Abstract
The cadherin–catenin complex is the major building block of the adherens junction. It is responsible for coupling Ca2+-dependent intercellular junctions with various intracellular events, including actin dynamics and signaling pathways. Determination of three-dimensional structures of cadherins, p120 catenin, β-catenin and α-catenin at atomic-level resolution has allowed us to examine how the structure and function of cell adhesion molecules are further modulated by protein–protein interactions. Structural studies of cadherins revealed the strand-swap-dependent and -independent trans-dimerization mechanisms, as well as a potential mechanism for lateral clustering of cadherin trans-dimers. Crystallographic and NMR analyses of p120 catenin revealed that it regulates the stability of cadherin-mediated cell–cell adhesion by associating with the majority of the E-cadherin juxtamembrane domain, including residues implicated in clathrin-mediated endocytosis and Hakai-dependent ubiquitination. Crystal structures of the β-catenin/E-cadherin complex and the β-/α-catenin chimera revealed extensive interactions necessary to form the cadherin/β-catenin/α-catenin ternary complex. Structural characterization of α-catenin has revealed conformational changes within the N-terminal and modulatory domains that are crucial for its role as a mechanosensor of cell–cell adhesion. Further insights into the connection between the cadherin–catenin complex and the actin cytoskeleton are integral to better understand how adjoining cells communicate through cell–cell adhesion.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Actin Filament
- Intercellular Junction
- Hereditary Diffuse Gastric Cancer
- Classical Cadherins
- Catenin Complex
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
The multi-protein complex consisting of cadherin, a cell adhesion receptor, and its cytosolic binding partners, the catenins, is the major building block of intercellular junctions, such as the adherens junction (AJ). Adjoining cells can be physically connected when the extracellular regions of cadherins on adjacent cell surfaces form Ca2+ -dependent homophilic interactions (Hirano et al. 1987; Yoshida and Takeichi 1982). However, the adhesion of the extracellular regions of cadherin alone is insufficient to establish a stable, mature cell–cell contact; the intracellular region of cadherin establishes functional linkages to the actin cytoskeleton and various signaling pathways through catenins (Meng and Takeichi 2009). The resulting architecture of cadherin-catenin-mediated cell–cell junctions is stable enough to support tissue structure and integrity, yet remains sufficiently dynamic to quickly dissolve obsolete connections and foster new connections among neighbouring cells during embryogenesis and wound healing (Takeichi 1995). In contrast, the loss of cadherin expression or dissociation between cadherin and catenins can be induced by a number of factors, including transcriptional regulation, mutation and aberrant cadherin internalization (Mosesson et al. 2008), and has been associated with tumour invasiveness and metastasis (Hanahan and Weinberg 2000; Takeichi 1993).
To better understand how cadherins and catenins regulate cell–cell adhesion mechanisms, cell adhesion molecules found in AJs, namely classical cadherins, p120 catenin, β-catenin and α-catenin , have been subjected to extensive structural characterization for over 15 years. Biophysical techniques, such as X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy and electron microscopy (EM), have been successfully employed to yield invaluable atomic-level views of their three-dimensional structures, and more importantly the details regarding the protein–protein interactions that are indispensable for the structure and function of the cell adhesion complex. In this chapter, the roles of cadherins and catenins in the regulation of cell–cell adhesion will be discussed based on recently obtained structural information regarding how cadherins and catenins assemble into an entire cell adhesion complex.
2 Cadherin
2.1 The Overall Structure of Cadherin
Classical (Type I) cadherins, such as E- and N-cadherins , engage in Ca2+ -dependent homophilic interactions important for embryogenesis and development (Harris and Tepass 2010; Nishimura and Takeichi 2009). These cadherins comprise the most well characterized subfamily of the cadherin superfamily, which consists of a large number of cell surface receptors recognized by the presence of multiple extracellular cadherin (EC) domains (or ectodomains), ranging from 2 EC domains in the worm cadherin HMR-1 to 34 EC domains in Fat cadherins found in flies to mammals (Nollet et al. 2000). Classical cadherins are single-pass transmembrane proteins that contain five EC domains (EC1 -5) on the extracellular side and highly conserved catenin-associating domains on the intracellular side (Fig. 3.1a). Cadherins are essential for connecting intercellular junctions, such as AJs, to the actin cytoskeleton and various signaling pathways.
Three-dimensional structures of cadherin ectodomains. a Scheme of E-cadherin structure. E-cadherin consists of the extracellular cadherin domains 1–5 (EC1-5), the transmembrane region (T) and the cytoplasmic tail, which contains the juxtamembrane domain (JMD) and the catenin-binding domain (CBD). Amino acid residue numbers of cadherin are based on the mature form unless indicated otherwise. b Overlay of crystal structures of EC1-5 trans-dimers of E-cadherin (blue; PDB code 3Q2V) (Harrison et al. 2011), N-cadherin (green; PDB code 3Q2W) (Harrison et al. 2011) and C-cadherin (magenta; PDB code 1L3W) (Boggon et al. 2002). Structures of EC1-5 monomers from all three cadherins are virtually identical (see the superposed bottom chains), however their trans-dimer arrangements vary slightly due to small differences in the strand-swap EC1-EC1 dimer formation. c Close-up view of the strand-swap dimer interface of C-cadherin. The surface of one of two EC1 protomers is shown (gray) to highlight the hydrophobic pocket where the side chain of W2 from the adjacent protomer is buried. d The X-dimer interface of T-cadherin EC1-2 (PDB code 3K5S) (Ciatto et al. 2010) involves the Ca2+-binding sites. e Model of cadherin trans-dimerization and cis-interaction of cadherins during cell–cell adhesion
2.2 Extracellular Cadherin Domains
The first three-dimensional structures of N- and E-cadherin EC1 domains determined by crystallographic and NMR studies, respectively, revealed that the individual EC domain consists of ~110 residues forming seven anti-parallel β-strands arranged into an immunoglobulin-like β-sandwich fold (Overduin et al. 1995; Shapiro et al. 1995). More recently, high-resolution structures of the entire extracellular region (EC1-5) have been determined for three classical cadherins: E-, N- and C-cadherin s, and revealed a conserved arch-shaped overall structure, which stretches out ~190 Å from the cell surface (Fig. 3.1b) (Boggon et al. 2002; Harrison et al. 2011). All three EC1-5 structures are in the Ca2+ -bound state and five EC domains are rigidified by coordinating three Ca2+ ions between any two consecutive EC domains connected by a short linker region (Nagar et al. 1996). The binding of Ca2+ to cadherin is known to make its extracellular region resistant to proteolytic degradation (Takeichi 1988). In contrast, the Ca2+-free state of EC1-5 is more prone to proteolytic cleavage by trypsin, and has been shown to lose its rigidity and adopt a globular shape by EM (Pokutta et al. 1994).
The first EC (EC1 ) domain of cadherin is crucial for the affinity and specificity of its homophilic interaction (Nose et al. 1990; Tomschy et al. 1996). The molecular basis of the underlying dimerization mechanism was first unveiled when the crystal structure of the N-cadherin EC1 domain was determined in a dimeric state (Shapiro et al. 1995). The dimerization of EC1 domains involves the ‘strand-swap’ mechanism which involves two protomers exchanging the first β-strand so that the side chain of Trp2 is firmly buried into the hydrophobic pocket of the adjacent molecule (Fig. 3.1c) (Shapiro and Weis 2009). The presence of Trp as the second residue on the N terminus is critical for this process and is attained by proteolytic cleavage of the cadherin prodomain (discussed later) (Häussinger et al. 2004). The EC1 domains of type II-subfamily of cadherins, such as cadherin-8, cadherin-11 and VE-cadherin , have been also shown to engage in strand-swap dimerization similar to the ones observed for classical cadherins, but their interactions involve two N-terminal Trp residues (Trp2 and Trp4) being inserted into a larger hydrophobic pocket (Brasch et al. 2011; Patel et al. 2006). While the critical role of Trp2 in cadherin-mediated cell–cell adhesion was confirmed by mutagenesis/cell aggregation experiments (Tamura et al. 1998), the crystallographic (Shapiro et al. 1995) and cross-linking (Brieher et al. 1996; Troyanovsky et al. 2003) studies suggested the possibility of strand-swap interaction involved in cis -dimerization of cadherins on the same cell surface. Surprisingly weak affinity displayed by the strand-swap interaction of cadherins (K D = 80−720 μM) (Häussinger et al. 2004; Koch et al. 1997) also promoted the idea of cis-dimerization or lateral clustering playing a role in activating cadherins to form trans -dimerization between two adjoining cells (Leckband and Prakasam 2006; Stemmler 2008). However, single-molecule studies employing fluorescence resonance energy transfer and atomic force microscopy have shown that trans -dimerization of cadherin does not require prior formation of cis-dimers (Zhang et al. 2009). This is consistent with most currently available strand-swap dimer structures depicting cadherins as trans-dimers: the arch-shaped EC domains place the dimerized EC1 domains to be oriented in the similar direction while the C termini of protomers aim toward the opposite direction (Boggon et al. 2002; Brasch et al. 2011; Harrison et al. 2011; Patel et al. 2006).
2.3 Juxtamembrane and Catenin-Binding Domains of Cadherin
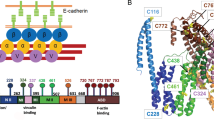
The cytoplasmic region of classical cadherin comprises of ~150 residues and contains several highly conserved sequence motifs (Fig. 3.1a) (Nollet et al. 2000). It can be further divided into two major domains: the juxtramembrane domain (JMD) and the catenin-binding domain (CBD). The JMD consists of ~50 residues immediately after the transmembrane domain and provides a specific binding site for p120 catenin and p120-related proteins, such as δ-catenin, ARVCF and p0071 (see below for further discussion) (Ishiyama et al. 2010; Thoreson et al. 2000). On the other hand, the CBD consists of the C-terminal ~100 residues and specifically binds to β-catenin and plakoglobin (Huber and Weis 2001) (see below for further discussion). Circular dichroism and proton NMR spectroscopic studies revealed that the recombinant form of the entire cadherin cytoplasmic region is largely unstructured in solution (Huber et al. 2001).
2.4 Other Dimerization Mechanisms of Cadherin
The strand-swap dimer mechanism alone cannot explain the observations from numerous biophysical studies of cadherin trans -dimerization suggesting that EC domains other than EC1 also contribute to adhesive forces produced by cadherins at different intermolecular bond distances (Shi et al. 2010; Sivasankar et al. 2001). One such example was recently revealed when the EC1-2 domains of T-cadherin were shown to dimerize through their Ca2+ -binding sites depicting a character ‘X’ (Fig. 3.1d) (Ciatto et al. 2010). T-cadherin is a divergent member of classical cadherin that is glycosylphosphatidylinositol-anchored, and lacks the N-terminal Trp required for the strand-swap dimer. Nonetheless, T-cadherin EC1-2 domains form X-dimer s with higher affinity (K D = ~40 μM) than the strand-swap dimer of E-cadherin EC1-2 domains (Ciatto et al. 2010). Interestingly, the X-dimer formation was previously observed with E-cadherin EC1-2 domains when they were crystallized with an N-terminal extension (Nagar et al. 1996). These observations suggest that other classical cadherins may be capable of facilitating trans -interaction via X-dimerization in addition to the strand-swap mechanism. Since there are multiple Ca2+-binding sites along the EC1-5 domains, it raises the possibility of other EC domain pairs, e.g., EC2-3, participating in trans-interactions of cadherins at intercellular junctions.
2.5 Clustering Through Cis -Interaction
The transformation of nascent cell–cell adhesion complexes into a stable intercellular junction is likely to involve lateral clustering of trans-dimers of cadherin (Fig. 3.1e). While the cytoplasmic region of cadherin has been implicated in cadherin clustering (Ishiyama et al. 2010; Yap et al. 1998), this process likely involves the cis -interaction of the EC domains as well (Yap et al. 1997). Visualization of the human epidermis by Cryo-EM tomography revealed a well organized architecture of the desmosome with the cell–cell junction mainly consisting of trans -dimers of desmosomal cadherins laterally packed at ~70 Å intervals (Al-Amoudi et al. 2007). The crystal packing contacts observed in the cadherin EC1 -5 domain crystals also offer additional clues to how trans -dimers of cadherins would participate in cis-interaction. In all three independently crystallized EC1-5 domains of E-, N- and C-cadherin s, it was observed that an EC1 surface opposite from the strand-swap interface interacts with the EC2 domain of an adjacent molecule, depicting cis-interaction of cadherin trans-dimer (Boggon et al. 2002; Harrison et al. 2011). Since the occurrence of a common crystal packing interface in three different crystals is extremely rare, it may be indicative of this cis-interface playing a physiological role in lateral clustering of cadherin trans -dimers. Indeed, mutations within the cis-interface (V81D/V175D) of E-cadheirn appears to interfere with proper formation of intercellular junctions (Harrison et al. 2011).
2.6 Post-Translational Modifications
Post-translational modifications of classical cadherins are a critical part of modulating the structure and function of cadherin adhesion receptors (Takeichi 1988). First, cadherins are N-glycosylated at numerous sites in the EC domains as part of the quality control process in the endoplasmic reticulum and Golgi network to ensure proper folding and stability (Boggon et al. 2002). Comparison of non-glycosylated and glycosylated EC1 -5 domains of VE-cadherin revealed that the glycosylation of the extracellular region affects the oligomeric state of VE-cadherin (Brasch et al. 2011). It was previously reported that the bacterially expressed recombinant protein of VE-cadherin EC1-4 domains (no posttranslational glycosylation) forms a hexamer made of two cis -trimers in trans-association (Legrand et al. 2001; Taveau et al. 2008). However, when VE-cadherin EC1-5 domains were produced using mammalian cell expression system, N-glycosylated recombinant proteins did not hexamerize, but instead formed strand-swap dimers in solution (Brasch et al. 2011), suggesting that the glycosyl moieties block the surface patches involved in the trimer/hexamer formation observed in vitro.
Second, a non-adhesive nascent cadherin molecule contains an N-terminal prodomain (residues 1–156 in mouse E-cadherin) (Takeichi 1988), which is cleaved by furin and other proprotein convertases in the trans-Golgi network to present the Asp-Trp pair as the first two residues of the EC1 domain (Ozawa and Kemler 1990; Stemmler 2008). The availability of these residues at the N terminus is critical for homophilic interaction of classical cadherins, as addition of one or two residues have been demonstrated to interfere with strand-swap dimerization (Ciatto et al. 2010; Häussinger et al. 2004; Nagar et al. 1996). Structure determination of the N-cadherin prodomain revealed that this region also has an EC-like fold despite a lack of sequence similarity with the rest of the EC domains (Koch et al. 2004).
Third, the adhesion function can be positively or negatively modulated by post-translational modifications of the cytoplasmic region of cadherin as well. Phosphorylation of Ser686 and Ser692 in the CBD promotes tighter binding with β-catenin , while tyrosine phosphorylation within the JMD residues Tyr599-Tyr600 recruits Hakai E3-ubiquitin ligase and induces ubiquitin-dependent internalization of cadherin (Fujita et al. 2002).
3 p120 Catenin
3.1 The Overall Structure of p120 Catenin
p120 catenin (p120) was first discovered as a prominent Src tyrosine kinase substrate (Reynolds et al. 1989), and subsequently recognized as an armadillo (arm) repeat containing protein which interacts with the cytoplasmic region of cadherin (Peifer et al. 1994; Reynolds et al. 1994; Reynolds et al. 1992). p120 specifically interacts with the juxtamembrane domain (JMD) that is located between the transmembrane domain and the β-catenin -binding domain of cadherin (Ishiyama et al. 2010; Thoreson et al. 2000). This interaction is critical for regulating the stability of cadherin–catenin cell–cell adhesion complexes at the cell surface, as downregulation of p120 results in aberrant internalization of cadherins (Davis et al. 2003; Xiao et al. 2003). Consistently, the loss, downregulation or mislocalization of p120 in tumors has been linked to poor prognoses (Thoreson and Reynolds 2002).
The p120 arm domain is flanked by an N-terminal regulatory region (NTR) and a C-terminal tail region (CTR), where the size of these regions depends on various isoforms resulting from multiple start codons (residues 1, 55, 102 and 324) and three alternatively spliced exons (A, B and C) (Fig. 3.2a) (Anastasiadis and Reynolds 2000). Numerous phosphorylation sites have been identified within both NTR and CTR (Mariner et al. 2001; Xia et al. 2003), but the functional consequence of these modifications remains unclear (Bauer et al. 1998). In addition, p120 binds to the Kaiso transcription repressor, which modulates non-canonical Wnt signaling (Daniel and Reynolds 1999; Kim et al. 2004; Park et al. 2006), through its arm domain. The NTR and a large insert within the arm domain of p120 have been shown to interact with Rho-GTPases, such as RhoA and Rac1 (Anastasiadis 2007; Yanagisawa et al. 2008). The recently determined crystal structure of the p120-E-cadherin complex has provided a first look at how this catenin specifically recognizes the cadherin JMD and regulates the internalization of cadherin–catenin complexes via endocytosis (Ishiyama et al. 2010).
The p120 catenin/E-cadherin juxtamembrane domain (JMD) complex. a Scheme of p120 catenin structure. The N-terminal region (NTR) contains a coiled-coil region (Coil) and four alternate start sites (residues 1, 55, 102 and 324). The arm domain contains nine arm repeats with exon C and a long insert between repeats 5 and 6. The C-terminal region (CTR) contains exon A and B. Binding sites for various p120-binding partners are indicated. b Crystal structure of the p120 catenin/E-cadherin JMD complex. p120 contains nine arm repeats (R1–9) with most repeats consisting of three α-helices (H1, H2 and H3), except for R1 and R8 (H3 shown in different colours; PDB codes 3L6X & 3L6Y) (Ishiyama et al. 2010). The JMDcore is shown in magenta (stick and space filling representation). c The surface electrostatic potential of the JMDcore-binding site of p120 with positively and negatively charged regions in blue and red, respectively (Ishiyama et al. 2010). 2F o -F c electron density map (green mesh; contoured at σ = 1.5) of the JMDcore (magenta) is shown. In the electrostatic interface, basic residues of p120 (e.g., K401 & K444) pair up with acidic residues of JMDcore (e.g., E604 & E608) to form several salt bridges. In the hydrophobic interface, the side chain of L618 is buried in the hydrophobic pocket of R1. d Model of dynamic and static interactions between p120 and the E-cadherin JMD. NMR studies revealed that JMD residues 580–590 (containing the endocytic LL motif (orange)) constitute the dynamic p120-binding site (yellow). JMD residues 591–625 (containing the JMDcore, the R593W cancer-associated mutation site in human E-cadherin (green) and the YY phosphorylation/Hakai-binding site (blue)) constitute the static p120-binding site. (cyan) (Ishiyama et al. 2010)
3.2 Armadillo Domain of p120
p120 was initially thought to contain as many as 10 arm repeats based on its amino acid sequence (Anastasiadis and Reynolds 2000; Reynolds and Roczniak-Ferguson 2004). A typical arm repeat consists of a ~40-residue motif forming three helices (H1, H2 and H3) arranged into a triangular shape (Huber et al. 1997). The crystal structure of a modified form of human p120 isoform 4A (containing a deletion in the arm insert region) in complex with the mouse E-cadherin JMD core fragment (JMDcore; residues 602–619) revealed that p120 contains a central arc-shaped arm domain (residues 368–825) with 9 arm repeats accompanied by a mostly disordered NTR (residues 324–367) and CTR (residues 826–933) (Fig. 3.2b) (Ishiyama et al. 2010). The p120 arm domain is similar to that of a closely related desmosomal molecule plakophilin-1 (PKP1) (Choi and Weis 2005) as both arm domains contain a long unstructured insert region between arm repeats 5–6, and the two structures can be superposed with a root mean square distance of 1.2 Å over 324 Cα atoms (Ishiyama et al. 2010). In addition, the CTR of p120 forms two α-helices that fold over the hydrophobic surface of arm repeat 9 (Fig. 3.2b). Though other p120 isoforms contain longer NTR, CTR and insert region than p120-4 A, the structure of the 9-repeat arm domain is likely conserved in all p120 isoforms, as well as other members of the p120-subfamily (p120, ARVCF, δ-catenin and p0071) and the PKP-subfamily (PKP-1,-2 and -3) of arm repeat proteins (McCrea and Gu 2010; McCrea and Park 2007).
3.3 p120-E-Cadherin Interfaces
Previous deletion and mutagenesis studies have determined that the 18-residue JMDcore region is critical for the binding of E-cadherin with p120, and more importantly for epithelial compaction (Ireton et al. 2002; Thoreson et al. 2000). In the crystal structure of the p120-JMD complex, the bound JMDcore peptide is stretched over the N-terminal half of the p120 arm domain in the opposite orientation (Fig. 3.2b) (Ishiyama et al. 2010). The JMDcore occupies part of the basic groove of the p120 arm domain formed by H3 helices of arm repeats 1–7 (Fig. 3.2c). The interaction between p120 and the JMDcore involves approximately 2400 Å2 of occluded solvent accessible surface area, and this large interface can be further divided into two different types of intermolecular interactions. The N-terminal acidic region (residues 602–610) of the JMDcore and p120 arm repeats 1–5 are involved in extensive electrostatic interactions, including the formation of five salt bridges formed between acidic residues (e.g., Glu604 and Glu608) from the cadherin JMDcore and basic residues (e.g., Lys401 and Lys444) from p120 (Fig. 3.2c) (Ishiyama et al. 2010). In the middle, the triple Gly motif (residues 605–607) of the JMDcore forms a turn that fits into a trough formed by p120, where the backbone of the JMDcore forms critical hydrogen bonds with p120 residues, including Asn478 (Fig. 3.2c). In comparison, the C-terminal half of the JMDcore (residues 611–619) and the N terminus of p120 are mainly involved in hydrophobic interactions (Fig. 3.2c). This region of the JMDcore is wedged between arm repeat 1 and the NTR of p120, resulting in the insertion or anchoring of the Leu618 side chain into an N-terminal hydrophobic pocket of the p120 arm domain. In VE-cadherin , Src -induced phosphorylation of Tyr611 (F613 in E-cadherin) in this region has been shown to prevent the binding of p120 (Potter et al. 2005). The importance of both electrostatic and hydrophobic interactions between p120 and the cadherin JMD is underscored by the strict conservation of the triple Gly motif with two flanking Glu residues (EGGGE) and the anchoring Leu in the JMDcore sequence from fly DE-cadherin to human E-cadherin (Ishiyama et al. 2010; Nollet et al. 2000).
The significance of the crystallographically determined JMD binding site of p120 was confirmed when single-residue mutations of p120, K401M and N478A, were shown to completely abolish the interactions of p120 with E- and N-cadherins by in vitro pull-down assays (Ishiyama et al. 2010). Compared to control p120, expression of these p120 mutants in p120-downregulated Madin-Darby canine kidney and MCF-7 cells resulted in complete loss of the p120-E-cadherin interaction and significantly reduced expression of E-cadherin at the cell perimeter. Furthermore, NMR and isothermal titration calorimetry studies revealed that p120 tightly binds to the majority of the JMD (residues 591–625), including the core, residues associated with a hereditary diffuse gastric cancer mutation of human E-cadherin (R593W) (Kaurah et al. 2007) and residues associated with Hakai -dependent ubiquitination /internalization of E-cadherin (Fujita et al. 2002), with a sub-micromolar affinity (Ishiyama et al. 2010). At the same time, it participates in a weak, dynamic interaction with the N-terminal clathrin-dependent endocytic motif (L587–L588), protecting it from endocytic proteins, such as the AP2 clathrin adaptor complex (Fig. 3.2d) (Kelly et al. 2008; Miyashita and Ozawa 2007a; Miyashita and Ozawa 2007b). These observations strongly suggest that these interfaces between p120 and E-cadherin JMDcore are crucial for p120 to colocalize with E-cadherin at the cell surface and to regulate the stability of cadherin–catenin complexes by countering cadherin internalization mechanisms at AJs (Ishiyama et al. 2010).
4 β-Catenin
4.1 The Overall Structure of β-Catenin
β-Catenin is an archetypal member of the armadillo repeat protein family, and plays an integral role in establishing adherens junctions by directly interacting with cadherin (McCrea and Gumbiner 1991; McCrea et al. 1991). It is also a critical transcriptional coactivator in the canonical Wnt signaling pathway that controls cell fate and proliferation when it forms a complex with members of Tcf/LEF-1 transcription factors in the nucleus (Graham et al. 2000). As β-catenin is involved in various protein–protein interactions that are crucial for embryogenesis, development and tumorigenesis, its population is tightly regulated by a dedicated degradation mechanism (Angers and Moon 2009). In the cadherin–catenin cell adhesion complex, β-catenin can simultaneously interact with cadherin, a cell adhesion molecule, and α-catenin , an actin-binding protein. The formation of a cadherin/β-catenin /α-catenin ternary complex is essential for linking cadherin-mediated cell–cell adhesion with actin dynamics (Meng and Takeichi 2009). The primary sequence of β-catenin is highly conserved from insects to humans, and its critical biological role in vertebrates is especially highlighted by strict conservation (> 95% sequence identity) of a 781-residue sequence from frogs to humans. The overall structure of β-catenin consists of three distinct domains, an N-terminal tail containing the α-catenin -binding site, a central arm domain that binds to the cytoplasmic region of cadherin and a C-terminal tail (Fig. 3.3a) (Shapiro and Weis 2009).
The β-catenin/E-cadherin catenin-binding domain (CBD) complex. a The full-length structure of β-catenin (PDB codes 1I7W, 1I7X & 2Z6G) consists of the central arm-repeat domain with 12 repeats (R1–12; orange) flanked by the N-terminal tail containing the α-catenin-binding site (blue) and the C-terminal tail that forms an additional α-helix (Huber et al. 1997; Huber and Weis 2001; Xing et al. 2008). The crystal structure of β-catenin/E-cadherin complex revealed that the entire arm domain is involved in associating with the CBD (rainbow coloured tube). The N-terminal tail helix is expected to change its conformation to accommodate the binding of α-catenin. b The arm repeat 5 of β-catenin contains three α-helices (H1–3) with a hydrophobic core. c Superposition of the p120/juxtamembrane domain (JMD)core and β-catenin/CBD complexes. p120 (purple) and β-catenin (navy) arm domains are shown as cylinders. JMD (green) and CBD (cyan) are shown as tubes. d Comparison of the basic arm grooves of p120 and β-catenin. p120 arm repeats 2–4 and β-catenin arm repeats 4–6 (PDB codes 1I7X & 3L6X) are superposed (Ishiyama et al. 2010). The Cα atoms of the JMDcore (green) and CBD (cyan) are shown as spheres
4.2 Armadillo Domain of β-Catenin
The arm domain (residues 146–662) of β-catenin is comprised of 12 arm repeats, which are sequentially packed together through hydrophobic interfaces into a superhelical architecture (Fig. 3.3a) (Huber et al. 1997). Structural determination of this domain was facilitated by crystallizing a protease-resistant, structurally stable fragment of β-catenin , containing residues 134–671, determined by limited trypsin digestion (Huber et al. 1997). Most of the arm repeats are comprised of ~40 residues forming three α-helices, H1, H2 and H3, that are arranged into a triangular shape (Fig. 3.3b). The β-catenin arm domain also contains atypical repeats: repeats 1 and 7 are missing H1 and repeat 10 contains a 15-residue insert between H2 and H3. The arm domain is slightly twisted and this results in consecutively ordered H3 helices forming a concave groove that is 95 Å long and 20 Å wide (Huber et al. 1997). Multiple basic residues from H3 and the first turn of H1 in arm repeats 1–10 give this groove a large positively charged surface critical for interacting with various ligands, including E-cadherin (Huber and Weis 2001). Besides the basic arm groove, the arm domain has an exposed hydrophobic pocket on repeat 1, which is also involved in ligand binding. More recently, structure determination of a full-length β-catenin from zebrafish has revealed that the C-terminal tail forms an additional α-helix that packs against the hydrophobic patch of repeat 12 (Xing et al. 2008).
4.3 β-Catenin -E-Cadherin Interfaces
Crystal structures of the β-catenin arm domain bound to either an unphosphorylated or phosphorylated E-cadherin cytoplasmic tail revealed the molecular basis of the intimate interaction between β-catenin and E-cadherin (Huber and Weis 2001). Although the crystallized β-catenin-E-cadherin complex contained the entire E-cadherin cytoplasmic region (residues 577–728), the structure of β-catenin -bound cadherin was limited to the CBD (residues 628–728) with the JMD (residues 577–627) remaining unbound and disordered (Huber et al. 2001). This is consistent with previous observation that β-catenin specifically interacts with the CDB of E-cadherin (Aberle et al. 1994; Yap et al. 1998). The interaction between β-catenin and E-cadherin involves all 12 armadillo repeats of β-catenin and the majority of the E-cadherin CBD. Similar to the p120-JMD complex (Ishiyama et al. 2010), the bound CBD peptide generally follows the concave groove of the arm domain in the opposite orientation so that the C terminus of CBD is bound to the N-terminal hydrophobic patch of β-catenin (Fig. 3.3a) (Huber and Weis 2001). The extensive binding interfaces can be further divided into five different regions (regions I–V) (Huber and Weis 2001). Most notably, region III forms the central interface involving residues 667–684 of E-cadherin forming critical electrostatic and hydrogen bonding interactions with arm repeats 4–9 of β-catenin (Fig. 3.3c). The transcription factor Tcf3 also utilizes the same binding interface to interact with β-catenin (Graham et al. 2000). Single-residue mutation of β-catenin residues, K312 and K435, involved in intermolecular salt bridge formation at this interface has been shown to abolish the interaction with the E-cadherin CBD (Graham et al. 2000). Interestingly, similar salt bridges are essential for the interaction between p120 and the E-cadherin JMD (Fig. 3.3d) (Ishiyama et al. 2010). On the other hand, region IV involves β-catenin -E-cadherin interactions that depend on the phosphorylation state of CBD residues 684–699. While the unphosphorylated CBD displayed disordered structure in region IV, phosphorylation of Ser684, Ser686 and Ser692 resulted in a stable interface, with phosphorylated Ser686 and Ser692 involved in additional ionic and hydrogen bonding interactions (Huber and Weis 2001). β-catenin has been shown to bind unphosphorylated E-cadherin cytoplasmic tail with a KD of 36 nM, but phosphorylation of CBD by casein kinase II increases the affinity to a KD of 52 pM (Choi et al. 2006). In contrast, phosphorylation of β-catenin residue Tyr654 located in arm repeat 12 by Src kinase has been shown to disrupt the binding of β-catenin to E-cadherin (Roura et al. 1999). The binding state of region IV also affects region V, which involves hydrophobic interaction between two anti-parallel α-helices formed at the C terminus of the CBD and the hydrophobic patch of β-catenin arm repeat 1 (Huber and Weis 2001).
4.4 α-Catenin -Binding Site
The α-catenin -binding site of β-catenin is located in the N-terminal tail (residues 118–149) immediately adjacent to region V of the β-catenin -cadherin interface (Aberle et al. 1994). The structure of the α-catenin -binding site in β-catenin in the absence of α-catenin has been observed as disordered (Huber et al. 1997) or as a long α-helix that further extends H2 of arm repeat 1 (Xing et al. 2008). When the α-catenin -binding site of β-catenin binds to α-catenin , this region forms two helices: a long helix (residues 120–141) and a short helix (residues 145–149) connected by a 3-residue linker (Pokutta and Weis 2000). These observations suggest that the cadherin-bound β-catenin could bind to α-catenin without any steric hindrance by forming a discrete α-helix within the α-catenin binding site (Huber and Weis 2001). A closely related plakoglobin (γ-catenin) also associates with E-cadherin and α-catenin at AJs, but does not recruit α-catenin to desmosome where it associates with desmosomal cadherins (Witcher et al. 1996). The crystal structure of a plakoglobin-E-cadherin CBD complex showed that observed interactions are virtually identical to the interactions between β-catenin and E-cadherin CBD (Choi et al. 2009). However, further biochemical studies reveal that the α-catenin -binding site is part of the desmosomal cadherin binding site, explaining the mutually exclusive nature of plakoglobin localization at the desmosome and its association with α-catenin (Choi et al. 2009).
5 α-Catenin
5.1 The Overall Structure of a-Catenin
α-catenin is a 102 kDa cytosolic protein implicated in anchoring the cadherin–catenin cell adhesion complex to the actin cytoskeleton at adherens junctions (Kobielak and Fuchs 2004). Unlike β-catenin and p120 catenin, it does not contain any armadillo repeat motifs and does not directly bind to the cadherin cytoplasmic region (Nagafuchi et al. 1991; Ozawa and Kemler 1992). Instead it is closely related to an actin-binding protein vinculin and it indirectly associates with cadherin by binding to the N-terminal segment of β-catenin bound to cadherin (Aberle et al. 1994; Ozawa et al. 1990). In addition, an αE-catenin homodimer has been shown to cross-link actin filaments (Rimm et al. 1995) as well as interfere with Arp2/3 -dependent actin polymerization/branching (Drees et al. 2005).
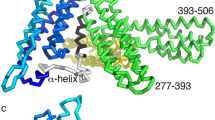
There are three known α-catenin subtypes in mammals, E (epithelial), N (neuronal), and T (prevalent in heart and testis), but invertebrates only express one homolog of α-catenin (Costa et al. 1998; Oda et al. 1993). Previous studies have revealed that α-catenin contains three major domains: an N-terminal (N) domain involved in β-catenin-binding and homodimerization (Aberle et al. 1994; Pokutta and Weis 2000); a modulatory (M) domain involved in binding to vinculin (Yang et al. 2001; Yonemura et al. 2010); and a C-terminal (C) domain involved in binding and bundling of actin filaments (Rimm et al. 1995) (Fig. 3.4a). All three domains contain vinculin homology regions (VH1, VH2 and VH3 in N, M and C domains, respectively) where α-catenin and vinculin share 25–35% sequence identity (Herrenknecht et al. 1991; Nagafuchi et al. 1991). As the N and C domains of α-catenin contain discrete binding sites for β-catenin and actin filaments, respectively, it was assumed that α-catenin would act as a stable linker between the cadherin–catenin complex and actin filaments (Gates and Peifer 2005; Weis and Nelson 2006). However, this ‘traditional’ model was called into question when αE-catenin was shown to interact with actin filaments only as a homodimer and not while being part of the cadherin–catenin complex by binding to β-catenin (Drees et al. 2005; Yamada et al. 2005). The Caenorhabditis elegans α-catenin homolog HMP-1, on the other hand, does not homodimerize and is auto-inhibited for actin binding as part of the complex or as a monomer (Kwiatkowski et al. 2010). These observations led to three models with different conformational states of α-catenin regulating its ability to interact with actin filaments (Fig. 3.4b) (Drees et al. 2005). The first model involves α-catenin dissociating from the cadherin–catenin complex to form homodimers to interact with actin filaments. The second model involves α-catenin bound to the cadherin–catenin complex adopting an active conformation to directly bind to actin filaments. The third model involves α-catenin bound to the cadherin–catenin complex binding to other actin-binding proteins to indirectly interact with actin filaments. Consistent with these models, several recent studies have reported that α-catenin acts as a mechanosensor at AJs: actomyosin-dependent forces trigger the conformational change in auto-inhibited α-catenin , which then recruits vinculin to cell–cell contact sites and links the cadherin–catenin complex with actin filaments both directly and indirectly (le Duc et al. 2010; Smutny et al. 2010; Yonemura et al. 2010).
The molecular architecture of α-catenin. a Scheme of the αE-catenin structure. It consists of N, M and C domains, which contain vinculin homology regions 1, 2 and 3, respectively. The N domain (blue) facilitates its homodimerization as well heterodimerization with β-catenin. The M domain (yellow) includes the vinculin binding site (Yonemura et al. 2010) and the adhesion modulation region (Yang et al. 2001). The C domain (red) is responsible for interacting with actin filaments (Rimm et al. 1995). b Models of α-catenin-mediated connections between the cadherin–catenin complex and actin filaments (F-actin). Model 1 shows auto-inhibited α-catenin dissociating from the cadherin–catenin complex resulting in its homodimerization, which induces the conformational change necessary to interact with F-actin (i). Model 2 shows auto-inhibited α-catenin bound to the cadherin–catenin complex changing its conformation to facilitate the direct connection to F-actin (ii). Model 3 shows auto-inhibited α-catenin bound to the cadherin–catenin complex changing its conformation to recruit other actin-binding proteins (brown) to the complex, allowing indirect connection to F-actin (iii). c Crystal structures of the α-catenin N domain in the homodimer arrangement (PDB code 1DOV) and the β-α-catenin chimeric protein depicting the heterodimer arrangement (PDB code 1DOW) (Pokutta and Weis 2000). d Crystal structures of the M fragments have been determined in the open and closed conformation. (PDB codes 1H6G & 1L7C)
5.2 N-Terminal Dimerization Domain
The structure of the N domain (residues 1–264) of αE-catenin has been determined in two dimeric states: a homodimer and a β- /α-catenin heterodimer (Pokutta and Weis 2000). The K D values for both αE-catenin homodimer formation and β-catenin binding have been estimated to be in the single micromolar range (Drees et al. 2005; Shapiro and Weis 2009). The heterodimer structure was determined by crystallizing a chimeric protein consisting of the α-catenin binding region of β-catenin (residues 118–151) fused to the N domain fragment of αE-catenin starting at residue 57. It consists of two sets of four-helix bundles connected by a long central helix. The N-terminal helical bundle contains an α-helix formed by β-catenin residues 120–141 (Fig. 3.4c). Interestingly, this chimeric structure of the α-catenin N domain highly resembles the N-terminal D1 domain structure of vinculin (its VH1 region shares 27% sequence identity) (Bakolitsa et al. 2004; Borgon et al. 2004), except the β-catenin helix is replaced by vinculin residues 9–33 forming its first N-terminal helix. Structural studies also suggest α-catenin and vinculin employ different heterodimerization mechanisms, as the vinculin/talin heterodimer complex structure resulted in a five-helix N-terminal bundle (with a talin fragment forming the fifth α-helix) instead of a mixed four-helix bundle observed in the β-/α-catenin chimera structure (Izard et al. 2004).
In comparison, the homodimer structure of the α-catenin N domain was determined by crystallizing a proteolysis-resistant fragment (residue 82–279) containing a region (residues 96–226) necessary for homodimerization (Koslov et al. 1997). N domain residues 82–258 in the homodimer state virtually adopt the same structure as the heterodimer, except for two α-helices in the N termini (residues 86–142) of two protomers which form an intermolecular four-helix bundle (Fig. 3.4c) (Pokutta and Weis 2000). Differences in the homodimer and heterodimer structures of the N domain suggests that the first α-helix (residues 57–83) of α-catenin observed in the β/α-catenin heterodimer is likely to pivot between open and closed conformations, making the homo- and hetero-dimerization of α-catenin mutually exclusive events (Fig. 3.4c) (Pokutta and Weis 2000). This is consistent with observations from other studies suggesting that αE-catenin bound to the cadherin-β-catenin complex does not directly associate with the actin cytoskeleton (Drees et al. 2005; Yamada et al. 2005).
5.3 Modulatory Domain
The modulatory (M) domain of αE-catenin consists of residues 277–631, which can be further divided into three subdomains: I, II and III (Fig. 3.4a). This region contains the VH2 region (residues 377–585) and shares 31% sequence identity with residues 582–796 of vinculin (Herrenknecht et al. 1991). Previously determined crystal structures of M domain fragments mostly consist of subdomains II (MII, residues 391–506) and III (MIII, residues 507–631) (Pokutta et al. 2002; Yang et al. 2001), and form a pair of four-helix bundles connected by a hinge region (Fig. 3.4d). Comparison of five independent crystal structures of the M fragment reveals that the hinge region connecting MII and MIII appears to be quite flexible as the angle between two bundles ranges from 57–100°. Recently, αE-catenin subdomain I (MI) was determined to contain the vinculin-binding site (residues 325–360), but the association of αE-catenin with vinculin is normally inhibited by MIII (Yonemura et al. 2010). These observations have led to a proposal that actomyosin-dependent conformational change within the M domain of αE-catenin attenuates the MIII-inhibition, resulting in the recruitment of vinculin to AJs (Yonemura et al. 2010).
5.4 C-Terminal Actin-Binding Domain
The C-terminal (C) domain of α-catenin is responsible for actin filament binding, and shares considerable sequence similarity with the D5 actin-binding domain of vinculin in the VH3 region (34% sequence identity) (Fig. 3.4a) (Herrenknecht et al. 1991). Previous studies have estimated the αE-catenin-F-actin interaction to have a K D value of 0.3 ± 0.4 μM, which is in the same affinity range between vinculin and F-actin (Johnson and Craig 1995), with the stoichiometry of one α-catenin dimer to 14 actin monomers (equivalent to an actin filament helical repeating unit) (Rimm et al. 1995). Nevertheless, α-catenin appears to have a distinct actin-binding mechanism involving an additional 42-residue tail (residues 865–906) that is not present in the C terminus of vinculin (Pokutta et al. 2002). A larger isoform of αN-catenin present during development has been shown to contain a 48-residue insertion after Gly810 in the C domain (Uchida et al. 1994). Although a high-resolution structure of the C domain of α-catenin remains elusive, VH3 region is expected have a similar fold as the five-helix bundle found in the D5 of vinculin (Bakolitsa et al. 1999). Determination of full-length vinculin structures revealed that one of two critical actin-binding interfaces within the D5 domain is occluded when vinculin is in its inactive closed conformation (Bakolitsa et al. 2004; Borgon et al. 2004). As vinculin has been shown to adopt an open conformation upon binding to various ligands, e.g., talin and phosphatidylinositol-4,5-bisphosphate (Bakolitsa et al. 2004; Winkler et al. 1996), it is tempting to speculate that activation of α-catenin could also involve modulation of inter-domain interactions (Fig. 3.4b).
6 The Cadherin–Catenin Cell Adhesion Complex
6.1 Hypothetical Model of the Cadherin–Catenin Complex
Since the determination of first high-resolution structures of E- and N-cadherin EC1 domains by NMR and X-ray crystallography over 15 years ago (Overduin et al. 1995; Shapiro et al. 1995), a nearly complete collection of three-dimensional structures of cadherins, catenins and their complexes have been determined, and more importantly, have provided invaluable atomic-level details about cadherin-catenin-dependent cell–cell adhesion mechanisms. To gain further insights into the multimeric arrangement of the cadherin–catenin complex in its entirety, a hypothetical model of the cadherin–catenin cell adhesion complex was constructed (Fig. 3.5). The core cell–cell adhesion complex consists of the E-cadherin ectodomain (PDB code 3Q2V; Harrison et al. 2011), the p120/JMD complex (PDB code 3L6X; Ishiyama et al. 2010), the β-catenin /CBD complex (PDB codes 1I7W and 1I7X; Huber and Weis 2001), and α-catenin fragments including the β-/α-catenin complex and the M domain (PDB codes 1DOW and 1H6G; Pokutta and Weis 2000; Yang et al. 2001). The model indicates that a single cadherin–catenin complex could take up an intracellular space with the dimensions of ~140 Å × ~140 Å × ~180 Å (length × width × height). However, cadherin–catenin complexes found in AJs are likely to occupy less space per complex by facilitating lateral clustering of both extracellular and intracellular components (Fig. 3.1e). In a mature intercellular junction, the presence of cadherin-bound p120, β-catenin and α-catenin in a tight space between the plasma membrane and the actin filament would restrict endocytic machineries and kinases from gaining access to the cytoplasmic tail of E-cadherin. The model also indicates the close proximity of the N terminus of p120 to the arm domain of β-catenin. This is consistent with the role of p120 in recruiting Fer kinase through its NTR to modulate the cadherin-β-catenin interaction (Lee et al. 2008; Xu et al. 2004). Additional structural studies are still pending to decipher whether α-catenin could interact with F-actin (PDB code 3B63; Cong et al. 2008) directly and/or indirectly via vinculin (PDB code 1ST6; Bakolitsa et al. 2004) and other actin-binding proteins (Fig. 3.4b).
Hypothetical model of the cadherin–catenin cell adhesion complex. The cadherin–catenin cell–cell adhesion complex consists of E-cadherin (PDB code 3Q2V), p120-catenin (PDB code 3L6X), β-catenin (PDB code 1I7W) and α-catenin (PDB codes 1DOW & 1H6G). α-catenin could either directly interact with F-actin (PDB code 3B63) or indirectly via vinculin (PDB code 1ST6) or other actin-binding molecules
7 Conclusion
Cadherin-mediated cell–cell adhesion requires intimate and intricate interactions between cadherins, catenins, and the actin cytoskeleton network. The structure and function of individual cell adhesion molecules are further modulated by protein–protein interactions, sometimes involving only a few amino acid residues. Three-dimensional structures of multiple classical cadherins, p120 catenin, β-catenin and parts of α-catenin have now been determined at atomic-level resolution, bringing considerable advantages to researchers in the field to further explore the relationships between the cadherin–catenin complex and various intracellular networks, including the actin cytoskeleton and numerous signaling pathways. With the recent recognition of α-catenin as a mechanosensor of cell–cell adhesion, precise structural information regarding the intermolecular relationships among cadherins, catenins and the actin cytoskeleton is indispensable to understand how adjoining cells communicate through cell–cell adhesion.
References
Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H (1994) Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J Cell Sci 107:3655–3663
Al-Amoudi A, Díez D, Betts M, Frangakis A (2007) The molecular architecture of cadherins in native epidermal desmosomes. Nature 450:832–837
Anastasiadis PZ (2007) p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim Biophys Acta 1773:34–46
Anastasiadis PZ, Reynolds AB (2000) The p120 catenin family: complex roles in adhesion, signaling and cancer. J Cell Sci 113:1319–1334
Angers S, Moon RT (2009) Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol 10:468–477
Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC (2004) Structural basis for vinculin activation at sites of cell adhesion. Nature 430:583–586
Bakolitsa C, de Pereda JM, Bagshaw CR, Critchley DR, Liddington RC (1999) Crystal structure of the vinculin tail suggests a pathway for activation. Cell 99:603–613
Bauer A, Lickert H, Kemler R, Stappert J (1998) Modification of the E-cadherin-catenin-complex in mitotic Madin-Darby Canine Kidney epithelial cells. J Biol Chem 273:28314–28321
Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L (2002) C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science 296:1308–1313
Borgon RA, Vonrhein C, Bricogne G, Bois PRJ, Izard T (2004) Crystal structure of human vinculin. Structure 12:1189–1197
Brasch J, Harrison OJ, Ahlsen G, Carnally SM, Henderson RM, Honig B, Shapiro L (2011) Structure and binding mechanism of vascular endothelial cadherin: a divergent classical cadherin. J Mol Biol 408:57–73
Brieher WM, Yap AS, Gumbiner BM (1996) Lateral dimerization is required for the homophilic binding activity of C-cadherin. J Cell Biol 135:487–496
Choi HJ, Weis WI (2005) Structure of the armadillo repeat domain of plakophilin 1. J Mol Biol 346:367–376
Choi HJ, Huber AH, Weis WI (2006) Thermodynamics of β-catenin-ligand interactions: the roles of the N- and C-terminal tails in modulating binding affinity. J Biol Chem 281:1027–1038
Choi HJ, Gross JC, Pokutta S, Weis WI (2009) Interactions of plakoglobin and β-catenin with desmosomal cadherins: basis of selective exclusion of α- and β-catenin from desmosomes. J Biol Chem 284:31776–31788
Ciatto C, Bahna F, Zampieri N, Vansteenhouse HC, Katsamba PS, Ahlsen G, Harrison OJ, Brasch J, Jin X, Posy S, Vendome J, Ranscht B, Jessell TM, Honig B, Shapiro L (2010) T-cadherin structures reveal a novel adhesive binding mechanism. Nat Struct Mol Biol 17:339–348
Cong Y, Topf M, Sali A, Matsudaira P, Dougherty M, Chiu W, Schmid MF (2008) Crystallographic conformers of actin in a biologically active bundle of filaments. J Mol Biol 375:331–336
Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR (1998) A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol 141:297–308
Daniel JM, Reynolds AB (1999) The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol 19:3614–3623
Davis MA, Ireton RC, Reynolds AB (2003) A core function for p120-catenin in cadherin turnover. J Cell Biol 163:525–534
Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI (2005) α-catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell 123:903–915
Fujita Y, Krause G, Scheffner M, Zechner D, Leddy HE, Behrens J, Sommer T, Birchmeier W (2002) Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol 4:222–231
Gates J, Peifer M (2005) Can 1000 reviews be wrong? Actin, α-Catenin, and adherens junctions. Cell 123:769–772
Graham TA, Weaver C, Mao F, Kimelman D, Xu W (2000) Crystal structure of a β-catenin/Tcf complex. Cell 103:885–896
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Harris TJC, Tepass U (2010) Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol 11:502–514
Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, Wu Y, Vendome J, Felsovalyi K, Hampton CM, Troyanovsky RB, Ben-Shaul A, Frank J, Troyanovsky SM, Shapiro L, Honig, B (2011) The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure 19:244–256
Häussinger D, Ahrens T, Aberle T, Engel J, Stetefeld J, Grzesiek S (2004) Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J 23:1699–1708
Herrenknecht K, Ozawa M, Eckerskorn C, Lottspeich F, Lenter M, Kemler R (1991) The uvomorulin-anchorage protein α catenin is a vinculin homologue. Proc Natl Acad Sci USA 88:9156–9160
Hirano S, Nose A, Hatta K, Kawakami A, Takeichi M (1987) Calcium-dependent cell-cell adhesion molecules (cadherins): subclass specificities and possible involvement of actin bundles. J Cell Biol 105:2501–2510
Huber AH, Nelson WJ, Weis WI (1997) Three-dimensional structure of the armadillo repeat region of β-catenin. Cell 90:871–882
Huber AH, Stewart DB, Laurents DV, Nelson WJ, Weis WI (2001) The cadherin cytoplasmic domain is unstructured in the absence of β-catenin. A possible mechanism for regulating cadherin turnover. J Biol Chem 276:12301–12309
Huber AH, Weis WI (2001) The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell 105:391–402
Ireton RC, Davis MA, van Hengel J, Mariner DJ, Barnes K, Thoreson MA, Anastasiadis PZ, Matrisian L, Bundy LM, Sealy L, Gilbert B, van Roy F, Reynolds AB (2002) A novel role for p120 catenin in E-cadherin function. J Cell Biol 159:465–476
Ishiyama N, Lee S-H, Liu S, Li G-Y, Smith MJ, Reichardt LF, Ikura M (2010) Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell 141:117–128
Izard T, Evans G, Borgon RA, Rush CL, Bricogne G, Bois PRJ (2004) Vinculin activation by talin through helical bundle conversion. Nature 427:171–175
Johnson RP, Craig SW (1995) F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature 373:261–264
Kaurah P, MacMillan A, Boyd N, Senz J, De Luca A, Chun N, Suriano G, Zaor S, Van Manen L, Gilpin C, Nikkel S, Connolly-Wilson M, Weissman S, Rubinstein WS, Sebold C, Greenstein R, Stroop J, Yim D, Panzini B, McKinnon W, Greenblatt M, Wirtzfeld D, Fontaine D, Coit D, Yoon S, Chung D, Lauwers G, Pizzuti A, Vaccaro C, Redal MA, Oliveira C, Tischkowitz M, Olschwang S, Gallinger S, Lynch H, Green J, Ford J, Pharoah P, Fernandez B, Huntsman D (2007) Founder and recurrent CDH1 mutations in families with hereditary diffuse gastric cancer. JAMA 297:2360–2372
Kelly BT, McCoy AJ, Späte K, Miller SE, Evans PR, Höning S, Owen DJ (2008) A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature 456:976–979
Kim SW, Park J-I, Spring CM, Sater AK, Ji H, Otchere AA, Daniel JM, McCrea PD (2004) Non-canonical Wnt signals are modulated by the Kaiso transcriptional repressor and p120-catenin. Nat Cell Biol 6:1212–1220
Kobielak A, Fuchs E (2004) α-catenin: at the junction of intercellular adhesion and actin dynamics. Nat Rev Mol Cell Biol 5:614–625
Koch AW, Pokutta S, Lustig A, Engel J (1997) Calcium binding and homoassociation of E-cadherin domains. Biochemistry 36:7697–7705
Koch AW, Farooq A, Shan W, Zeng L, Colman DR, Zhou M-M (2004) Structure of the neural (N-) cadherin prodomain reveals a cadherin extracellular domain-like fold without adhesive characteristics. Structure 12:793–805
Koslov ER, Maupin P, Pradhan D, Morrow JS, Rimm DL (1997) α-catenin can form asymmetric homodimeric complexes and/or heterodimeric complexes with β-catenin. J Biol Chem 272:27301–27306
Kwiatkowski AV, Maiden SL, Pokutta S, Choi H-J, Benjamin JM, Lynch AM, Nelson WJ, Weis WI, Hardin J (2010) In vitro and in vivo reconstitution of the cadherin-catenin-actin complex from Caenorhabditis elegans. Proc Natl Acad Sci USA 107:14591–14596
le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, de Rooij J (2010) Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol 189:1107–1115
Leckband D, Prakasam A (2006) Mechanism and dynamics of cadherin adhesion. Annu Rev Biomed Eng 8:259–287
Lee S-H, Peng I-F, Ng YG, Yanagisawa M, Bamji SX, Elia LP, Balsamo J, Lilien J, Anastasiadis PZ, Ullian EM, Reichardt LF (2008) Synapses are regulated by the cytoplasmic tyrosine kinase Fer in a pathway mediated by p120catenin, Fer, SHP-2, and β-catenin. J Cell Biol 183:893–908
Legrand P, Bibert S, Jaquinod M, Ebel C, Hewat E, Vincent F, Vanbelle C, Concord E, Vernet T, Gulino D (2001) Self-assembly of the vascular endothelial cadherin ectodomain in a Ca2+-dependent hexameric structure. J Biol Chem 276:3581–3588
Mariner DJ, Anastasiadis P, Keilhack H, Böhmer FD, Wang J, Reynolds AB (2001) Identification of Src phosphorylation sites in the catenin p120ctn. J Biol Chem 276:28006–28013
McCrea PD, Gu D (2010) The catenin family at a glance. J Cell Sci 123:637–642
McCrea PD, Gumbiner BM (1991) Purification of a 92-kDa cytoplasmic protein tightly associated with the cell-cell adhesion molecule E-cadherin (uvomorulin). Characterization and extractability of the protein complex from the cell cytostructure. J Biol Chem 266:4514–4520
McCrea PD, Park JI (2007) Developmental functions of the P120-catenin sub-family. Biochim Biophys Acta 1773:17–33
McCrea PD, Turck CW, Gumbiner B (1991) A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science 254:1359–1361
Meng W, Takeichi M (2009) Adherens junction: molecular architecture and regulation. Cold Spring Harbor Perspect. Biol 1:a002899
Miyashita Y, Ozawa M (2007a) A dileucine motif in its cytoplasmic domain directs β-catenin-uncoupled E-cadherin to the lysosome. J Cell Sci 120:4395–4406
Miyashita Y, Ozawa M (2007b) Increased internalization of p120-uncoupled E-cadherin and a requirement for a dileucine motif in the cytoplasmic domain for endocytosis of the protein. J Biol Chem 282:11540–11548
Mosesson Y, Mills GB, Yarden Y (2008) Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer 8:835–850
Nagafuchi A, Takeichi M, Tsukita S (1991) The 102 kd cadherin-associated protein: similarity to vinculin and posttranscriptional regulation of expression. Cell 65:849–857
Nagar B, Overduin M, Ikura M, Rini JM (1996) Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature 380:360–364
Nishimura T, Takeichi M (2009) Remodeling of the adherens junctions during morphogenesis. Curr Top Dev Biol 89:33–54
Nollet F, Kools P, van Roy F (2000) Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol 299:551–572
Nose A, Tsuji K, Takeichi M (1990) Localization of specificity determining sites in cadherin cell adhesion molecules. Cell 61:147–155
Oda H, Uemura T, Shiomi K, Nagafuchi A, Tsukita S, Takeichi M (1993) Identification of a Drosophila homologue of α-catenin and its association with the armadillo protein. J Cell Biol 121:1133
Overduin M, Harvey TS, Bagby S, Tong KI, Yau P, Takeichi M, Ikura M (1995) Solution structure of the epithelial cadherin domain responsible for selective cell adhesion. Science 267:386–389
Ozawa M, Kemler R (1990) Correct proteolytic cleavage is required for the cell adhesive function of uvomorulin. J Cell Biol 111:1645–1650
Ozawa M, Kemler R (1992) Molecular organization of the uvomorulin-catenin complex. J Cell Biol 116:989–996
Ozawa M, Ringwald M, Kemler R (1990) Uvomorulin-catenin complex formation is regulated by a specific domain in the cytoplasmic region of the cell adhesion molecule. Proc Natl Acad Sci USA 87:4246–4250
Park J-I, Ji H, Jun S, Gu D, Hikasa H, Li L, Sokol SY, McCrea PD (2006) Frodo links Dishevelled to the p120-catenin/Kaiso pathway: distinct catenin subfamilies promote Wnt signals. Dev Cell 11:683–695
Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, Shapiro L (2006) Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell 124:1255–1268
Peifer M, Berg S, Reynolds AB (1994) A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 76:789–791
Pokutta S, Weis WI (2000) Structure of the dimerization and β-catenin-binding region of α-catenin. Mol Cell 5:533–543
Pokutta S, Herrenknecht K, Kemler R, Engel J (1994) Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur J Biochem 223:1019–1026
Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI (2002) Biochemical and structural definition of the l-afadin- and actin-binding sites of α-catenin. J Biol Chem 277:18868–18874
Potter MD, Barbero S, Cheresh DA (2005) Tyrosine phosphorylation of VE-cadherin prevents binding of p120- and β-catenin and maintains the cellular mesenchymal state. J Biol Chem 280:31906–31912
Reynolds AB, Roczniak-Ferguson A (2004) Emerging roles for p120-catenin in cell adhesion and cancer. Oncogene 23:7947–7956
Reynolds AB, Roesel DJ, Kanner SB, Parsons JT (1989) Transformation-specific tyrosine phosphorylation of a novel cellular protein in chicken cells expressing oncogenic variants of the avian cellular src gene. Mol Cell Biol 9:629–638
Reynolds AB, Herbert L, Cleveland JL, Berg ST, Gaut JR (1992) p120, a novel substrate of protein tyrosine kinase receptors and of p60v-src, is related to cadherin-binding factors β-catenin, plakoglobin and armadillo. Oncogene 7:2439–2445
Reynolds AB, Daniel J, Mccrea PD, Wheelock MJ, Wu J, Zhang Z (1994) Identification of a new catenin: the tyrosine kinase substrate p120cas associates with E-cadherin complexes. Mol Cell Biol 14:8333–8342
Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS (1995) α1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci USA 92:8813–8817
Roura S, Miravet S, Piedra J, García de Herreros A, Duñach M (1999) Regulation of E-cadherin/Catenin association by tyrosine phosphorylation. J Biol Chem 274:36734–36740
Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grübel G, Legrand JF, Als-Nielsen J, Colman DR, Hendrickson WA (1995) Structural basis of cell-cell adhesion by cadherins. Nature 374:327–337
Shapiro L, Weis WI (2009) Structure and biochemistry of cadherins and catenins. Cold Spring Harbor Perspect. Biol 1:a003053
Shi Q, Maruthamuthu V, Li F, Leckband D (2010) Allosteric cross talk between cadherin extracellular domains. Biophys J 99:95–104
Sivasankar S, Gumbiner B, Leckband D (2001) Direct measurements of multiple adhesive alignments and unbinding trajectories between cadherin extracellular domains. Biophys J 80:1758–1768
Smutny M, Cox HL, Leerberg JM, Kovacs EM, Conti MA, Ferguson C, Hamilton NA, Parton RG, Adelstein RS, Yap AS (2010) Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat Cell Biol 12:696–702
Stemmler MP (2008) Cadherins in development and cancer. Mol Biosyst 4:835–850
Takeichi M (1988) The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development 102:639–655
Takeichi M (1993) Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol 5:806–811
Takeichi M (1995) Morphogenetic roles of classic cadherins. Curr Opin Cell Biol 7:619–627
Tamura K, Shan WS, Hendrickson WA, Colman DR, Shapiro L (1998) Structure-function analysis of cell adhesion by neural (N-) cadherin. Neuron 20:1153–1163
Taveau J-C, Dubois M, Le Bihan O, Trépout S, Almagro S, Hewat E, Durmort C, Heyraud S, Gulino-Debrac D, Lambert O (2008) Structure of artificial and natural VE-cadherin-based adherens junctions. Biochem Soc Trans 36:189–193
Thoreson MA, Anastasiadis PZ, Daniel JM, Ireton RC, Wheelock MJ, Johnson KR, Hummingbird DK, Reynolds AB (2000) Selective uncoupling of p120ctn from E-cadherin disrupts strong adhesion. J Cell Biol 148:189–202
Thoreson MA, Reynolds AB (2002) Altered expression of the catenin p120 in human cancer: implications for tumor progression. Differentiation 70:583–589
Tomschy A, Fauser C, Landwehr R, Engel J (1996) Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. EMBO J 15:3507–3514
Troyanovsky RB, Sokolov E, Troyanovsky SM (2003) Adhesive and lateral E-cadherin dimers are mediated by the same interface. Mol Cell Biol 23:7965–7972
Uchida N, Shimamura K, Miyatani S, Copeland NG, Gilbert DJ, Jenkins NA, Takeichi M (1994) Mouse αN-catenin: two isoforms, specific expression in the nervous system, and chromosomal localization of the gene. Dev Biol 163:75–85
Weis WI, Nelson WJ (2006) Re-solving the cadherin-catenin-actin conundrum. J Biol Chem 281:35593–35597
Winkler J, Lünsdorf H, Jockusch BM (1996) The ultrastructure of chicken gizzard vinculin as visualized by high-resolution electron microscopy. J Struct Biol 116:270–277
Witcher LL, Collins R, Puttagunta S, Mechanic SE, Munson M, Gumbiner B, Cowin P (1996) Desmosomal cadherin binding domains of plakoglobin. J Biol Chem 271:10904–10909
Xia X, Mariner DJ, Reynolds AB (2003) Adhesion-associated and PKC-modulated changes in serine/threonine phosphorylation of p120-catenin. Biochemistry 42:9195–9204
Xiao K, Allison DF, Buckley KM, Kottke MD, Vincent PA, Faundez V, Kowalczyk AP (2003) Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J Cell Biol 163:535–545
Xing Y, Takemaru K, Liu J, Berndt JD, Zheng JJ, Moon RT, Xu W (2008) Crystal structure of a full-length β-catenin. Structure 16:478–487
Xu G, Craig AW, Greer P, Miller M, Anastasiadis PZ, Lilien J, Balsamo J (2004) Continuous association of cadherin with β-catenin requires the non-receptor tyrosine-kinase Fer. J Cell Sci 117:3207–3219
Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ (2005) Deconstructing the cadherin-catenin-actin complex. Cell 123:889–901
Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, Copland JA, Anastasiadis PZ (2008) A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J Biol Chem 283:18344–18354
Yang J, Dokurno P, Tonks NK, Barford D (2001) Crystal structure of the M-fragment of α-catenin: implications for modulation of cell adhesion. EMBO J 20:3645–3656
Yap AS, Brieher WM, Pruschy M, Gumbiner BM (1997) Lateral clustering of the adhesive ectodomain: a fundamental determinant of cadherin function. Curr Biol 7:308–315
Yap AS, Niessen CM, Gumbiner BM (1998) The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J Cell Biol 141:779–789
Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M (2010) α-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol 12:533–542
Yoshida C, Takeichi M (1982) Teratocarcinoma cell adhesion: Identification of a cell-surface protein involved in calcium-dependent cell aggregation. Cell 28:217–224
Zhang Y, Sivasankar S, Nelson WJ, Chu S (2009) Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc Natl Acad Sci USA 106:109–114
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Ishiyama, N., Ikura, M. (2012). The Three-Dimensional Structure of the Cadherin–Catenin Complex. In: Harris, T. (eds) Adherens Junctions: from Molecular Mechanisms to Tissue Development and Disease. Subcellular Biochemistry, vol 60. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-4186-7_3
Download citation
DOI: https://doi.org/10.1007/978-94-007-4186-7_3
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-4185-0
Online ISBN: 978-94-007-4186-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)