably also been secondarily lost in a tunicate lineage, the appendicularians. In vertebrates, the rar genes have been duplicated, with, for example, Homo sapiens possessing three rar paralogs (rarα, rarβ and rarγ). In contrast, RXRs are present in all metazoan lineages. In addition, rxr genes have also been duplicated in vertebrates as, for example, Homo sapiens possessing three rxr paralogs (rxrα, rxrβ and rxrγ). The presence of a functional retinoic acid (RA) signaling pathway is indicated by a + or − sign, however, in many lineages this has not been determined (?). Numbers indicate the number of RAR and RXR coding genes in a given lineage. The red dot indicates the probable origin of rar and the yellow triangle indicates the whole genome duplication that took place in the vertebrate lineage and the purple x indicates the loss of RAR. Protein accession numbers: (source http://www.ncbi.nlm.nih.gov/, excepting the Capitella teleta sequence, which is accessible at: http://genome.jgi-psf.org/Capca1/Capca1.home.html): Homo sapiens RARα (P10276), RARβ (P10826), RARγ (P13631), RXRα (P19793.1), RXRβ (P28702.2) and RXRγ (P48443.1), Ciona intestinalis RAR (NP_001072037) and RXR (NP_001071809), Polyandrocarpa misakiensis RAR (BAA25569) and RXR (BAA82618), Botrylloides leachi RAR (DQ523226) and RXR, Branchiostoma floridae RAR (AAM46149) and RXR (AAM46151), Saccoglossus kowalevskii RAR (XP_002742241) and RXR (ADB22634), Strongylocentrotus purpuratus RAR (XP_779976) and RXR (XP_784246), Capitella teleta RAR (168520) and RXR (186691), Thais (Reisha) clavigera RAR (BAN82614.1) and RXR (E9RHD8), Drosophila melanogaster USP (AAF45707), Dirofilaria immitis USP (AAM08269) and Tripedalia cystophora RXR (AAC80008)
Current State of the Field: RA Signaling in Non-vertebrate Animals
Presence or Absence of RA-Signaling in Metazoan Genomes: Chordates
The RA signaling pathway was long thought to be vertebrate-specific. However, it is now well established that most of the components of the RA machinery are present and functional in all chordate phyla. More particularly, single rar homologs have been identified and functionally characterized in the following chordates (Fig. 4.2): Ciona intestinalis [31], Botrylloides leachi [80] and Polyandrocarpa misakiensis [40] (all of which are ascidian tunicates), as well as in the cephalochordate Branchiostoma floridae also referred to as amphioxus [28].
As previously mentioned, vertebrates possess at least three RARs, RARα, RARβ and RARγ (Fig. 4.2), but as indicated, only one RAR is present in chordates (cephalochordates and ascidian tunicates) (Fig. 4.2). The evolution of the chordate RA signaling pathway has been studied extensively [27, 30, 31, 88]. In amphioxus it has been shown that RAR and RXR are able to heterodimerize, bind RA and activate the transcription of genes upon RA binding [28]. Moreover, it was shown that the amphioxus RAR has a ligand-binding pocket structure similar to RARβ [27]. RA functions have been described in the embryonic development of amphioxus. For example, it has been shown that RA regulates the anteroposterior patterning of the CNS and neuronal specification in a hox-dependent manner [88]. RA also mediates the patterning of the ectoderm and the formation of the tail fin in this specie [14]. In addition, orthologs of cyp26, rdh and raldh have been identified in the genome of amphioxus, however, their function remains to be assessed. In contrast, there is no evidence of the existence of genes coding for proteins responsible for retinol storage, transport and cellular uptake, suggesting that these might have appeared specifically in the vertebrate lineage [14].
Components of the RA machinery have been identified in tunicates. It was shown that RARs are present in Ciona intestinalis [31], Botrylloides leachi [80] and Polyandrocarpa misakiensis [40] that bind and are activated by RA. However, the function of the enzymes involved in RA synthesis and metabolism has not been assessed yet in these animals [14]. In addition, RA signaling has been secondarily modified in different lineages [14]. For example, there is a partial loss of the RA-dependent regulation of the hox code in ascidians [14], but the appendicularian tunicate, Oikopleura dioica, stands out as an exception because it lacks the genes coding for the main actors of the RA signaling pathway, such as, rar, aldh1 and cyp26 [13]. In the course of evolution, these genes were very likely secondarily lost in this animal since these gene are present in all the other urochordate species studied, such as Ciona intestinalis [31], Botrylloides leachi [80] and Polyandrocarpa misakiensis [40]. Of interest, it seems that the anterioposterior patterning of O. dioica during embryonic development may be RA-independent, although detailed characterization of the developmental pathways used by this species is still lacking. These observations suggest that anteroposterior patterning can be achieved using many different signaling pathways and is not necessarily dependent on RA.
In conclusion, the RA signaling pathway is present and functional in chordates, however, it should be noted that, compared to the situation in vertebrates, very little is known about the regulation of this signaling cascade and its function in invertebrate-chordates.
Presence or Absence of RA-Signaling in Metazoan Genomes: Non-chordates
Bioinformatic analyses have revealed that genes involved in the RA signaling machinery are present in the genomes of a variety of non-chordates (ambulacrarians and lophotrochozoans) [1, 12, 13, 65, 92]. Based on phylogenetic analysis of the nuclear hormone receptor superfamily, it was hypothesized that a proto-RAR might have been present in Urbilateria, the last common ancestor of all bilaterians [4].
Homologous genes of rar have so far been identified in the genomes of the following non-chordate invertebrates (Fig. 4.2): the ambulacrarians Saccoglossus kowalevskii (a hermichordate) and Strongylocentrotus purpuratus (an echinoderm) [13, 65] and the lophotrochozoans Lottia gigantea (a mollusk) and Capitella teleta (an annelid, formerly Capitella capitata) [12]. The phylogenetic analyses performed and the surprisingly high degree of amino acid conservation of the RARs found in these species indicate that these receptors may be bona-fide RARs that might be able to bind RA [12]. These findings allow us to conclude that the gene encoding RAR has a much more ancient evolutionary origin than previously believed. In particular, the fact that a clear rar ortholog is found in lophotrochozoans (annelids and mollusks) strongly suggests that a rar gene was already present in the ancestor of all bilaterians. This has important implications for the evolution and diversification of metazoan animals, since it indicates that functional roles for RA in developmental patterning might be evolutionarily much more ancient than originally thought (reviewed in [12]). However, recently, the characterization of a RAR from the mollusk Thais clavigera showed that this receptor is unable to bind RA [97], raising the question of the function of RAR and RA in non-chordate animals. Thus, the unambiguous experimental demonstration of a functional RAR and of a functional RA signaling cascade outside the chordate phylum still remains elusive.
In contrast, previous in silico analyses did not identify rar homologs in appendicularian tunicates (see above), ecdysozoans (arthropods and nematodes), and cnidarians (Fig. 4.2). These results allow two tentative conclusions: (1) if the absence of rar from cnidarian genomes is confirmed, for example by the analysis of other genomes, the origin of RAR and RA signaling can probably firmly be placed at the base of bilaterian animals and not at the base of metazoan animals. Given that anteroposterior patterning is an important feature of bilaterians, it is tempting to speculate that RAR might have played a role in the emergence of bilaterian body axes and, particularly, in the evolution of anteroposterior patterning systems, but this remains speculative given the very limited amount of data currently available on the role of RA outside chordates (see below); (2) the absence of rar in appendicularian tunicates and ecdysozoans suggests that these genes have been independently lost in these animal lineages and indicate that a complex anteroposterior patterning system can be established in the absence of RAR.
Homologs of the enzymes involved in RA metabolism, such as RALDH and CYP26 have also been identified in non-chordate animals. For example, raldh homologs have been identified in the genome of ambulacrarians (Strongylocentrotus purpuratus and Saccoglossus kowalevskii), lophotrochozoans (Lottia gigantea and Capitella teleta), ecdyzosoans (Caenorhabditis elegans, Daphnia pulex and Drosophila melanogaster) and cnidarian (Nematostella vectensis) [1, 12, 13, 65, 92]. In contrast, cyp26 homologs have only been identified in lophotrochozoans and ambulacrarians [1, 12, 13, 65, 92]. Altogether, these data reinforce the notion that the RA signaling pathway might have been present in Urbilateria, the last common ancestor of protostomes and deuterostomes. However, the unambiguous experimental demonstration of a functional RAR and of a functional RA signaling cascade outside the chordate phylum still remains elusive.
In contrast, rxr homologs have been identified in the genomes of various non-chordate phyla including lophotrochozoans [9], ecdysozoans [104] and basal metazoan species, such as cnidarians [29, 55] and sponges [100]. Therefore, RXR seems to have a more ancient evolutionary origin than RAR. Apparently, the RXR receptors have also experienced a complex evolutionary history. For example, in vertebrates, several paralogs of RXR were acquired following whole genome duplication events [77] and in insects, ligand recognition by USP (i.e. RXR) has been altered independently in different lineages [16, 44, 46]. For instance, it was shown that the USP from Tribolium castaneum does not have a ligand-binding pocket and, thus, does not bind and is not activated by RXR ligands despite the high degree of identity between these receptors [46]. In addition, in mecopteridan USPs, the ligand-binding pocket is very large and is occupied by a phospholipid [6, 17]. These data reveal the evolutionary plasticity of the ligand binding pocket of nuclear hormone receptors and highlight the requirement for experimental analyses of divergent receptors.
RA Derivatives in Non-chordate Invertebrates
In vertebrates, RAR is able to bind at least two different isomers of RA, all-trans RA and 9-cis RA, whereas RXR only binds to 9-cis RA in vitro. Moreover, many studies have shown that treatment of vertebrate species with all-trans RA or 9-cis RA causes developmental defects and malformations [79]. In an effort to better understand the biological roles of each RA isomer, efforts have been made to measure the presence of these compounds in different organs of vertebrates. The presence of all-trans RA during development was detected, for example, in mouse, rat and human serum, liver, kidney, brain and testis [39, 48, 87]. The concentration of all-trans RA in these tissues was between 7 and 10 nM, however, it was lower in the serum [48].
In contrast, far less is known about the metabolism of 9-cis RA and the presence of 9-cis RA in vivo. Until recently, the biological significance of this isomer remained controversial. In fact, 9-cis RA was first detected by high performance liquid chromatography (HPLC) in mouse liver and kidney [39] and rat epididymal tissue [75] at concentrations two or three times lower than those of all-trans RA. Subsequent analytical assays, however, failed to detect this isomer in liver, kidney and other murine tissues (reviewed in [47]). This failure to detect 9-cis RA in vivo has contributed to the controversy about the biological relevance of this RA isomer. In contrast, by using liquid chromatography/tandem mass spectrometry a recent study has revealed the presence of 9-cis RA in mouse pancreas [49]. This work further established that 9-cis RA is able to attenuate glucose-stimulated insulin secretion in the pancreas [49]. These data not only show that 9-cis RA is a naturally-occurring RA isomer in the pancreas, but also that 9-cis RA is biologically active. In fact, it is possible, and even likely, that 9-cis RA can simply not be detected in other tissues, because its levels are below the limit of detection of the employed analysis methods.
In addition to mice, rats and humans, endogenous RA levels have also been measured in a number of other animals. But before reviewing these data, it has to be mentioned that derivatives of retinol, generally termed retinoids, are also involved in the visual cycle and that in particular 11-cis retinaldehyde plays a crucial role in vision. This aspect is not considered in the following discussion, but it is nonetheless important to be aware of the retinoid derivatives that participate in vision as it helps to explain the wide distribution in metazoan animals of various enzymes able to act on retinoid compounds. For example, 9-cis RA was detected in the regenerating limb of urodele amphibians [98], all-trans RA was detected in budding tunicates [52] and both all-trans RA and 9-cis RA were detected in the chordate amphioxus [20].
In non-chordates, endogenous all-trans RA and 9-cis RA were detected by HPLC in the central nervous system and the hemolymph of the mollusk Lymnea stagnalis [23]. All-trans RA concentrations were higher than those of 9-cis RA with all-trans RA concentrations estimated at 693 nM in the central nervous system and 155 nM in the hemolymph, whereas, for 9-cis RA, the estimated concentrations were 380 nM in the central nervous system and 120 nM in the hemolymph [23]. These values are thus relatively high, when compared to the values typically reported in mammals, but the significance of this difference is still unclear. Similarly, in the locust embryo, both all-trans RA and 9-cis RA were detected [74]. The all-trans RA concentration in the whole embryo was estimated to be 3 nM and the 9-cis RA concentration in the locust embryo was around 1.5 nM [74]. All-trans RA and 9-cis RA were also detected in limb blastemas of the fiddler crab during regeneration. Here, all-trans RA concentrations were 19 pg/µg protein/blastema and 9-cis RA concentrations were 83 pg/µg protein/blastema [41].
There are other RA isomers, such as 9,13-di-cis RA and 13-cis RA, that, at least in vertebrates, are characterized by reduced or absent biological activity [43, 95]. Interestingly, these RA isomers are distributed relatively widely in different mammalian tissues, for example, serum, brain, liver, kidney, adipose tissue, muscle, spleen and testis [50]. Moreover, they have also been detected in invertebrates, including sponges, which are basal metazoans. Sponges contain both retinyl esters and 13-cis RA and are able to accumulate β-carotene [5, 71]. In addition, retinol and retinyl esters have been detected in mollusks, which also possess some kind of capacity to endogenously store retinoids [33].
In nematodes, the only evidence of a possible action of RA has been reported in the parasitic nematodes Brugia malayi and Onchocerca volvulus [94, 101]. In both worm species only retinol could be detected [94, 101] and it was shown that Brugia malayi worms were able to take up and accumulate all-trans RA in their tissues [101]. In addition, it was shown that the development of the parasitic nematode Litomosoides carinii in vitamin A-deficient cotton rats was delayed [93]. Together, these data suggest that retinoids may play a role in the development of parasitic nematodes, however, it is important to stress that there is no RAR in nematodes and that a homolog of USP (i.e. RXR) has only been identified in a single species of parasitic nematodes [90].
Role of RA Derivatives in Non-chordate Invertebrates
As discussed above, in vertebrates, both RA isomers, all-trans RA and 9-cis RA, are not only present, but also biologically active. In fact, it is known that RA controls many developmental processes of vertebrates [79]. Indeed, exposure of developing embryos to exogenous RA causes severe defects in vertebrates and more generally in chordates. However, outside chordates, evidence for biological functions of RA is scarce and only a few studies have addressed the impact of RA on embryonic development. For example, in many mollusk species, such as, Lymnaea stagnalis, Physa fontinalis and Bithynia tentaculata, constant treatment starting at gastrulation with all-trans RA (100 nM) affects eye formation [19]. In addition, the use of higher concentrations of all-trans RA (1 μM) on Lymnaea stagnalis embryos causes various malformations, for example, eye and shell defects and, at times, growth arrest [19]. Moreover, it was shown that RA treatments were only effective during gastrulation as treatment of embryos after gastrulation did not cause any evident developmental defects [19]. This suggests that RA might control specific developmental events in this species, which is reminiscent of the role played by RA during vertebrate development.
Treatment of cultured neurons from the mollusk Lymnaea stagnalis with all-trans RA (100 nM) induces neurite outgrowth and growth cone turning [23, 24]. A comparable effect was observed in vertebrates with a similar RA concentrations [18]. 9-cis RA treatments also have an effect on mollusks. For example, in the mollusk Thais clavigera, the injection of 9-cis RA (1 μM) causes imposex, i.e. the development of male organs in females [73]. Taken together, although the available evidence is still very fragmentary and based mainly on morphological observations, these data on effects of RA treatments in mollusks are nonetheless indicative of possible functional roles of RA derivatives in mollusks. However, recently a RAR from the mollusk Thais clavigera has been characterized and it was shown that this receptor is unable to bind RA [97], thus suggesting that effects of RA on mollusks might be RAR independent.
Data are also available that describe the effects of treatments with RA derivatives in echinoderms. For example, treatment of micromere-derived cells from the sea urchin Hemicentrotus pulcherrimus with all-trans RA (100 nM) induces pseudopodial cable growth [56]. Intriguingly, no effect was observed after treatment of embryos of the sea urchin Paracentrotus lividus with all-trans RA (20 μM). The embryos seem to develop normally, although their development is delayed [89].
Finally, there is growing evidence for a role of RA derivatives in tissue formation and regeneration of a wide variety of animals, including platyhelminthes [83], insects [38], crustaceans [42], cnidarians [70] and sponges [45]. This role might be related to the known effect of retinoids in controlling fin regeneration in zebrafish [7] and limb regeneration in urodeles [98]. In addition, retinoids appear to be implicated in regulating tissue regeneration in the fruit fly Drosophila melanogaster [38]. Mutations in the genes coding for enzymes responsible of retinoid metabolism, such as BCO (β-carotene 15,15′-monooxygenase) and a retinaldehyde dehydrogenase, generated a delay in tissue regeneration in this species. A similar effect was observed when flies were deprived of caroteinoids, a natural source of retinoids, and the normal timing was restored by supplementation of retinoids [38]. These results are very surprising because in the fruit fly retinoids were thought to be involved exclusively in vision. Moreover, there is no RAR in the fruit fly genome and USP (i.e. RXR) is unable to bind retinoids (see [46] and references therein).
Moreover, in the platyhelminthe Girardia tigrina, treatment with exogenous all-trans RA (0.5 mM) disrupts anterior but not posterior regeneration [83]. In the fiddler crab Uca pugilator, treatment of regenerating limb buds with high concentrations of all-trans RA (50 μM) causes growth delay and malformations of the limb bud [42]. In the cnidarian Hydractinia echinata, treatments with retinol, retinaldehyde and all-trans RA (1 nM to 1 μM each) increase the regeneration rate, number of tentacles and budding rate [70]. Sponges are also affected by treatment with all-trans RA (50 μM), which causes morphogenetic malformations of buds and gemmules [45]. In addition, it was shown that treatment with all-trans RA (1 mM) upregulates the expression of the gene encoding the BMP-1 (bone morphogenetic protein-1) homolog in the sponge Suberites domuncula [71] and downregulates the expression of the c-myb gene in the sponge Geodia cydonium [5]. Altogether, these observations suggest that RA might have various pleiotropic effects in metazoan animals that may range from organ formation to regeneration.
Relevance
It is important to point out that from these experiments we cannot formally conclude that endogenous retinoids play physiological roles in these different animals. In fact, some of the effects observed upon RA exposure might be caused exclusively by the toxicity of the molecule at high dose or by its transformation into a different active compound. In addition, even if, in a given animal, the molecule acts on a specific target at a relatively weak concentration, one cannot necessarily conclude that the relevant metabolic pathway exists endogenously in that animal [62]. The case of bisphenol A is exemplary for this situation: bisphenol A is an artificially produced molecule that has been shown to exert very strong hormonal effects in mammals by regulating the activity of estrogen receptors [84]. Although these effects inadvertently exist in vivo, bisphenol A is certainly not an endogenously-produced hormone.
Coming back to RA, its derivatives and their potential biological functions, the solution of the problem seems rather straightforward: the receptors need to be characterized in full, including both molecular capacities and physiological functions, and the data hence obtained need to be correlated with the in vivo retinoid content of a given species. Of course, this work approach will not yield insights into retinoid functions that are not mediated by RAR and/or RXR. Even so, to date we completely lack a functional characterization of non-chordate RARs and we have thus no indication whatsoever of the possible ligand of these RARs. It could be all-trans RA, another RA isomer or some other compound of unknown nature. Therefore, we cannot conclude that the effects observed upon RA treatment are indeed RAR-dependent or even RA signaling-dependent.
Future Directions
With the sequencing of the genomes of many different animal species, it has become evident that the RA signaling pathway, that was initially thought to be chordate specific, has a much wider phylogenetic distribution than originally thought. In fact, some of the components of this pathway are found in the genomes of a wide variety of non-chordate taxa. For example, many components of this signaling pathway, such as the homologs of rar, raldh and cyp26 have been identified in the genomes of ambulacrarians and lophotrochozoans [1, 12, 13, 65, 92]. Although an experimental validation of the biological functions of these receptors and metabolic enzymes is still lacking, these data nonetheless suggests that some kind of RA signaling pathway was probably already present in the last common ancestor of all bilaterian animals: Urbilateria [12].
Recent data have highlighted that RAR, in addition to being a classical nuclear receptor regulated by the binding of its ligand is also a protein whose activity can be controlled by phosphorylation [86]. Several studies have shown that these phosphorylation events are important for the full regulatory potential of RARs. It is still unclear how this level of regulation evolved but a recent phylogenetic analysis has shown that some, but not all, of those sites, are conserved between RARs from various taxa, including vertebrates and invertebrates [85]. This is, in particular, the case of the serine phosphorylation site located in the N-terminal A/B region of the receptor that is present in the RARs of vertebrates, amphioxus and ascidian tunicates (such as Ciona intestinalis) and that may also be present in protostome RARs. It will therefore be of great interest to study how the regulatory logic controlling these invertebrate receptors has been set up. Similarly, recent work has revealed the evolutionary conservation in vertebrates of RAR targets through the conservation of direct repeats DR5, DR2 and related elements in target gene promoter regions [58]. Again, almost nothing is known about RAR target genes outside of vertebrates, but it can be anticipated that studies in non-vertebrates will yield exciting insights into the evolutionary diversification of the gene regulatory network controlled by RA and its receptors.
The RA signaling pathway has evolved differently in chordates, lophotrochozoans, ambulacrarians and ecdysozoans. The receptor controlling this pathway in vertebrates, RAR, is present in all these groups except in ecdysozoans where it seems to have been lost. However, it should be noted that, given the limited number of species studied and the intriguing biological effects of RA derivatives in insects and crustaceans, one should not conclude that this pathway was lost altogether in ecdysozoans. Moreover, although lophotrochozoans and ambulacrarians posses an evident rar homolog, there is still no experimental evidence showing that these genes encode bona-fide RARs that are capable of binding RA and of activating transcription in a ligand-dependent fashion.
Thus, it is important to be cautious and to remember that the presence of the ortholog of a given vertebrate gene in a distant organism does not necessarily imply the presence of the identical function known for the product of this gene in vertebrates [62]. For example, an ortholog of the estrogen receptor is present in annelids and mollusks, but several studies have shown that the mollusk orthologs do not bind estrogens [96], whereas the annelid othologs are able to bind estrogens at high concentrations [53]. In addition, it seems that estrogens might not even be present endogenously in these species [26, 63, 64]. Therefore, future research should focus on a detailed characterization of the functions of the components of the RA signaling pathway in a wide variety of different metazoan species.
Abbreviations
- ADH:
-
Alcohol dehydrogenase
- AF:
-
Activation function
- BCO:
-
β-carotene 15,15′-monooxygenase
- BMP-1:
-
Bone morphogenetic protein-1
- CRABP:
-
Cellular retinoic acid binding protein
- CRBP:
-
Cellular retinol binding protein
- CYP:
-
Cytochrome P450
- CYP26:
-
Cytochrome P450 subfamily 26
- DBD:
-
DNA binding domain
- DR:
-
Direct repeat
- FRET:
-
Fluorescence resonance energy transfer
- HPLC:
-
High performance liquid chromatography
- LBD:
-
Ligand binding domain
- RA:
-
Retinoic acid
- RALDH:
-
Retinal dehydrogenase
- RAR:
-
Retinoic acid receptor
- RARE:
-
Retinoic acid response element
- RDH10:
-
Retinol dehydrogenase 10
- RXR:
-
Retinoid X receptor
- SDR:
-
Short-chain dehydrogenase/reductase
- USP:
-
Ultraspiracle
References
Albalat R, Cañestro C (2009) Identification of Aldh1a, Cyp26 and rar orthologs in protostomes pushes back the retinoic acid genetic machinery in evolutionary time to the bilaterian ancestor. Chem-Biol Interact 178:188–196
Balmer JE, Blomhoff R (2005) A robust characterization of retinoic acid response elements based on a comparison of sites in three species. J Steroid Biochem Mol Biol 96:347–354
Baek SH, Rosenfeld MG (2004) Nuclear receptor coregulators: their modification codes and regulatory mechanism by translocation. Biochem Biophys Res Co 319:707–714
Bertrand S, Brunet FG, Escriva H, Parmentier G, Laudet V, Robinson-Rechavi M (2004) Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol 21:1923–1937
Biesalski HK, Doepner G, Tzimas G, Gamulin V, Schroder HC, Batel R, Nau H, Muller WE (1992) Modulation of myb gene expression in sponges by retinoic acid. Oncogene 7:1765–1774
Billas IM, Moulinier L, Rochel N, Moras D (2001) Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects. J Biol Chem 276:7465–7474
Blum N, Begemann G (2012) Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration. Development 139:107–116
Boleda MD, Saubi N, Farrés J, Parés X (1993) Physiological substrates for rat alcohol dehydrogenase classes: aldehydes of lipid peroxidation, omega-hydroxyfatty acids, and retinoids. Arch Biochem Biophys 307:85–90
Bouton D, Escriva H, de Mendonça RL, Glineur C, Bertin B, Noël C, Robinson-Rechavi M, de Groot A, Cornette J, Laudet V, Pierce RJ (2005) A conserved retinoid X receptor (RXR) from the mollusk Biomphalaria glabrata transactivates transcription in the presence of retinoids. J Mol Endocrinol 34:567–582
Bruck N, Vitoux D, Ferry C, Duong V, Bauer A, de The H, Rochette-Egly C (2009) A coordinated phosphorylation cascade initiated by p38MAPK/MSK1 directs RARα to target promoters. EMBO J 28:34–47
Calleja C, Messaddeq N, Chapellier B, Yang H, Krezel W, Li M, Metzger D, Mascrez B, Ohta K, Kagechika H, Endo Y, Mark M, Ghyselinck NB, Chambon P (2006) Genetic and pharmacological evidence that a retinoic acid cannot be the RXR-activating ligand in mouse epidermis keratinocytes. Genes Dev 20:1525–1538
Campo-Paysaa F, Marlétaz F, Laudet V, Schubert M (2008) Retinoic acid signaling in development: tissue-specific functions and evolutionary origins. Genesis 46:640–656
Cañestro C, Postlethwait JH, Gonzàlez-Duarte R, Albalat R (2006) Is retinoic acid genetic machinery a chordate innovation? Evol Dev 8:394–406
Carvalho JE, Schubert M (2013) Retinoic acid: metabolism, developmental functions and evolution. In: Dakshinamurti K, Dakshinamurti S (eds) Vitamin-binding proteins: their functional consequences. CRC Press/Taylor & Francis Group, Boca Raton, Florida, USA (in press)
Chambon P (1996) A decade of molecular biology of retinoic acid receptors. FASEB J 10:940–954
Chaumot A, da Lage JL, Maestro O, Martin D, Iwema T, Brunet F, Belles X, Laudet V, Bonneton F (2012) Molecular adaptation and resilience of the insect’s nuclear receptor USP. BMC Evol Biol 12(1):199
Clayton GM, Peak-Chew SY, Evans RM, Schwabe JW (2001) The structure of the ultraspiracle ligand-binding domain reveals a nuclear receptor locked in an inactive conformation. Proc Natl Acad Sci USA 98:1549–1554
Corcoran J, Maden M (1999) Nerve growth factor acts via retinoic acid synthesis to stimulate neurite outgrowth. Nat Neurosci 2:307–308
Creton R, Zwaan G, Dohmen R (1993) Specific developmental defects in molluscs after treatment with retinoic acid during gastrulation. Dev Growth Differ 35:357–364
Dalfó D, Albalat R, Molotkov A, Duester G, Gonzàlez-Duarte R (2002) Retinoic acid synthesis in the prevertebrate amphioxus involves retinol oxidation. Dev Genes Evol 212:388–393
Dehal P, Boore JL (2005) Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3:e314
de Urquiza AM, Liu S, Sjöberg M, Zetterström RH, Griffiths W, Sjövall J, Perlmann T (2000) Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science 290:2140–2144
Dmetrichuk JM, Carlone RL, Jones TRB, Vesprini ND, Spencer GE (2008) Detection of endogenous retinoids in the molluscan CNS and characterization of the trophic and tropic actions of 9-cis retinoic acid on isolated neurons. J Neurosci 28:13014–13024
Dmetrichuk JM, Carlone RL, Spencer GE (2006) Retinoic acid induces neurite outgrowth and growth cone turning in invertebrate neurons. Dev Biol 294:39–49
Duester G (2008) Retinoic acid synthesis and signaling during early organogenesis. Cell 134:921–931
Eick GN, Thornton JW (2011) Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Mol Cell Endocrinol 334:31–38
Escriva H, Bertrand S, Germain P, Robinson-Rechavi M, Umbhauer M, Cartry J, Duffraisse M, Holland L, Gronemeyer H, Laudet V (2006) Neofunctionalization in vertebrates: the example of retinoic acid receptors. PLoS Gene 2(7):e102
Escriva H, Holland ND, Gronemeyer H, Laudet V, Holland LZ (2002) The retinoic acid signaling pathway regulates anterior/posterior patterning in the nerve cord and pharynx of amphioxus, a chordate lacking neural crest. Development 129:2905–2916
Escriva H, Safi R, Hänni C, Langlois MC, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V (1997) Ligand binding was acquired during evolution of nuclear receptors. PNAS 94:6803–6808
Fujiwara S (2006) Retinoids and nonvertebrate chordate development. J Neurobiol 66:645–652
Fujiwara S, Kawamura K (2003) Acquisition of retinoic acid signaling pathway and innovation of the chordate body plan. Zool Sci 20:809–818
Germain P, Iyer J, Zechel C, Gronemeyer H (2002) Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature 415:187–192
Gesto M, Castro LF, Reis-Henriques MA, Santos MM (2012) Retinol metabolism in the mollusk Osilinus lineatus indicates an ancient origin for retinyl ester storage capacity. PLoS One 7:e35138
Glass CK, Rosenfeld MG (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14:121–141
Glover JC, Renaud JS, Rijli FM (2006) Retinoic acid and hindbrain patterning. J Neurobiol 66:705–725
Gronemeyer H, Gustafsson JA, Laudet V (2004) Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 3:950–964
Guengerich FP, Cheng Q (2011) Orphans in the human cytochrome P450 superfamily: approaches to discovering functions and relevance in pharmacology. Pharmacol Rev 63:684–699
Halme A, Cheng M, Hariharan IK (2010) Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr Biol 20:458–463
Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C (1992) 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell 68:397–406
Hisata K, Fujiwara S, Tsuchida Y, Ohashi M, Kawamura K (1998) Expression and function of a retinoic acid receptor in budding ascidians. Dev Genes Evol 208:537–546
Hopkins P (2001) Limb regeneration in the fiddler crab, Uca pugilator: hormonal and growth factor control. Amer Zool 398:389–398
Hopkins PM, Durica DS (1995) Effects of all-trans retinoic acid on regenerating limbs of the fiddler crab. Uca pugilator. J Exp Zool 272:455–463
Horst RL, Reinhardt TA, Goff JP, Nonnecke BJ, Gambhir VK, Fiorella PD, Napoli JL (1995) Identification of 9-cis,13-cis-retinoic acid as a major circulating retinoid in plasma. Biochemistry 34:1203–1209
Hult EF, Tobe SS, Chang BS (2011) Molecular evolution of ultraspiracle protein (USP/RXR) in insects. PLoS One 6:e23416
Imsiecke G, Borojevic R, Custudio M, Müller WEG (1994) Retinoic acid acts as a morphogen in freshwater sponges. Invertebr Reprod Dev 26:89–98
Iwema T, Billas IM, Beck Y, Bonneton F, Nierengarten H, Chaumot A, Richards G, Laudet V, Moras D (2007) Structural and functional characterization of a novel type of ligand-independent RXR-USP receptor. EMBO J 26:3770–3782
Kane MA (2012) Analysis, occurrence, and function of 9-cis-retinoic acid. Biochim Biophys Acta 1821:10–20
Kane MA, Chen N, Sparks S, Napoli JL (2005) Quantification of endogenous retinoic acid in limited biological samples by LC/MS/MS. Biochem J 388:363–369
Kane MA, Folias AE, Pingitore A, Perri M, Obrochta KM, Krois CR, Cione E, Ryu JY, Napoli JL (2010) Identification of 9-cis-retinoic acid as a pancreas-specific autacoid that attenuates glucose-stimulated insulin secretion. PNAS 107:21884–21889
Kane MA, Folias A, Napoli J (2008) HPLC/UV quantitation of retinal, retinol, and retinyl esters in serum and tissues. Anal Biochem 378:71–79
Kastner P, Leid M, Chambon P (1994) The role of nuclear retinoic acid receptors in the regulation of gene expression. Vitamin A in health and disease. Marcel Dekker, New York, pp 189–238
Kawamura K, Hara K, Fujiwara S (1993) Developmental role of endogenous retinoids in the determination of morphallactic field in budding tunicates. Development 117:835–845
Keay J, Thornton JW (2009) Hormone-activated estrogen receptors in annelid invertebrates: implications for evolution and endocrine disruption. Endocrinology 150:1731–1738
Kim CI, Leo MA, Lieber CS (1992) Retinol forms retinoic acid via retinal. Arch Biochem Biophys 294:388–393
Kostrouch Z, Kostrouchova M, Love W, Jannini E, Piatigorsky J, Rall JE (1998) Retinoic acid X receptor in the diploblast, Tripedalia cystophora. PNAS 95:13442–13447
Kuno S, Kawamoto M, Okuyama M, Yasumasu I (1999) Outgrowth of pseudopodial cables induced by all-trans retinoic acid in micro- mere-derived cells isolated from sea urchin embryos. Dev Growth Differ 41:193–199
Kuraku S (2011) Hox gene clusters of early vertebrates: do they serve as reliable markers for genome evolution? Genomics Proteomics Bioinform 9:97–103
Lalevée S, Anno YN, Chatagnon A, Samarut E, Poch O, Laudet V, Benoit G, Lecompte O, Rochette-Egly C (2011) Genome-wide in silico identification of new conserved and functional retinoic acid receptor response elements (direct repeats separated by 5 bp). J Biol Chem 286:33322–33334
Laudet V, Gronemeyer H (2002) The nuclear receptors factsbook. Academic Press, London
Lemotte PK, Keidel S, Apfel CM (1996) Phytanic acid is a retinoid X receptor ligand. Eur J Biochem 236:328–333
Marill J, Capron CC, Idres N, Chabot GG (2002) Human cytochrome P450s involved in the metabolism of 9-cis- and 13-cis-retinoic acids. Biochem Pharmacol 63:933–943
Markov GV, Paris M, Bertrand S, Laudet V (2008) The evolution of the ligand/receptor couple: a long road from comparative endocrinology to comparative genomics. Mol Cell Endocrinol 293:5–16
Markov GV, Laudet V (2011) Origin and evolution of the ligand-binding ability of nuclear receptors. Mol Cell Endocrinol 334:21–30
Markov GV, Tavares R, Dauphin-Villemant C, Demeneix BA, Baker ME, Laudet V (2009) Independent elaboration of steroid hormone signaling pathways in metazoans. PNAS 106:11913–11918
Marlétaz F, Holland L, Laudet V, Schubert M (2006) Retinoic acid signaling and the evolution of chordates. Int J Biol Sci 2:38–47
McKenna NJ, O’Malley BW (2002) Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465–474
Mendoza-Parra MA, Walia M, Sankar M, Gronemeyer H (2011) Dissecting the retinoid-induced differentiation of F9 embryonal stem cells by integrative genomics. Mol Syst Biol 7:538
Mic FA, Molotkov A, Benbrook DM, Duester G (2003) Retinoid activation of retinoic acid receptor but not retinoid X receptor is sufficient to rescue lethal defect in retinoic acid synthesis. PNAS 100:7135–7140
Moutier E, Ye T, Choukrallah MA, Urban S, Osz J, Chatagnon A, Delacroix L, Langer D, Rochel N, Moras D, Benoit G, Davidson I (2012) Retinoic acid receptors recognize the mouse genome through binding elements with diverse spacing and topology. J Biol Chem 287:26328–26341
Müller W (1984) Retinoids and pattern formation in a hydroid. J Embryol Exp Morph 271:253–271
Müller WE, Binder M, von Lintig J, Guo YW, Wang X, Kaandorp JA, Wiens M, Schröder HC (2011) Interaction of the retinoic acid signaling pathway with spicule formation in the marine sponge Suberites domuncula through activation of bone morphogenetic protein-1. Biochim Biophys Acta 1810:1178–1194
Napoli JL (1999) Retinoic acid: its biosynthesis and metabolism. Prog Nucleic Acid Res Mol Biol 63:139–188
Nishikawa J, Mamiya S, Kanayama T, Nishikawa T, Shiraishi F, Horiguchi T (2004) Involvement of the retinoid X receptor in the development of imposex caused by organotins in gastropods. Environ Sci Technol 38:6271–6276
Nowickyj SM, Chithalen JV, Cameron D, Tyshenko MG, Petkovich M, Wyatt GR, Jones G, Walker VK (2008) Locust retinoid X receptors: 9-Cis-retinoic acid in embryos from a primitive insect. PNAS 105:9540–9545
Pappas RS, Newcomer ME, Ong DE (1993) Endogenous retinoids in rat epididymal tissue and rat and human spermatozoa. Biol Reprod 48:235–247
Petkovich PM (2001) Retinoic acid metabolism. J Am Acad Dermatol 45:136–142
Philip S, Castro LF, da Fonseca RR, Reis-Henriques MA, Vasconcelos V, Santos MM, Antunes A (2012) Adaptive evolution of the retinoid X receptor in vertebrates. Genomics 99:81–89
Pittlik S, Begemann G (2012) New sources of retinoic acid synthesis revealed by live imaging of an Aldh1a2-GFP reporter fusion protein throughout zebrafish development. Dev Dyn 241:1205–1216
Rhinn M, Dollé P (2012) Retinoic acid signalling during development. Development 139:843–858
Rinkevich Y, Paz G, Rinkevich B, Reshef R (2007) Systemic bud induction and retinoic acid signaling underlie whole body regeneration in the urochordate Botrylloides leachi. PLoS Biol 5:e71
Roberts ES, Vaz AD, Coon MJ (1992) Role of isozymes of rabbit microsomal cytochrome P-450 in the metabolism of retinoic acid, retinol, and retinal. Mol Pharmacol 41:427–433
Robinson-Rechavi M, Boussau B, Laudet V (2004) Phylogenetic dating and characterization of gene duplications in vertebrates: the cartilaginous fish reference. Mol Biol Evol 21:580–586
Romero R, Bueno D (2001) Disto-proximal regional determination and intercalary regeneration in planarians, revealed by retinoic acid induced disruption of regeneration. Int J Dev Biol 45:669–673
Rubin BS (2011) Bisphenol A: an endocrine disruptor with widespread exposure and multiple effects. J Steroid Biochem 127:27–34
Samarut E, Amal I, Markov GV, Stote R, Dejaegere A, Laudet V, Rochette-Egly C (2011) Evolution of nuclear retinoic acid receptor alpha (RARα) phosphorylation sites. Serine gain provides fine-tuned regulation. Mol Biol Evol 7:2125–2137
Samarut E, Rochette-Egly C (2011) Nuclear retinoic acid receptors: conductors of the retinoic acid symphony during development. Mol Cell Endocrinol 348:348–360
Schmidt CK, Brouwer A, Nau H (2003) Chromatographic analysis of endogenous retinoids in tissues and serum. Anal Biochem 315:36–48
Schubert M, Holland ND, Laudet V, Holland LZ (2006) A retinoic acid-hox hierarchy controls both anterior/posterior patterning and neuronal specification in the developing central nervous system of the cephalochordate amphioxus. Dev Biol 296:190–202
Sciarrino S, Matranga V (1995) Effects of retinoic acid and dimethylsulfoxide on the morphogenesis of the sea urchin embryo. Cell Biol Int 19:675–680
Shea C, Hough D, Xiao J, Tzertzinis G, Maina CV (2004) An rxr/usp homolog from the parasitic nematode, Dirofilaria immitis. Gene 324:171–182
Shimozono S, Iimura T, Kitaguchi T, Higashijima SI, Miyawaki A (2013) Visualization of an endogenous retinoic acid gradient across embryonic development. Nature 496:363–366
Simões-Costa MS, Azambuja AP, Xavier-Neto J (2008) The search for non-chordate retinoic acid signaling: lessons from chordates. J Exp Zoo B Mol Dev Evol 310:54–72
Storey DM (1982) Vitamin A deficiency and the development of Litomosoides carinii (Nematoda, Filarioidea) in cotton rats. Z Parasitenk 67:309–315
Sturchler D, Wyss F, Hanck A (1981) Retinol, onchocerciasis and Onchocerca volvulus. T Roy Soc Trop Med H 75:617
Tang GW, Russell RM (1990) 13-cis-retinoic acid is an endogenous compound in human serum. J Lipid Res 31:175–182
Thornton JW, Need E, Crews D (2003) Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science 301:1714–1717
Urushitani H, Katsu Y, Ohta Y, Shiraishi H, Iguchi T, Horiguchi T (2013) Cloning and characterization of the retinoic acid receptor-like protein in the rock shell, Thais clavigera. Aquat Toxicol 142–143:403–413
Viviano CM, Horton C, Maden M, Brockes JP (1995) Synthesis and release of 9-cis retinoic acid by the urodele wound epidermis. Development 121:3753–3762
White RJ, Schilling TF (2008) How degrading: Cyp26 s in hindbrain development. Dev Dyn 237:2775–2790
Wiens M (2003) Retinoid X receptor and retinoic acid response in the marine sponge Suberites domuncula. J Exp Biol 206:3261–3271
Wolff KM, Scott AL (1995) Brugia malayi: retinoic acid uptake and localization. Exp Parasitol 80:282–290
Wu BX, Moiseyev G, Chen Y, Rohrer B, Crouch RK, Ma JX (2004) Identification of RDH10, an all-trans retinol dehydrogenase, in retinal Müller cells. IOVS 45:3857–3862
Yang ZN, Davis GJ, Hurley TD, Stone CL, Li TK, Bosron WF (1994) Catalytic efficiency of human alcohol dehydrogenases for retinol oxidation and retinal reduction. Alcohol Clin Exp Res 18:587–591
Yao TP, Segraves WA, Oro AE, McKeown M, Evans RM (1992) Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell 71:63–72
Acknowledgment
This work was supported by funds from ANR (ANR-09-BLAN-0129-01, ANR-09-BLAN-0262-02, ANR-11-JSV2-002-01) and CNRS to Michael Schubert and Vincent Laudet.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Gutierrez-Mazariegos, J., Schubert, M., Laudet, V. (2014). Evolution of Retinoic Acid Receptors and Retinoic Acid Signaling. In: Asson-Batres, M., Rochette-Egly, C. (eds) The Biochemistry of Retinoic Acid Receptors I: Structure, Activation, and Function at the Molecular Level. Subcellular Biochemistry, vol 70. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-9050-5_4
Download citation
DOI: https://doi.org/10.1007/978-94-017-9050-5_4
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-017-9049-9
Online ISBN: 978-94-017-9050-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)