Abstract
Superficially, evolutionary engineering is a paradoxical field that balances competing interests. In natural settings, evolution iteratively selects and enriches subpopulations that are best adapted to a particular ecological niche using random processes such as genetic mutation. In engineering desired approaches utilize rational prospective design to address targeted problems. When considering details of evolutionary and engineering processes, more commonality can be found. Engineering relies on detailed knowledge of the problem parameters and design properties in order to predict design outcomes that would be an optimized solution. When detailed knowledge of a system is lacking, engineers often employ algorithmic search strategies to identify empirical solutions. Evolution epitomizes this iterative optimization by continuously diversifying design options from a parental design, and then selecting the progeny designs that represent satisfactory solutions. In this chapter, the technique of applying the natural principles of evolution to engineer microbes for industrial applications is discussed to highlight the challenges and principles of evolutionary engineering.
The authors Niti Vanee and Adam B. Fisher are contributed equally.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Cellular objectives
- Directed evolution
- Diversity
- Engineering objectives
- Evolutionary engineering
- Fitness landscapes
- Industrial microbiology
- Screening and selection
1 Introduction
The interaction of humans and microorganisms has a long history including the domestication of microbes as early as the fifth century BC for a variety of fermentation processes such as baking and viticulture. With the progression of time and scientific knowledge, microbiology has been applied to many industrial sectors including food, waste treatment, health and medicine, and more recently, energy. The microbes used in these processes have all been advantageous over other production methodologies due to the unique properties of life: a self-replicating system, capable of organizing highly complex, chaotic chemistry in response to constraints imposed by the surrounding state or environment. In this light, industrial microbiology utilizes the beneficial properties of microorganisms by programming cellular biochemistry to maximize production of a target chemical.
An additional interesting and potentially complicating aspect of microorganisms is that they evolve. Evolution, under natural conditions, is the process of change throughout a population over time: cycling between generation of diversity, and subsequent selection for the most ‘fit’ subpopulations. Classically, biology generates diversity through genetic mutation while natural selection acts as an evaluator of competitive fitness. In its most basic form, evolution can be considered to be an optimization function, finding maxima through iterative trial-and-error. Nature changes the genotypic composition of the cell through various non-directed phenomena such as point mutations, genome rearrangements and recombination (as well as proposed non-genetic mechanisms, i.e. epigenetics). These inherited changes are key determinants to the ultimately displayed phenotypic variations. However, the staggering complexity of even the smallest genomes makes it difficult to draw conclusive cause-and-effect relationships between the genotype and phenotype. Such unpredictability occludes ambitions of rational design and engineering of complex phenotypes without significant a priori knowledge; an important goal for industrial microbiology.
Historically, one of the most powerful methods to produce a desired phenotype has involved leveraging the innate optimization properties of evolution. Often industrial microbiology goals are at odds with normal cellular function, so aligning evolutionary and industrial goals can lead to productive results. By utilizing a diverse population and selecting subpopulations for incorporation in subsequent rounds of evolution, it is possible to employ evolution as a search function for optimal genotypes with respect to the desired phenotype. This practice is known as evolutionary engineering, and will be the focus of the following chapter. As alluded by the name, evolutionary engineering is conceptually characterized by parameters of both the evolutionary process and the engineering principles (Fig. 3.1). As such several specific evolutionary engineering (EvoEng) methodologies and examples will be discussed with particular emphasis placed upon theory and applications to the design of industrially-relevant microbes. Additionally, EvoEng will be evaluated for its implications in systems biology and synthetic biology, as well as the implications of advancements in DNA sequencing and synthesis technologies on the future of EvoEng.
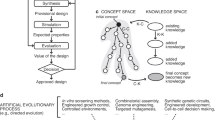
The work flow for Evolutionary Engineering draws on two different, but related cycles – the evolutionary optimization cycle and the engineering cycle. First a wild-type strain or seed sequence is selected as a platform to begin the Evolutionary engineering workflow. The seed sequence proceeds one of two ways: it can proceed to the engineering cycle, where alterations will be made rationally, or the seed sequence can be optimized through the evolutionary cycle. Both cycles return to the screening/selection node at which the resulting characterization of fitness confers either a satisfactory production strain or a seed sequence for further iteration
2 Optimization
In connecting evolutionary concepts to industrial microbiology design, an objective (e.g. production titer of biofuel) can be represented by a relative measure of fitness. This would allow for a visual and mathematical means of monitoring increases in a desired objective. The preferred method for visualizing adaptation toward fitness is a fitness landscape (Fig. 3.2), a three-dimensional fitness map where the X- and Y-axes are chosen to represent underlying contributing factors to fitness. The possible cellular functions that are depicted on a fitness landscape represent the solution space (SS) or the design space. With accurate fitness landscapes models can describe the topological landscape of the correlation between the X and Y projection to fitness (Kauffman and Levin 1987). These models are then compiled and used to make predictions about the behavior of future designs. Thereby, it becomes impossible to achieve accuracy or precision in the de novo design of optimized systems without, first, accurate data regarding the behavior of parts within the system.
Fitness landscapes. The three-dimensional fitness landscapes projected here represent the relationship between genotype and phenotype, with the two-dimensional Solution Space (SS) representing the genotype and all of its possible variations. During the evolutionary optimization process, genotypes diversify from a starting or seed sequence (Black dots) to related genotypes surrounding the seed sequence in the SS, while selection pressures to improve fitness provide a directionality to the evolutionary paths (Black arrows). As the topology is traversed, the global landscape and seed sequence will directly impact the final convergence. In (a) the fitness landscape is very homogenous, with one central peak, so all starting points will converge upon this global maxima, however, in (b) the multitude of local maximas allows many different paths of convergence even for the same seed sequence, although not every peak is accessible to the same seed sequence. In (b) this diversity of possible evolutionary paths is caused by the homogeneity of the rugged landscape; the slopes and peak heights are all identical, so trajectories have no bias in directionality and the different optima have no selectable fitness bias
If a framework can be developed to describe the fitness landscape/SS, then it may also be possible to utilize mathematical or algorithmic approaches for interrogating the space for design purposes. There are a variety of different approaches that could be employed including expectation-maximization algorithms, simulated annealing algorithms, or cost-benefit analysis modeling approaches (Dekel and Alon 2005; Gillespie 1984). These types of approaches could not only help identify a global maximum in a fitness landscape, but also help direct the best methods to evolutionarily reach that maximum.
2.1 The Fitness Landscape
The fitness landscape is classically a three-dimensional plot, which has a “fitness” score displayed on the Z-axis as a dependence of the X and Y variables. First introduced by Wright (1988, 1931), the fitness landscape in evolution uses the genome sequence as the X- and Y- axes, where the area of the two-dimensional plane formed by these two parameters represents the total SS for that particular genome. Within the total SS for an organism, each point represents one particular variant of the genome sequence. This projection of the fitness landscape can manifest as a monotonic (Fig. 3.2a) or a rugged landscape (Fig. 3.2b), which will greatly affect the movements around the SS. Each immediately neighboring point then represents one individual change in the genomic sequence. Given an Escherichia coli genome consists of ∼4.6 million base pairs of DNA, which have four possible outcomes (or informational units), the possible SS would be 44,600,000, as calculated by:
Where N is the number of possible sequences, λ is the number of informational units, and L is the length of the genome. Obviously, this is a fantastically large number that is impossible to explore by empirical experimental characterization of each possible variants.
2.1.1 Parameters of Fitness Landscapes
Fitness landscapes are an extremely useful tool in evolutionary analysis when they are created using tightly controlled constants. These fitness landscapes are generated under the assumption that a phenotype is correlated only to changes in genotype, however this is clearly untrue. Abiotic environmental factors can drastically change cellular dynamics and thereby, phenotype. From an only slight increase in incubation temperature, the global phenotype of a microbe could radically switch to a heat-shock response – which will create a very different projection of cellular fitness. Indeed, many of these global changes of phenotypes will also change the evolutionary parameters of the microbe; in times of stress such as heat-shock or starvation microbial populations will increase the rate of mutation (through error-prone DNA repair systems). These changes to the evolutionary parameters alters the topology of the fitness landscape, as well as the movement of subpopulations around the SS. Thus, the fitness landscape is fundamentally linked to genotype, but the detailed fitness landscape project can change based upon any number of factors that can influence gene expression.
Similarly, fitness landscapes are susceptible to the influences of coevolution. It is always important to remember that the process of evolution happens on a population-scale: any measure of an individual’s fitness is a relative quantification based upon the competitiveness of that particular variant against the other populations present. However, relationships may emerge between two variants that cause the exhibited phenotype to depend upon the presence of both strains. To observe such an evolutionary event it becomes important to maintain a level of heterogeneity of the culture, but have distinct separation of variants during the screening process.
In short, the parameters of the fitness landscape encompass a variety of factors that can alter the phenotype (fitness). These parameters that uncouple phenotype from genotype, have become the subject Epigenetics, coined by Conrad Waddington in the 1940s and conceptually illustrated using fitness landscapes (Siegal and Bergman 2002; Waddington 1942, 1959, 1960) Focusing on instances of non-coupled phenotypic and genotypic variation, epigenetics can alter an overall cellular fitness landscape, acting as a sort of underlying epigenetic landscape. After all, each cell in the human body contains identical genetic information, yet liver tissue is very different from gut mucosal tissue. For phenotypic optimization it is important to remain cognizant of these epigenetic parameters, which may be underlying the perceived genetic changes.
2.1.2 Traversing the Fitness Landscape Through Evolution
Optimization in a fitness landscape becomes the process of climbing peaks to find fitness maxima. This process is possible through the capability of evolution to (i) move about the SS, (ii) sense improvements in fitness and (iii) select for subpopulations with an improved fitness. The movement around the SS is dependent upon the rate of diversification, the topology of the landscape, and the seed sequences – the starting points in the SS. For a moment, imagine being a blind mountain climber with the goal of climbing to the highest peak in an uncharted mountain range. Without direction, the climber would attempt to achieve their goal by continually climbing upward. In this way we could climb upward in any direction until we reach a peak – however – much to our chagrin, we reach the top of what we thought was the highest peak, only to realize there was a taller peak hidden behind the peak we just climbed! Now we have no way of reaching the higher peak without descending into a valley. Without changing the process (always moving upward) one way to reach a different peak would be to have a different starting point.
This is a simple analogy, but it is easy to see what we have encountered is the problem of local maxima – we will not be able to reach the global maximum from our initial conditions. If we had, perhaps, started in a different position or not had the first mountain blocking the larger peak behind it, we could’ve climbed the tallest peak the first time. Another possible solution is to be able to move greater distances with each successive step – possibly allowing us to “jump” across valleys. These changes in simulation conditions reflect favorable selection of seed sequences or a tuning of the mutational rate. Either a priori knowledge of the system or utilizing many starting points can drastically improve identification of seed sequences; for many phenotypes this can be as general as selecting the proper organism (or chassis) for evolution. Changing the search movements around the SS by tuning mutational rates can be done through many in vivo (i.e. sexuality) and in vitro (i.e. in vitro recombination) methodologies, which will be covered in Sect. 3.4.
2.2 Length Scales
The shape and step-size for traversing a fitness landscape can change in relation to different types of mechanisms of generating genetic diversity. While the foundation for building a fitness landscape is DNA, the conceptual framework discussed to this point has focused on variations that occur within the context of an organism’s wild-type genome. All organisms have a basal genetic mutation rate for point mutations. Point mutations constitute the smallest length-scale of genetic change. Point mutations will generally make small, if any, change to the shape of the fitness landscape and represents the smallest step size for traversing the fitness landscape. Different length-scales of genetic modifications can be used to make larger changes to the shape of the fitness landscape and to more quickly traverse through the landscape. Whole gene deletions or additions represent a next level of genetic change followed by introduction/deletion of pathways. Recent approaches to industrial microbiology have explored the design and use of microbial consortia for directed production. This represents a broad scale approach where the fitness landscape would be defined by the capabilities of two independent genomes. Looking ahead, the broadest scope would be to consider design and the fitness landscape from a metagenomic perspective where any genetic information is possible and synthetic biology can be used to experimentally implement completely novel gene combinations.
3 Design Parameters
3.1 Rationality vs. Randomness
Adaptation over a period of time to confer a favorable and stable functional state is at the core of biological evolution. As mentioned previously, natural biological evolution occurs by the generation of genetic diversity (via random mutations) and selection according to improvements in fitness associated with survival and replication. The genetic information of the organism not only codes for the information related to how the organism should function in the current environment but also the potential for evolving with changing environments. Genotypic alterations that are positively selected by the environmental factors/selection are at the core of Adaptive Evolution (AE) (Atwood et al. 1951). Besides environment, engineering designs can be developed to implement evolution for real-world applications to attain desired characteristic traits or engineering objectives in parallel to maintaining the basic cellular objectives. This approach is one form of EvoEng and is defined as a rational approach toward the design and fabrication of cells to obtain a stable phenotype. These phenotypic objectives play a primary role in quantification of evolution for respective genotypic changes. In other words, there exists a one-way relationship between the genotypic and phenotypic components (i.e. the changes at the genetic level are translated into phenotype). EvoEng, attempts to simultaneously maximize engineering objectives and cellular objectives by incorporating rational design to select for the engineering objective and randomness of evolution to search across the SS.
Ultimately, the source of genetic variation is mutation. Mutations may occur spontaneously or be induced by external mutagens to achieve diversity to address desired cellular and or metabolic objective. Spontaneous mutations that occur naturally in the form of point mutations, genome rearrangements or horizontal gene transfers are considered to be relatively stable and occur at a low basal rate (Drake 1999). Natural environmental adversities such as nutrient deficiency or metabolic stress modulate the rate of such mutations. On the other hand there are external mutagens that can cause considerable changes in the environment of the organism leading to the phenomenon of frame shifts, deletion or insertions of the nucleotides.
Past researchers have proven through environment-dependent mutagenesis that engineering with evolution is sensitive to two major drawbacks. First, the rate of adaptation may not directly correlate to the rate of mutation. While comparing sexually reproducing populations and asexually reproducing populations it is known that the rate of mutation is elevated in asexual populations, yet does not alone accelerate the speed of evolutionary adaptation, for then sexuality would be outcompeted as a phenotype. Second, isolation of a mutant strain with desirable traits is highly dependent upon the selection and screening for the desired trait, which requires traits that are differential and quantifiable. To deal with these concerns, it is necessary to incorporate rationality in the design of an efficient and successful EvoEng investigation. When introducing rationality to evolution, there is a trade-off as rationality can constrain the possible evolutionary trajectories.
Metabolic engineering and Synthetic biology approaches utilize available gene-function information to take a fully rational approach to the design and construction of desired strains (Yokobayashi et al. 2002). In particular, Synthetic biology strives to establish bioengineering as a classical engineering discipline by developing methods for standardization, modularity and abstraction of biological parts (Valente and Fong 2011). This will enable biological design to occur in a high-throughput, rational fashion with all the benefits of a forward-engineering discipline.
Ideally, a completely rational approach would be taken in improving and altering production strains. In a complete rational design approach every cellular function could be designed to attain the optimal balance of cellular objectives with engineering objectives. However, this level of complete rational design requires absolute knowledge of the biological system – given the limitations of biological knowledge, this is not currently possible. At its core, EvoEng blends rationale with randomness by attempting to direct function towards a goal (rationale) by using the native cellular processes involved with evolution (randomness).
3.2 Establishing the Solution Space
The SS represents possible phenotypes based upon biological parameters. In Sect. 3.2, fitness landscapes were described as a method for visualizing and modeling local and global maxima of cell physiology, with the genotype considered as the underlying parameter that dictates the shape of the landscape. Establishing the shape of the fitness landscape is contingent upon a satisfactory knowledge of the underlying parameters. In this case, comprehensive evaluations of fitness landscapes were difficult prior to whole genome sequencing. However, a breadth of experiments probing AE without full genome sequencing has contributed rough fitness projections. In the study by Lenski et al., where Escherichia coli was evolved in a laboratory setting for more than 10,000 generations by simple serial dilutions of batch culture. In these studies, the average fitness (growth) of the derived genotype was increased by approximately 50% relative to the ancestor (Lenski et al. 1998). The shape of the fitness landscape is determined by the genomic sequence of E. coli movement across the landscape is related to mutations arising in the population, which are selected and characterized to explain the nature of the improved phenotype (Fong et al. 2005a, b). This process of AE and strain improvement is predicated by the starting genotype – what we have considered the seed sequence.
The best seed sequence is the one that produces a phenotype closest to the desired phenotypic maxima. This way the prospects for achieving an improved outcome are greatly increased as the required time to achieve the desired outcome is decreased. One proven method to improve seed sequence is to choose a fitting chassis. Chassis selection plays an important role in defining the SS as well as the culture conditions, which will be developed to underlying the industrial conditions. This way a design is optimized for certain environments, for example: enzymes have been evolved for thermostability by heterologous expression and selection in a thermostable chassis (Steipe 1999). As more AE experiments are conducted, results can be collectively analyzed to extrapolate genotype-phenotype relationships. In addition, collection of high-throughput “-omics” data provides detailed molecular-level gene expression data related to the global landscape. Improved knowledge of mechanisms leading to a desired trait can then be used to facilitate bottom-up specification of seed sequences for subsequent EvoEng or rational design approaches.
Besides favoring the improved starting point there is also a need to monitor the process of evolution with time to keep a track of each stage and collect data from each evolved generation. Keeping a record of the process allows us to monitor progress towards objectives as well as possibly collect information about major changes in trajectories. One particular 10,000 generation adaptation experiment illustrates a plot of competitive fitness generated through exemplary step measurement and monitoring (Lenski et al. 1998). With the rapid development of DNA technologies, sequencing has become much more feasible even allowing full genome re-sequencing projects to identify genetic mutations that occur across the genome as a result of AE (Atsumi et al. 2010; Barrick et al. 2009; Conrad et al. 2009; Gresham et al. 2008; Gabriel et al. 2006; Herring et al. 2006).
3.3 Competing Objectives
When considering the fitness landscape, the highest fitness is related to the phenotypic function with respect to a specific objective. In terms of EvoEng, there are normally two different objectives that often are at odds with each other, a cellular objective (e.g. growth) and an engineering objective (e.g. production of a target chemical).
3.3.1 Cellular Objective
In natural evolution organisms strive to maximize their competitiveness for an ecological niche – for microbes, this is usually through growth. We will refer to growth here as the cellular objective (CO), which is to maximize representation through increased reproduction or efficiency of biomass utilization. A cell must first incorporate nutrients to fuel cellular metabolic pathways, to finally carry out replication functions. For an entire cell, using the CO acts a measure of global fitness, which as we will see, may not always be the most optimal production strain.
3.3.2 Engineering Objective
Designing for industry requires the definition of an engineering objective (EO), which is the fitness score for industrial potential. Using either selection or screening (Sect. 3.4) the EO must be increased in each round. This objective can abstractly represent any phenotype, from resistance to a toxin to production of a useful intermediate, but the EO must overlap the CO if evolutionary selection toward the EO is to occur (Valente and Fong 2011). Further, maximizing both the CO and EO maximizes the fitness of a strain, but can be difficult to accomplish. When the target molecule for production is not a critical biomass component, the EO will not sufficiently overlap the CO and the cell will attempt to utilize its cellular resources to maximize its CO. Many of the target products of industry are secondary metabolites, which, by definition, fall into this latter category of targets. Hereby, in EvoEng the challenge is to design a selection system which can tie the EO to the CO. This may seem a difficult task, but consider the potential advantages of approaching engineering in a whole-cell optimization using the cells natural ability to grow and evolve.
3.4 Evolutionary Selection
Natural evolution processes achieve the diversity and complexity in the biosphere however the stability in this diversity is a function of natural selection. By having a continuous selection pressure, there is ongoing selection of beneficial characteristics, so a genotype with a fitness advantage will be maintained. Explaining in terms of the most simple AE, an evolved species acclimates to an environment and thrives, eventually replacing less adapted subpopulations (Hardin 1960). A major challenge faced by EvoEng is overlapping cellular COs that occur during evolution with EOs. By manipulating the growth environment and cellular interactions, natural selection is coupled to an engineering objective to perform selection in either in step-wise batch culture or continuous culture.
3.4.1 Selection Considerations
Using step-wise batch culturing, the culture is susceptible to several phenomena such as “Müller’s ratchet”. “Müller’s ratchet” appears in asexual populations through the accumulation of deleterious mutations as a side effect of nonspecific mutagenic targeting. This creates a strain overfit to its selection; the strain may satisfy the EO and be selected for, but becomes severely crippled from accumulation of deleterious mutations towards the cellular phenotype permitted to “hitchhike” along (Müller 1964). Competition-based selection may also create mutational dynamics in which several subpopulations rise to existence, each possessing different beneficial alleles. In a process known as “clonal interference” (Müller 1964) one subpopulation may gain a slight advantage and overrun the other populations – preventing incorporation of possible other beneficial mutations. By dominating these other mutations, there may be a perceived reduction in the mutational rates, as noted by Luria and Delbruck (1943). Step-wise batch culture is susceptible to the effects of genetic drift – as cultures are restarted in fresh media from a small portion of the original culture subpopulations may be lost, including adapted variants. Clonal interference and genetic drift can be minimized in chemostat-driven experimentation when a higher number of mutants remain in the population (Muller 1932). In contrast, serial dilution of batch cultures causes homogeneity as selective pressures favor sweeps from clonal interference and drift, purging the diversity from culture (Conrad et al. 2011).
While some phenomena (clonal interference and drift) can promote population homogeny, other phenomena such as cross-feeding, can lead to stable subpopulation diversity. Cross-feeding features an evolved, stable commensalism between the metabolism of two or more subpopulations. For instance, it has been shown that E. coli mutants grown in glucose-limited media over 773 generations will yield a cross-feeding adaptation where a majority of the population represents a glucose-feeder/acetate-excreting phenotype, while a smaller slower-growing part of the population uptakes and metabolizes acetate (Rosenzweig et al. 1994; Helling et al. 1987). This particular cross-feeding example has been replicated, and shown to sometimes evolve as a diauxic switch in the acetate-feeding strain. Here the acetate-feeder metabolizes glucose at a slower rate than the native glucose-scavengers, but has an advantage of switching to an acetate-based metabolism more quickly than the wild-type glucose-respiring strain (Friesen et al. 2004). In industry, this form of co-metabolism is usually unfavorable as the culture phenotype will be a synthesis of two separate genomic sequences – meaning there is no individual variant with the desired phenotype. However there does exist circumstances for the design of microbial consortia. Cross-feeding adaptations that arise from random mutations are often undesired, but rationally-designed cross-feeding between populations can be advantageous as part of system design. Examples include a study where a hydrogen-consuming Methanococcus maripaludis was evolved along the hydrogen-excreting, lactate-fermenter, Desulfovibrio vulgaris. The consumption of hydrogen by M. maripaludis fueled the thermodynamic driving force for the growth of D. vulgaris through lactate-fermentation (Hillesland and Stahl 2010).
3.4.2 Continuous-Culture Selection
Extended culture growth in chemostats has resulted in a variety of phenotypic adaptations (Helling et al. 1987; Sorgeloos et al. 1976; Atwood et al. 1951; Novick and Szilard 1950). In E. coli and S. cerevisiae carbon-limited chemostats have been used to increase biomass yield, growth rate and resistance to adverse conditions (Parekh et al. 2000; Vinci and Byng 1999; Rowlands 1984). The choice of limiting nutrient will dictate the evolutionary paths, and are therefore, central to selection. The results of limitation by nitrogen, phosphate, potassium, sulfur and other non-carbon source nutrients have been shown to be linked to the overproduction of various metabolic by-products (Dawson 1985). A very good review of adaptive laboratory evolution and continuous culture by U. Sauer, details the various formats of continuous culture including chemostats and their variants, batch culture, and microcolonization (Sauer 2001).
Since Environmental conditions can alter the evolutionary landscape so radically, the repeatability of EvoEng projects rely considerably on an experimenter’s ability to control and report culture conditions (selection pressure). Furthermore, in industrial-scale fermentations repeatability can be the most essential quality of a robust production strain. This starts by cultivating investigations as close to industrial conditions as possible. This includes, but is not limited to; aeration, carbon sources, nutrient sources, pH, osmolarity, temperature, light-exposure, or cell density.
Eventually, advances in synthetic biology may address some of the current challenges with screening and selection. There continues to be a challenge in EvoEng of having two different objectives (cellular and engineering) to consider. It may be possible to utilize different synthetic constructs (RNA apatmers, ribozymes, etc.), designed biosensors, or genetic circuits to detect multiple desired inputs and convey selective fitness advantages.
4 Methodology
Traditionally, EvoEng has been strongly associated with “Classical Strain Improvement”, through continuous culturing of a production-associated organism under selective conditions – usually paralleling industrial production conditions (Santos and Stephanopoulos 2008). However, like any science or engineering field, EvoEng has enjoyed significant advancements correlated with increases in technology. Recent improvements in sequencing, cloning, and high-throughput technologies have opened doors for researchers to investigate fitness landscapes and cellular optimization in unprecedented ways. This section will cover specific methodologies that have “evolved” to expand the EvoEng toolbox.
4.1 Generating Diversity
4.1.1 Point Mutations
Genetic diversity can occur by different mutagenic processes (SNPs, Insertions, and Deletions) that result in movement through the fitness landscape. If relying upon spontaneous mutagenesis as the main mechanism for phenotypic improvement, it is possible to conduct an evolutionary experiment with little foreknowledge of the system. A seminal piece of work, using only spontaneous improved penicillin titers 4,000-fold through selection on solid-media plates (Rowlands 1984). Naturally-occurring mutations actually occur at a nominally low and stable rate in normal cellular physiology, with DNA replicating faithfully (Drake 1999). This rate can be increased through the use of environmental influences or genetically-modified ‘mutator’ strains. Environmental factors can range from conditions which induce a stress-state in the cell (stationary-phase, glucose-limitation) to chemical mutagens: ethyl methane sulfonate (EMS) and nitroso-methyl guanidine (NTG) to ultraviolet irradiation (UV); all of which enhance various specific mutations (Rowlands 1982). For instance, it is known that NTG typically mutates close to the replication fork while UV irradiation is known to cause pyrimidine-dimers (Witkin 1976) – therefore mutagens are often varied in long-term experimentation. Utilizing these classic methods of spontaneous mutation has great utility as a fine-adjustment to the genotypic sequences. A mutation can occur at any part of the genome, the changes are completely independent of each other, and the rates of mutation can be controlled.
The original method of directing evolution has been most successful through the application of polymerase chain reaction (PCR). Using either ‘leaky’ DNA polymerase or a substitution of catalyzing ions (manganese as opposed to magnesium) to cause error-prone DNA polymerization. By designing primers to target your gene of interest, a large diversity of the enzyme is created which may be screened for desired phenotypes. More than likely, a wild-type enzyme will not be optimized for commercial purposes, but by utilizing PCR mutagenesis properties have been engineered into enzymes, such as thermostability, tolerance, novel catabolic activites, enantioselectivity, and substrate/product inhibition (Luetz et al. 2008).
4.1.2 Gene Modifications
Spontaneous mutagenesis can result in the silencing or overexpression of genes, but it is often difficult or costly to identify these mutations after they have occurred. For the purpose of quickly and inexpensively tracking genetic changes, transposons are widely available for use (de Lorenzo et al. 1998). These DNA elements are capable of self-catalyzing their insertion and movement across the genome or extra-chromosomal elements. Very often this will lead to inactivation of a gene on the chromosome, but some of these transposons feature highly expressive promoters, so it is also possible for transposon movement to result in gene overexpression if a transposon insertion is properly aligned next to a coding sequence (Schneider and Lenski 2004). However, their real faculty lies in the fact that these transposons represent unique sequence, which can be used to trace observed phenotype back to the cognate genotypic change.
Another way the genome can be randomly overexpressed is through a collection of an overexpression library. First, the genome is sheared into smaller pieces that are inserted into plasmid vectors. These vectors can then be reinserted into strains to select for variants advantaged by overexpression of the inserted genomic sequence. This is conceptually similar to the common genetics technique, complementation. Methods of utilizing knockouts or overexpressions are commonly used to establish the seed sequence for an evolutionary or metabolic engineering investigation. Some of the recent work that has demonstrated the utility of augmenting gene expression was conducted in E. coli for the production of lycopene (Jin and Stephanopoulos 2007). In fact, it was shown that screening a library of plasmid-encoded genomic segments inside of a previously evolved knockout strain of E. coli increased production of lycopene further over application of either method in isolation (Jin and Stephanopoulos 2007).
Recombination is a powerful genetic tool for generating general genetic diversity or for specifically targeting desirable traits. In EvoEng, the potential for recombination during sexual reproduction can be a useful tool to produce recombinant phenotypes between parental strains bearing beneficial phenotypes. Eukaryotic organisms such as S. cerevisiae, which can exists in diploid or haploid states, are capable of in vivo recombination by the fusion of two haploids to create a chimeric diploid. For example, attributes of a highly-specialized production strain might be combined with a fast-growing industrial strain. Indeed, industrial fungal production has utilized the mating of yeast to reintroduce accelerated growth rates and more efficient biomass conversion in previously crippled strains (Rowlands 1982). More importantly, recombination can allow two mutually beneficial mutations to recombine – possibly resulting in a strain with compounded effects to the beneficial traits.
We have already seen that in vivo recombination can occur in clonal populations from the self-excising movements of transposons. However, the normal methods by which new DNA sequences arrive inside a prokaryote involves conjugation, transformation, or transduction. Most in vitro expansion of diversity will utilize transformation to introduce desired genetic information on an overexpression plasmid, but other constructs can be used. Conjugative plasmids have been used in the dairy industry (Vinci and Byng 1999) and phage-based transduction has been utilized for allelic replacement (Esvelt et al. 2011).
In vitro recombination allows for generation of PCR products that are recombinant versions of the parental template strains. In one of the more simple and widely used processes, staggered extension process (StEP), the templates are amplified by outer primers, but is staggered by abbreviated elongation cycles (Zhao et al. 1998). These abbreviated cycles yield only partially extended templates that can bind to heterologous template strands with limited homology yielding final products that are chimeric versions of the original templates. In the original study using this method by Zhao et al. the authors were able to isolate a version of Subtilin E that showed thermostability 25–50 times that of the wild-type enzyme (Zhao et al. 1998).
One benefit of these methodologies lies in their ability to address the phenomena of epistasis – for instance, a scenario were two mildly deleterious mutations on their own can combine to render a beneficial phenotype (Conrad et al. 2011, 2009; Dykhuizen and Hartl 1980). Also, allelic replacement can be used as a means of influencing the outcome of movement through a fitness landscape by altering the starting point of a strain. While it would be difficult to try and isolate these mutations together in a spontaneous manner, in allelic replacement these options are fully investigable. Epistatic interactions manifest as complexities in an organism’s network topology and constrain the viable paths of evolution (Poelwijk et al. 2007).
4.1.3 Large-Scale Modifications
Genome shuffling can be viewed as a form of forced, accelerated recombination, as the process is leveraged for its ability to generate chimeras from a set of parental strains. This process, known as recursive protoplast fusion, relies upon the removal of the cell wall by polyethylene glycol (PEG) – a process used to prepare Gram-positive organisms for transformation of DNA. Protoplasts can also be fused, resulting in a diploid state in which the genomes exchange loci through homologous-crossing over. Since both genomes come from the same species the high level of homology will permit significant recombination, but by recursively fusing progeny of previous selections the SS searched grows considerably. When recursive protoplast fusion experiments were conducted in Streptomyces fradiae to investigate improved yield of the macrolide, tylosin, the authors we able to generate the same titers in two rounds of genome shuffling as accomplished by 20 rounds of classical mutagenesis (Zhang et al. 2002). In this way genome shuffling offers a strategy to do in vivo recombination in a variety of clonal populations to yield “progeny” with possible additive adaptive mutations (Petri and Schmidt-Dannert 2004).
4.1.3.1 Global Transcriptional Machinery Engineering
In adaptive laboratory evolution experimentation, mutations in the global transcriptional machinery can appear spontaneously (Conrad et al. 2010, 2009). Cultures of MG1655 E. coli grown in minimal glycerol M9 media showed mutations to rpoC of the β’ subunit of the RNA polymerase holoenzyme in ∼80 % sequenced mutants (Conrad et al. 2010).
While the core holoenzyme of RNA polymerase is the most conserved transcriptional machinery, targeting the sigma factors or factors giving the RNA polymerase its DNA specificity provide a greater modularity of control over distinct phenotypes. Using PCR mutagenesis, minor alterations in the DNA recognition motifs of these sigma factors (or their homologs) can greatly affect the RNA polymerase transcriptional kinetics toward the genes under control of the sigma factor. Using this approach the sigma 70 subunit in E. coli (normally controls housekeeping genes) was mutated to generate strains with increased ethanol tolerance, simultaneous sodium dodecyl sulfate and ethanol tolerance, and lycopene overproduction (Alper and Stephanopoulos 2007). Similarly, in yeast the genes encoding the TATA-binding protein, SPT15, and its associated factors, TAF25, were mutated to yield a strain of yeast with the industrially-relevant profile of high-glucose/ethanol tolerance, with an 70 % improvement in volumetric ethanol production (Alper et al. 2006). To emphasize the validity of targeting transcriptional hubs further, this process was repeated in a strain of Lactobacillus plantarum. Using growth/colony size as the phenotypic measurement, the approach of targeting transcriptional machinery was compared directly to mutagenesis by nitrosoguanidine chemical mutagenesis. In this study the investigators showed that targeting transcriptional machinery was able to create a greater diversity of phenotypic profiles than PCR mutagenesis alone. The greater degree of diversity generated by modifying aspects of transcriptional machinery made it possible to more rapidly isolate a strain of L. plantarum with an increased tolerance to malic acid.
4.1.3.2 Ribosomal Engineering
Just as the transcriptional machinery could be mutated to generate altered cellular phenotypic landscapes, the ribosomal machinery can be mutated to alter the global translational profile. A long history of studying the ribosome utilizing inhibitory antibiotics targeting the ribosome has continued as the preferred method for generating ribosomal variants. Termed “ribosome engineering”, variants are isolated by plating resistant cultures on differing concentrations of ribosomal targeting antibiotics. This application has yielded strains of Streptomyces capable of increased antibiotics production, increased α-amylase and protease production in Bacillus subtilis, and increased tolerance to aromatic compounds in Pseudomonas putida (Ochi 2007; Ochi et al. 2004). The varying phenotypes due to mutated ribosomes are postulated to correlate with an activation of the ‘stringent response’ when stress signals of stationary phase lead to increased protein production from select stationary-phase loci such as sporulation, alternative carbon source utilization, and production of secondary metabolites.
4.1.3.3 MAGE
Multiplex Automated Genome Engineering, or MAGE, has received considerable attention recently for the speed and scope of genetic changes that can be achieved. MAGE represents an extremely high-throughput methodology to perform major alterations across the entire genome. Mediated by homologous recombination, the process utilizes a fairly complex cultivation system featuring a series of growth chambers, an electrophoresis machine, a computer controller and a library of synthetic oligos (Wang et al. 2009). The synthetic oligo library contains the target mutations that are moved into the cell by electrotransformation, and incorporated through homologous recombination. This continuous culture system can run indefinitely to eventually generate all possible variants represented in the oligo library. In its initial application, MAGE was used to create E. coli pools containing over 15 billion genetic variants, targeting 24 separate genes to increase lycopene production 500% over wild-type lycopene producing strains (Wang et al. 2009). Ultimately, MAGE may be a powerful tool to modulate multiple genome targets simultaneously and with complete control. One of the current limiting factors of MAGE is the price of the technology, which currently still proves restrictive to academia, but may represent a viable opportunity for large-scale industrial projects.
4.1.3.4 TRMR
A variant of allelic replacement, TRMR (pronounced tremor) or trackable multiplex recombineering, has been used to construct libraries of up- and down-regulated variants of ∼96 % of the entire E. coli genome in less than a single week and for less than $1 per target (Warner et al. 2010). Yet this staggering diversity only represents half of the full potential of this approach. A specific strength of this approach is that all of the generated mutations were completely trackable by the incorporation of DNA barcodes. By hybridizing the each of the generated mutant strains to a DNA microarray, the relative levels of each mutation can be quantitatively measured. TRMR is then capable of rapidly identifying gene interactions that could then be used to structure further optimization projects. It has been pointed out that combining TRMR as an initial coarse-grained investigation of optimization, with the MAGE approach as a fine-grained adjustment could generate genome-wide, highly precise optimization (Tipton and Dueber 2010).
4.2 Functional Characterization
4.2.1 Whole-Genome [Re] Sequencing
Ultimately, the assumptions of EvoEng boil down to the relationship between sequence information and the resultant phenotype. Accordingly, the most fundamental method of screening relies on investigating the sequence information of the resulting mutants. Many clever selection schemes exist to screen populations by automatically removing undesired populations, but eventually an isolated population must be sequenced to identify genetic changes that may have arisen by natural random mutation. Again, directed evolution can help constrain the final sequencing requirements, but this means possible beneficial mutations inherited in the cellular background may be ignored.
As next-generation sequencing technologies have expanded the feasible limits on data collection and screening have expanded accordingly. Many recent EvoEng explorations have utilized next-gen sequencing to find changes on a genomic scale by comparing whole-genome sequencing of evolved strains to their ancestral strain. Evolutionary paths have been tracked by uncovering changes through single-nucleotide substitutions, insertions, deletions, and genomic rearrangements (Araya et al. 2010; Atsumi et al. 2010; Charusanti et al. 2010; Kishimoto et al. 2010; Lee and Palsson 2010; Lee et al. 2010; Barrick et al. 2009; Conrad et al. 2009; Gresham et al. 2008; Friedman et al. 2006; Herring et al. 2006; Velicer et al. 2006; Albert et al. 2005). Information gained by whole genome re-sequencing can be a useful source of information to feed into methods like MAGE or TRMR for a rational investigation into recombination adaptation. Further, whole genome sequencing preserves information and data that may be useful to the elucidation of cellular physiology to provide a holisitic systems biology view of cellular function (Conrad et al. 2011; Fong 2009).
Another approach to identifying genetic changes is a reapplication of DNA microarrays for array-based discovery of adaptive mutations (ADAM). In an EvoEng context, Goodzari et al. utilized a selectable marker, linked to a functional mutation (such as an insertion inactivation). Minimally, the ADAM approach requires a library of selectable markers transposed throughout the parental strain’s DNA, a mechanism for transferring markers from the parental strain to the evolved strain in such a way that the sequence surrounding the marker replaces the corresponding DNA in the evolved strain, and a method for measuring the frequency of markers throughout the evolved population (Goodarzi et al. 2009). Basically, if the newly evolved strain replaced a beneficial mutation with the parental sequence – a reversion in effect – then this evolved/parental chimeric would show a decrease in fitness. By then hybridizing to separate microarrays the fitness of the evolved strains and the “revertants” could be compared, pinpointing advantageous mutations by a decrease in signal from the “revertant” populations.
4.2.2 Additional High-Throughput Data
As related to the EvoEng approach, high-throughput data can be useful as a means of characterizing the phenotypic state of a cell in detail. This provides a means of gaining information to connect genotypic changes to whole-cell phenotypic changes.
Transcriptomics ushered in the advent of system-level quantitation of fundamental cellular components. While gene expression microarrays are the more established method of measuring transcriptomics data, recent studies have shifted more towards using RNA sequencing (RNAseq), especially since RNASeq data has been shown to correlate with microarray hybridization techniques in reproducibility and relative quantification (Alexeyev and Shokolenko 1995). Due to their lower cost, gene expression arrays are still advantageous for revealing the enrichment or depletion of clones as a consequence of selection (Kao 1999); accessing genes which confer a selective advantage or disadvantage.
In addition to transcriptomic data, proteomic and metabolomics data would prove valuable for investigating and understanding cellular function. Both of these data types have improved as technological advances have increased the reliability and scope of measurable data. For instance, a proteome analysis of Sacchromyces cerevisiae response to carbon and nitrogen limitation was done using multidimensional protein identification technology (MudPIT), combined with the labeling of proteins showed an up-regulation of protein in response to glucose limitation that was transcriptionally controlled, while the up-regulation in the presence of nitrogen occurred from regulation of a post-transcriptional nature (Kolkman et al. 2006). An example of a metabolomics study using a coupled detection of electrospray ionization in tandem with mass spectrometry (ESI-MS) was used to identify and measure of up to 84 % the metabolome of S. cerevisiae (Højer-Pedersen et al. 2008). In fact, recently a group published a high-throughput metabolomics workflow for investigating yeast in a multi-well format (Ewald et al. 2009).
4.3 Synthetic Biology
As a field, synthetic biology allows for better-controlled modification of a biological system. This started with the demonstration of synthetically generated genetic circuits (toggle switch and repressilator) that behaved in a designed fashion (Elowitz and Leibler 2000; Gardner et al. 2000). One application of synthetic biology can be to utilize designed synthetic constructs to sense and modify cellular function by developing novel circuits. For EvoEng, it may become possible to more directly couple COs and EOs using synthetic circuits.
4.3.1 SELEX
RNA is an extremely unique molecule in that it has been found in nature to be functional as an informational molecule, but also as a catalyst. Importantly, RNA polymers have the ability to form complex tertiary structures capable of specifically binding to small molecules and performing catalytic functions, such as self-cleavage. In 1990, the laboratories of G.E. Joyce (1989), J.W. Szostak (Ellington and Szostak 1990), and L. Gold (Tuerk and Gold 1990) independently developed a technique which allows the simultaneous screening of more than 1015 individual nucleic acid molecules for different functionalities (1–3 below). The selectable evolution of ligands by exponential enrichment (SELEX) is an EvoEng process in itself. It works by expanding a DNA library of oligos by PCR mutagenesis then selecting for variants capable of binding a target molecule. These isolates can be amplified through PCR methods, and the process can be iteratively repeated. For more details the reader is directed to several good reviews on the process (Sinha et al. 2011; Klug and Famulok 1994), or Aptamer base (Cruz-Toledo et al. 2012) (www.aptamer.freebase.com), a database of apatmer experimentation performed to date.
4.3.2 Designing Riboregulators
SELEX is an excellent tool to create RNA that binds a target molecule, altering a relatively small amount of sequence to search through a complexity of tertiary structures. However, as an in vivo biosensor the ability to bind target molecules alone does not convey a measurable signal. The trans-acting aptamer, or riboregulator, still requires a mechanism to alter its expressional state (Roth and Breaker 2009; Henkin 2008). In short, the RNA needs to have trans-binding activity for the target molecule and cis-regulatory properties. Much work has been done in the field to determine methods by which to integrate RNA aptamers with a regulatory motif in order to create programmable, in vivo biosensors (Lucks et al. 2011; Win and Smolke 2007; Isaacs et al. 2004). Recently, Qi et al., reported a method by which to program non-coding RNAs to suppress the expression of a target mRNA when an upstream aptamer domain was bound to a target molecule – building a post-translational NOR gate (Qi et al. 2012). All these design mechanisms will allow the construction of target specific, tunable, and orthogonal in vivo biosensors or regulators.
5 Interpreting the Results
Having discussed the application and utility of the natural evolution process and EvoEng it is important to consider the process for utilizing results for two major reasons: (1) To monitor whether or not we are moving in the right direction and (2) to record the evolutionary paths through genotypic variations and their effect at each level of cellular organization.
Tracking results and traits during evolution is important as explained in Sect. 3.4 to ensure that the system and selection pressure is leading to an outcome consistent with desired cellular and engineering objectives. The second consideration provides details of genetic changes and allows them to be correlated to phenotype i.e. collecting the data for each stage of step-wise continuous process of evolution. (Lenski et al. 1998) Monitoring the history of adaptation and using this information to model a particular cellular system enables researchers to make hypotheses for subsequent experiments.
5.1 Systems Biology
Systems-level modeling, especially the constraint-based approach, has proven to be a robust method to account for biological complexity while integrating high-throughput data. Using this tool in the cyclic process of evolutionary adaptation is discussed by many authors in past (Valente and Fong 2011; Fong 2009; Sauer 2001). Metabolic modeling, in conjunction with experimental high-throughput data discussed in Sect. 3.2, can be used to analyze potential targets for strain improvements to yield optimized productivity. Modeling simulations can reduce the cost and time needed for the EvoEng process(Fong et al. 2005a, b) by employing algorithmic analyses including: OptKnock (Burgard et al. 2003), objective tilting (Feist et al. 2010), RobustKnock (Tepper and Shlomi 2010) and OptGene (Patil et al. 2005). For example, metabolic flux analysis was applied to help understand the carbon metabolism of several E.coli strains under different growth conditions(Sauer et al. 1999). As for industrial applications, Lewis et al. have discussed methods that have been developed to predict, in silico, the growth-coupled desired phenotypes by performing deletions or insertions in the metabolic networks (Lewis et al. 2010). By encompassing data from previous iterations, it is possible to infer evolutionary trajectories as pointers for subsequent targets of adaptation.
Currently, constraint-based modeling approaches have been limited to metabolism and the biomass-assimilating pathways, ignoring portions of cellular physiology. In order to add detail to these models, efforts have been put forth to incorporate transcriptional regulation. (Gianchandani et al. 2006; Chandrasekaran and Price 2010; Thiele et al. 2010) and other cellular processes (Molenaar et al. 2009; Covert et al. 2008). This however, faces a major challenge of extrapolating the altered binding kinetics and motifs of the proteins because of the mutations performed in the regulatory proteins and ultimately affecting the topology of the regulatory network.
High-Throughput Biologically Optimized Search Engineering (HT-BOSE) introduced by Valente and Fong (Valente and Fong 2011), describes a method of leveraging EvoEng in conjunction with Systems biology and Synthetic biology. Systems-level information can provide a foundation to plan and conduct three-part cyclic process of evolutionary engineering. This biologically optimized search engine starts from a “Registry of Seed Designs”, similar to the “Registry of Biological Parts” (partsregistry.org). This Seed Design Registry would include phylogenetic information on how designs were evolved, as well as on what subsequent designs originated from them. Fitness scores are assigned to these designing depending upon how far they are from the desired objectives, yet the selection of a seed sequence from the registry should retain evolutionary flexibility. High fitness assays quantifying seed sequences satisfying the EOs, may reflect local maximums. It is therefore, important to switch between the fitness assays and a “supporting assay”, where the later is inclined toward the identification of evolutionary flexibility. Finally, the search strategy should maximum biological information by exploring SS at multiple length-scales.
6 Patenting Evolutionary Engineering
Throughout collaboration between academia, research, and large-scale industrial sectors there exists a movement away from publishing results in journals to prioritizing the invention protection by filing a patent, particularly in engineering and “applied” sciences (Leimkühler and Meyers 2004, 2005). While the timeline of the patent application process (Fig. 3.3) is extensive, patentability requirements and regulation under European and U.S. patent laws prescribe the exclusivity of invention to provide protection for a span of 20 years (an additional 5 years for pharmaceutical drug molecules). The validity of patent claims are predicated upon three features: (1) Novelty as defined in European Patent Convention (EPC) Article 54 and US regulation 35 U.S.C. § 102; (2) Inventiveness as defined in EPC Article 56 and 35 U.S.C. § 103 (a); (3) Industrial application as defined in EPC Article 57 and 35 U.S.C. § 101.
The typical patent application consists of the claim itself, a precise description of specifications, and the rationale for protection. In the specific field of EvoEng patents filed to date have claimed produced molecules with known function and utility, state-of-the-art production processes of molecules with known utility, or a selection process for useful traits in an already characterized strain. As explained in Fig. 3.1, the process of EvoEng consists of 3 steps: amplification, diversification and selection. Protection claims may be directed towards the steps in the cycle, specific conditions of the cycle, or improvements to previous patents (e.g. the specific and defined mutation conditions used to obtain variation and/or definite selection with respect to fitness toward an EO). The penultimate example of EvoEng intellectual property is the SELEX technique (WO 91/19813), discussed in Sect. 3.3.1 and its follow-up patents regarding variations and improvements (Leimkühler and Meyers 2004, 2005).
7 Conclusion
Recent supportive technological advancements have played an integral role in aiding the application of EvoEng to industrial targets. Rapid and inexpensive whole genome sequencing and other omics analyses have made it feasible to characterize the genotype of the strain with high fidelity and to associate the genotype to the phenotype (Lee et al. 2011). In the effort to reach a $1,000-genome (DeFraccesco 2012), there has been exponential progress from Sanger sequencing to fluorophores (Braslavsky et al. 2003), to Nanopore (Church et al. 1998; Kasianowicz et al. 1996) and other next-generation sequencing technologies. This progress has been compared to “Moore’s Law” and is predicted to continue in this manner into the immediate future with promises of technologies allowing over five kilobase contiguous reads (Hayden 2012; Clarke et al. 2009). Similarly, the technology for synthesizing DNA improves every day, with synthesis companies currently offering synthesis and delivery of 500 base pair double-stranded DNA oligos in 3–4 business days and for less than 100 US dollars (www.idtdna.com). Concurrent advances in cloning have enable even the smallest laboratories to assemble synthesized oligos up to several kilobases in length in under an hour (Gibson et al. 2009). Laboratory automation, microfluidics and other emergent screening technologies and have enabled the extreme increases in throughput and precision of rate-limiting steps in EvoEng such as serial dilution enrichment and chemostat selection (Grabar et al. 2006; Tyo et al. 2006; Zhou et al. 2006; Sonderegger and Sauer 2003). These impressive technologies open possibilities for whole genome sequencing and editing, in conjugation with massive “-omics” data collection. However, the success of implementing these technologies will remain dependent on our ability to interpret our findings – it is nice to own a fast car, but it does you no good when there is a low speed limit.
Metabolic engineering and forward-engineering cannot be reliably pursued without concrete genotype-phenotype correlations which equivocate with a priori knowledge of the system. Currently our level of knowledge of even the most elucidated model systems proves limiting. Selection for robust phenotypes by evolutionary engineering helps minimize instabilities such as those posed by genetic drift and clonal interference pictured in Fig. 3.4. For example, in order to improve the production of aromatic compounds, the phosphotransferase system (PTS) of E. coli was deleted and spontaneous glucose-utilizing revertants with increased aromatic titers were selected (Flores et al. 1996). Evolving the non-PTS system in E coli presumably preserved more phosphoenolpyruvate, a precursor of the aromatic targets. However, when a non-PTS heterologous system was rationally engineered into the PTS knockout, the improvement in target titers were not observed (Chen et al. 1997). Even with the phenotype successfully capitulated by rational engineering, there is been no optimization for evolutionary stability – a trait absolutely necessary for industrial continuous-culture. To address this pitfall, a hybrid-approach known as “inverse metabolic engineering” has been proposed (Bailey et al. 2002). Employing a rational-approach, targeted phenotypes can be isolated rapidly as seed sequences for a subsequent Evolutionary engineering approach for optimization. This rational “constraint” on SS to display the phenotype significantly reduces the time invested in optimization and identification of a seed sequence, particularly when transferring heterologous traits to a new chassis. Evolutionary engineering, inverse metabolic engineering nor any other bioengineering approach represents a full-proof approach to the design of industrial microbes. Many of the evolutionary engineering techniques presented in this chapter could be conjugated into more powerful hybrid systems capable of improved search functionality. Ultimately however, the future lies in innovating upon current selection procedures, improving the knowledge base of genotype-phenotype correlations, and harnessing advances in technologies.
Natural evolutionary process support the growth of square colonies over the period of evolution. Performing enrichment screening to obtain circled colonies might temporarily endows the culture with larger amount of those colonies however, because of squares’ evolutionary advantage they will continually outcompete the circled colonies. In contrast to the enrichment screening approach, synthetic intervention or selection can be useful in isolating clones with optimized for both cellular and engineering objectives
Abbreviations
- ADAM:
-
array-based discovery of adaptive mutations
- CO:
-
cellular objectives
- EMS:
-
ethyl methane sulfonate
- EO:
-
engineering objectives
- EvoEng:
-
evolutionary engineering
- MAGE:
-
multiplex automated genome engineering
- NTG:
-
nitroso-methyl guanidine
- Oligo(s):
-
oligonucleotide(s)
- RNAseq:
-
RNA sequencing
- SELEX:
-
selectable evolution of ligands by exponential enrichment
- SS:
-
solution space
- StEP:
-
staggered extension process
- TRMR:
-
trackable multiplex recombineering
References
Albert TJ, Dailidiene D, Dailide G, Norton JE, Kalia A, Richmond TA, Molla M, Singh J, Green RD, Berg DE (2005) Mutation discovery in bacterial genomes: metronidazole resistance in helicobacter pylori. Nat Method 2:951–953
Alexeyev MF, Shokolenko IN (1995) Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of gram-negative bacteria. Gene 160:59–62
Alper H, Stephanopoulos G (2007) Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng 9:258–267
Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314:1565–1568
Araya CL, Payen C, Dunham MJ, Fields S (2010) Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomic 11:88
Atsumi S, Wu TY, Machado IM, Huang WC, Chen PY, Pellegrini M, Liao JC (2010) Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol 6:449
Atwood KC, Schneider LK, Ryan FJ (1951) Periodic selection in Escherichia coli. Proc Natl Acad Sci U S A 37:146–155
Bailey JE, Sburlati A, Hatzimanikatis V, Lee K, Renner WA, Tsai PS (2002) Inverse metabolic engineering: a strategy for directed genetic engineering of useful phenotypes. Biotechnol Bioeng 79:568–579
Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF (2009) Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247
Braslavsky I, Hebert B, Kartalov E, Quake SR (2003) Sequence information can be obtained from single DNA molecules. Proc Natl Acad Sci U S A 100:3960–3964
Burgard AP, Pharkya P, Maranas CD (2003) Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng 84:647–657
Chandrasekaran S, Price ND (2010) Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and mycobacterium tuberculosis. Proc Natl Acad Sci U S A 107:17845–17850
Charusanti P, Conrad TM, Knight EM, Venkataraman K, Fong NL, Xie B, Gao Y, Palsson BO (2010) Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene. PLoS Genet 6:e1001186
Chen R, Hatzimanikatis V, Yap WM, Postma PW, Bailey JE (1997) Metabolic consequences of phosphotransferase (PTS) mutation in a phenylalanine-producing recombinant Escherichia coli. Biotechnol Prog 13:768–775
Church GM, Deamer DW, Branton D, Baldarelli R, Kasianowicz J (1998) Characterization of individual polymer molecules based on monomer-interface interactions. Harvard College EP0815438
Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H (2009) Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol 4:265–270
Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, Gao Y, Palsson BO (2009) Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol 10:R118
Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, Lewis NE, Landick R, Palsson BO (2010) RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A 107:20500–20505
Conrad TM, Lewis NE, Palsson BO (2011) Microbial laboratory evolution in the era of genome-scale science. Mol Syst Biol 7:509
Covert MW, Xiao N, Chen TJ, Karr JR (2008) Integrating metabolic, transcriptional regulatory and signal transduction models in Escherichia coli. Bioinformatics 24:2044–2050
Cruz-Toledo J, McKeague M, Zhang X, Giamberardino A, McConnell E, Francis T, Derosa MC, Dumontier M (2012) Aptamer base: a collaborative knowledge base to describe aptamers and SELEX experiments. Database J Biol Databases Curation 2012: bas006
Dawson PSS (1985) Continuous cultivation of microorganisms. CRC Crit Rev 2:315–372
de Lorenzo V, Herrero M, Sánchez JM, Timmis KN (1998) Mini-transposons in microbial ecology and environmental biotechnology. FEMS Microbiol Ecol 27:211–224
DeFraccesco L (2012) Life technologies promises $1,000 genome. Nat Biotechnol 14:126
Dekel E, Alon U (2005) Optimality and evolutionary tuning of the expression level of a protein. Nature 436:588–592
Drake JW (1999) The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann N Y Acad Sci 870:100–107
Dykhuizen D, Hartl DL (1980) Selective neutrality of 6PGD allozymes in E. coli and the effects of genetic background. Genetics 96:801–817
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Elowitz MB, Leibler S (2000) A synthetic oscillatory network of transcriptional regulators. Nature 403:335–338
Esvelt KM, Carlson JC, Liu DR (2011) A system for the continuous directed evolution of biomolecules. Nature 472:499–503
Ewald JC, Heux S, Zamboni N (2009) High-throughput quantitative metabolomics: workflow for cultivation, quenching, and analysis of yeast in a multiwell format. Anal Chem 81:3623–3629
Feist AM, Zielinski DC, Orth JD, Schellenberger J, Herrgard MJ, Palsson BO (2010) Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab Eng 12:173–186
Flores N, Xiao J, Berry A, Bolivar F, Valle F (1996) Pathway engineering for the production of aromatic compounds in Escherichia coli. Nat Biotechnol 14:620–623
Fong SS (2009) Evolutionary engineering of industrially important microbial phenotypes. Metab Pathw Eng Handb 1:1–15
Fong SS, Burgard AP, Herring CD, Knight EM, Blattner FR, Maranas CD, Palsson BO (2005a) In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol Bioeng 91:643–648
Fong SS, Joyce AR, Palsson BO (2005b) Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res 15:1365–1372
Friedman L, Alder JD, Silverman JA (2006) Genetic changes that correlate with reduced susceptibility to daptomycin in staphylococcus aureus. Antimicrob Agents Chemother 50:2137–2145
Friesen ML, Saxer G, Travisano M, Doebeli M (2004) Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution 58:245–260
Gabriel A, Dapprich J, Kunkel M, Gresham D, Pratt SC, Dunham MJ (2006) Global mapping of transposon location. PLoS Genet 2:e212
Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 403:339–342
Gianchandani EP, Papin JA, Price ND, Joyce AR, Palsson BO (2006) Matrix formalism to describe functional states of transcriptional regulatory systems. PLoS Comput Biol 2:e101
Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Method 6:343–345
Gillespie JH (1984) Molecular evolution over the mutational landscape. Evolution 38:1116–1129
Goodarzi H, Hottes AK, Tavazoie S (2009) Global discovery of adaptive mutations. Nat Method 6:581–583
Grabar TB, Zhou S, Shanmugam KT, Yomano LP, Ingram LO (2006) Methylglyoxal bypass identified as source of chiral contamination in l(+) and d(−)-lactate fermentations by recombinant Escherichia coli. Biotechnol Lett 28:1527–1535
Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ (2008) The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet 4:e103
Hardin G (1960) The competitive exclusion principle. Science 131:1292–1297
Hayden EC (2012) Sequencing set to alter clinical landscape. Nature 482(7385):288
Helling RB, Vargas CN, Adams J (1987) Evolution of Escherichia coli during growth in a constant environment. Genetics 116:349–358
Henkin TM (2008) Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev 22:3383–3390
Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, Palsson BO (2006) Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet 38:1406–1412
Hillesland KL, Stahl DA (2010) Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci U S A 107:2124–2129
Højer-Pedersen J, Smedsgaard J, Nielsen J (2008) The yeast metabolome addressed by electrospray ionization mass spectrometry: initiation of a mass spectral library and its applications for metabolic footprinting by direct infusion mass spectrometry. Metabolomics 4(4):393–405
Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ (2004) Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol 22:841–847
Jin YS, Stephanopoulos G (2007) Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab Eng 9:337–347
Joyce GF (1989) Amplification, mutation and selection of catalytic RNA. Gene 82:83–87
Kao CM (1999) Functional Genomic technologies: creating new paradigms for fundamental and applied biology. Biotechnol Prog 15:304–311
Kasianowicz JJ, Brandin E, Branton D, Deamer DW (1996) Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A 93:13770–13773
Kauffman S, Levin S (1987) Towards a general theory of adaptive walks on rugged landscapes. J Theor Biol 128:11–45
Kishimoto T, Iijima L, Tatsumi M, Ono N, Oyake A, Hashimoto T, Matsuo M, Okubo M, Suzuki S, Mori K, Kashiwagi A, Furusawa C, Ying BW, Yomo T (2010) Transition from positive to neutral in mutation fixation along with continuing rising fitness in thermal adaptive evolution. PLoS Genet 6:e1001164
Klug SJ, Famulok M (1994) All you wanted to know about SELEX. Mol Biol Rep 20:97–107
Kolkman A, Daran-Lapujade P, Fullaondo A, Olsthoorn MM, Pronk JT, Slijper M, Heck AJ (2006) Proteome analysis of yeast response to various nutrient limitations. Mol Syst Biol 2:2006.0026
Lee DH, Palsson BO (2010) Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a nonnative carbon source, L-1,2-propanediol. Appl Environ Microbiol 76:4158–4168
Lee HH, Molla MN, Cantor CR, Collins JJ (2010) Bacterial charity work leads to population-wide resistance. Nature 467:82–85
Lee JW, Kim TY, Jang YS, Choi S, Lee SY (2011) Systems metabolic engineering for chemicals and materials. Trends Biotechnol 29:370–378
Leimkühler M, Meyers H (2005) Patenting in evolutionary biotechnology evolutionary methods in Biotechnology. WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, pp 191–209
Lenski RE, Mongold JA, Sniegowski PD, Travisano M, Vasi F, Gerrish PJ, Schmidt TM (1998) Evolution of competitive fitness in experimental populations of E. coli: what makes one genotype a better competitor than another? Antonie van Leeuwenhoek 73:35–47
Lewis NE, Hixson KK, Conrad TM, Lerman JA, Charusanti P, Polpitiya AD, Adkins JN, Schramm G, Purvine SO, Lopez-Ferrer D, Weitz KK, Eils R, Konig R, Smith RD, Palsson BO (2010) Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol 6:390
Lucks JB, Qi L, Mutalik VK, Wang D, Arkin AP (2011) Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc Natl Acad Sci U S A 108:8617–8622
Luetz S, Giver L, Lalonde J (2008) Engineered enzymes for chemical production. Biotechnol Bioeng 101:647–653
Luria SE, Delbruck M (1943) Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511
Molenaar D, van Berlo R, de Ridder D, Teusink B (2009) Shifts in growth strategies reflect tradeoffs in cellular economics. Mol Syst Biol 5:323
Muller HJ (1932) Some genetic aspects of Sex. Am Nat 66:118–138
Müller HJ (1964) The relation of recombination to mutational advance. Mutat Res 106:2–9
Novick A, Szilard L (1950) Description of the chemostat. Science 112:715–716
Ochi K (2007) From microbial differentiation to ribosome engineering. Biosci Biotechnol Biochem 71:1373–1386
Ochi K, Okamoto S, Tozawa Y, Inaoka T, Hosaka T, Xu J, Kurosawa K (2004) Ribosome engineering and secondary metabolite production. Adv Appl Microbiol 56:155–184
Parekh S, Vinci VA, Strobel RJ (2000) Improvement of microbial strains and fermentation processes. Appl Microbiol Biotechnol 54:287–301
Patil KR, Rocha I, Forster J, Nielsen J (2005) Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinform 6:308
Petri R, Schmidt-Dannert C (2004) Dealing with complexity: evolutionary engineering and genome shuffling. Curr Opin Biotechnol 15:298–304
Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ (2007) Empirical fitness landscapes reveal accessible evolutionary paths. Nature 445:383–386
Qi L, Lucks JB, Liu CC, Mutalik VK, Arkin AP (2012) Engineering naturally occurring trans-acting non-coding RNAs to sense molecular signals. Nucleic Acids Res 40:5775–5786
Rosenzweig RF, Sharp RR, Treves DS, Adams J (1994) Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137:903–917
Roth A, Breaker RR (2009) The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78:305–334
Rowlands RT (1982) Industrial fungal genetics and strain selection. In: Smith JE, Berry DR, Kristiansen B (eds) The filamentous fungi”, volume 4 of “fungal technology”. Edward Arnold (Publishers) Ltd, London, pp 346–372
Rowlands RT (1984) Industrial strain improvement: mutagenesis and random screening procedures. Enzyme Microb Technol 6:3–10
Santos CNS, Stephanopoulos G (2008) Combinatorial engineering of microbes for optimizing cellular phenotype. Curr Opin Chem Biol 12:168–176
Sauer U (2001) Evolutionary engineering of industrially important microbial phenotypes. Adv Biochem Eng Biotechnol 73:129–169
Sauer U, Lasko DR, Fiaux J, Hochuli M, Glaser R, Szyperski T, Wuthrich K, Bailey JE (1999) Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J Bacteriol 181:6679–6688
Schneider D, Lenski RE (2004) Dynamics of insertion sequence elements during experimental evolution of bacteria. Res Microbiol 155:319–327
Siegal ML, Bergman A (2002) Waddington’s canalization revisited: developmental stability and evolution. Proc Natl Acad Sci U S A 99:10528–10532
Sinha J, Topp S, Gallivan JP (2011) From SELEX to cell dual selections for synthetic riboswitches. Methods Enzymol 497:207–220
Sonderegger M, Sauer U (2003) Evolutionary engineering of saccharomyces cerevisiae for anaerobic growth on xylose. Appl Environ Microbiol 69:1990–1998
Sorgeloos P, Van Outryve E, Persoone G, Cattoir-Reynaerts A (1976) New type of turbidostat with intermittent determination of cell density outside the culture vessel. Appl Environ Microbiol 31:327–331
Steipe B (1999) Evolutionary approaches to protein engineering. Curr Top Microbiol Immunol 243:55–86
Tepper N, Shlomi T (2010) Predicting metabolic engineering knockout strategies for chemical production: accounting for competing pathways. Bioinformatics 26:536–543
Thiele I, Fleming RM, Bordbar A, Schellenberger J, Palsson BO (2010) Functional characterization of alternate optimal solutions of Escherichia coli’s transcriptional and translational machinery. Biophys J 98:2072–2081
Tipton KA, Dueber J (2010) Shaking up genome engineering. Nat Biotechnol 28:812–813
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Tyo KE, Zhou H, Stephanopoulos GN (2006) High-throughput screen for poly-3-hydroxybutyrate in Escherichia coli and Synechocystis sp. Strain PCC6803. Appl Environ Microbiol 72:3412–3417
Valente AXCN, Fong SS (2011) High-throughput biologically optimized search engineering approach to synthetic biology. eprint arXiv:1103.5490
Velicer GJ, Raddatz G, Keller H, Deiss S, Lanz C, Dinkelacker I, Schuster SC (2006) Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc Natl Acad Sci U S A 103:8107–8112
Vinci VA, Byng G (1999) Strain improvement by non recombinant methods. In: Demain AL, Davies JE (eds) Manual of industrial microbiology and biotechnology, second edition. ASM Press, Washington, DC, pp 103–113
Waddington CH (1942) Canalization of development and the inheritance of acquired characters. Nature 150:563–565
Waddington CH (1959) Evolutionary systems; animal and human. Nature 183:1634–1638
Waddington CH (1960) Experiments in canalising selection. Genet Res 1:140–150
Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM (2009) Programming cells by multiplex genome engineering and accelerated evolution. Nature 460:894–898
Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LB, Gill RT (2010) Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol 28:856–862
Win MN, Smolke CD (2007) A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci U S A 104:14283–14288
Witkin EM (1976) Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev 40:869–907
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Wright S (1988) Surfaces of selective value revisited. Am Nat 131:115–123
Yokobayashi Y, Weiss R, Arnold FH (2002) Directed evolution of a genetic circuit. Proc Natl Acad Sci U S A 99:16587–16591
Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WP, del Cardayre SB (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:644–646
Zhao H, Giver L, Shao Z, Affholter JA, Arnold FH (1998) Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat Biotechnol 16:258–261
Zhou S, Shanmugam KT, Yomano LP, Grabar TB, Ingram LO (2006) Fermentation of 12 % (w/v) glucose to 1.2 M lactate by Escherichia coli strain SZ194 using mineral salts medium. Biotechnol Lett 28:663–670
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Vanee, N., Fisher, A.B., Fong, S.S. (2012). Evolutionary Engineering for Industrial Microbiology. In: Wang, X., Chen, J., Quinn, P. (eds) Reprogramming Microbial Metabolic Pathways. Subcellular Biochemistry, vol 64. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-5055-5_3
Download citation
DOI: https://doi.org/10.1007/978-94-007-5055-5_3
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-5054-8
Online ISBN: 978-94-007-5055-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)