Abstract
Colorectal cancer (CRC) represents one of the most frequent causes of death for cancer. CRC has been recently defined as the third most common cancer. MicroRNAs (miRNAs) are a class of small interfering RNAs frequently involved in the pathogenesis of cancer. Polymorphisms within miRNAs binding regions have been described as new risk factors for CRC. Several genome-wide profiling studies have identified miRNAs deregulated in CRC tissue. A number of experimental studies on these miRNAs revealed insight into miRNA-mediated regulatory links to well-known oncogenic and tumor suppressor signaling pathways. Several investigations have also described the ability of specific miRNA expression profiles to predict prognosis and therapy response in CRC patients. In this chapter, we focus on the most significant findings of original studies on miRNAs involvement in CRC pathogenesis, focusing also on the potential of cancer-related miRNAs as biomarkers for diagnosis, prognosis, prediction, and therapeutical targets.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Epidermal Growth Factor Receptor
- Colon Cancer Cell
- miRNA Expression
- Adenomatous Polyposis Coli
- Fecal Occult Blood Test
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
4.1 Introduction
Deregulation of microRNAs (miRNAs) can affect carcinogenesis if their mRNA targets are encoded by oncogenes or tumor suppressor genes. Both, over-expression and silencing or switching off of specific miRNAs, have been described in the carcinogenesis of colorectal cancer (CRC). Up-regulation of mature miRNA may occur as a consequence of transcriptional activation or amplification of the miRNA encoding gene, whereas silencing or reduced expression may result from deletion of a particular chromosomal region, epigenetic silencing, or defects in their biogenesis (Rossi et al. 2010).
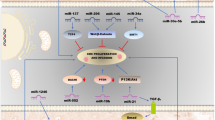
In principle, two approaches are applied today to reveal the connection between miRNAs and CRC. On one hand, miRNAs seem to regulate many known oncogenic and tumor suppressor pathways involved in the pathogenesis of CRC. Many proteins involved in key signaling pathways of CRC, such as members of the Wnt/β-catenin and phosphatidylinositol 3-kinase (PI3K) pathways, KRAS, p53, extracellular matrix regulators as well as epithelial-mesenchymal transition (EMT) transcription factors (Fearon and Vogelstein 1990), are altered and seem to be affected by miRNA regulation in CRC (Fig. 4.1). Analyses of these miRNAs in mechanistic studies are crucial to better understanding CRC pathogenesis (Aslam et al. 2009; Faber et al. 2009) and may provide exciting therapeutic opportunities (Cho 2010a). Findings from this area are discussed in the first part of this chapter. On the other hand, expression profiles of hundreds of miRNAs have been shown to have at least the same potential for identification of biomarkers as profiling of their mRNA or protein counterparts. This allows predicting prognosis and therapy response as well as distinguishing certain disease entities, including CRC, as discussed in the second part that focuses on miRNA expression profiling (Cho 2010b; Slaby et al. 2009).
MicroRNAs involvement in Vogelstein’s model of colorectal cancer development. (APC – adenomatous polyposis coli, CTGF – connective tissue growth factor, TSP1 – thrombospondin 1, EGFR – epidermal growth factor receptor, mTOR – mammalian target of rapamycin, PTEN – phosphatase and tensin homolog, DCC – deleted in colorectal carcinoma, TGFβR1/2 – transforming growth factor, beta receptor 1/2, CASP3 – caspase 3, SIRT1 – sirtuin 1, CDK4,6 – cyclin-dependent kinase 4,6, ECM – extracellular matrix, EMT – epithelial–mesenchymal transition, ICAMs – intercellular adhesive molecules, TIMP3 – tissue inhibitor of metalloproteinase 3, PDCD4 – programmed cell death 4, RECK – reversion-inducing-cysteine-rich protein with kazal motifs, uPAR – plasminogen activator, urokinase receptor, MMPs – matrix metallopeptidases, ZEB1/2 – zinc-finger E-box binding homeobox 1) (Slaby et al. 2009)
4.2 MiRNAs in Colorectal Cancer Signaling
CRC development has been linked to the progressive acquisition of mutations in genes with a crucial role in cell growth, proliferation and programmed cell death As shown in many different studies, miRNAs might perfectly fit and integrate this model initially postulated by Vogelstein by controlling several pathways involved in CRC development (Valeri et al. 2009).
4.2.1 Wnt/β-Catenin Pathway
The Wnt/β-catenin pathway plays a pivotal role in an early colorectal tumor development. Inactivation of the adenomatous polyposis coli (APC) gene is a major initiating event in colorectal carcinogenesis occurring in more than 60% of colorectal adenomas and carcinomas and leading to stimulation of the Wnt pathway via free β-catenin (Fearon and Vogelstein 1990). According to a recent study by Nagel et al. (2008), miRNAs represent a novel mechanism for APC regulation in CRC. MiR-135a and miR-135b decrease translation of the APC transcript in vitro. Of note, miR-135a and miR-135b were also found to be up-regulated in vivo in colorectal adenomas and carcinomas and correlated with low APC levels (Nagel et al. 2008). These observations suggest that alteration in the miR-135 family can be one of the early events in CRC’s molecular pathogenesis. On other hand, restoration of APC function in colorectal cancer cells led to the deregulation of several cancer-related miRNAs, such as miR-122a which was recognized as the liver-specific microRNA. MiR-122a was down-regulated in gastrointestinal cancer cell lines as well as primary carcinoma tissues. Inhibition of miR-122a could reverse wild-type APC-induced growth inhibition of gastrointestinal cancer cells while miR-122a mimic inhibited cell growth (Wang et al. 2009).
4.2.2 EGFR Signaling (KRAS and PI3K Pathways)
The epidermal growth factor receptor (EGFR) signaling significantly participates in promotion and progression of broad spectrum of solid tumors, and members of EGFR pathway constitute promising targets for anti-cancer therapy. Two different studies showed that miR-7 and miR-128 are able to down-regulate EGFR and its downstream pathways in breast, glioblastoma and lung cancer cell lines and in lung cancer patients (Webster et al. 2009; Weiss et al. 2008). Even though miR-128 and miR-7 have been shown to be deregulated in CRC (Motoyama et al. 2009; Ng et al. 2009) no study support a role for their down-regulation in EGFR over-expression. Stimulation of the EGFR and, subsequently, KRAS signaling lead to the activation of numerous signal transduction molecules initiating a cascade of downstream effectors that mediate tumor growth, survival, angiogenesis and metastasis (Ciardiello and Tortora 2008). KRAS oncogene has been reported to be a direct target of the let-7 miRNA family (Johnson et al. 2005). When let-7 low-expressing DLD-1 colon cancer cells were transfected with let-7a-1 precursor, significant growth suppression with concurrent decrease of the KRAS protein levels was observed while the levels of both of their mRNAs remained almost unchanged (Akao et al. 2006). Another miRNA associated with KRAS regulation in CRC is miR-143 (Chen et al. 2009). KRAS was deduced to be a miR-143 target not only by computational prediction but also by noting the inverse correlation between miR-143 and KRAS protein level in clinical samples. KRAS expression in vitro was significantly abolished by treatment with miR-143 precursor, whereas miR-143 inhibitor increased the KRAS protein level. Moreover, constitutive phosphorylation of MAPK was efficiently blocked through inhibition of KRAS expression by miR-143 (Chen et al. 2009). Recently, miR-18a was observed to directly regulate KRAS but not N- and HRAS levels in the colon adenocarcinoma HT-29 cells (Tsang et al. 2009).
Another central signaling pathway downstream from EGFR and important in CRC development is the phosphatidylinositol 3-kinase (PI3K) pathway. MiRNAs arrays-based studies revealed a ubiquitous loss of miR-126 expression in CRC cell lines when compared to normal human colon epithelium. Reconstitution of miR-126 resulted in a significant growth reduction (Guo et al. 2008). As a direct target of miR-126, the p85β regulatory subunit involved in stabilizing and propagating the PI3K signal was mechanistically proven. Furthermore, this p85β reduction mediated by miR-126 was accompanied by a substantial reduction in phosphorylated Akt levels in the cancer cells, suggesting an impairment in PI3K signaling. In a series of matched normal colon and primary colon tumor tissues, each of the tumors demonstrated miR-126 down-regulation together with an increase in the p85β protein level (Guo et al. 2008). Another important regulatory component of the PI3K pathway, the tumor suppressor gene PTEN, is strongly repressed by miR-21, which was demonstrated on a hepatocellular carcinoma model (Meng et al. 2007). However, miR-21 is the miRNA most frequently up-regulated in CRC (Schetter et al. 2008; Slaby et al. 2007; Krichevsky and Gabriely 2009). It seems that suppression of PTEN controlled by miR-21 is associated with augmentation of PI3K signaling and progression of CRC.
4.2.3 p53 Pathway
A well-known tumor suppressor gene, p53 is mutated in about 50–75% of all CRCs and many other human tumors. p53 responds to DNA damage or deregulation of mitogenic oncogenes through the induction of cell cycle checkpoints, apoptosis, or cellular senescence (Fearon and Vogelstein 1990). Although p53 is clearly a transcriptional activator, numerous reports have indicated that p53 also represses the expression of specific genes either directly or indirectly (Bailey et al. 2010; He et al. 2007). The manner in which this is achieved was obscure, with both transcriptional and post-transcriptional suppression as possible mechanisms. In the latter case, the discovery of an extensive regulatory network of miRNAs offered the possibility that p53-mediated control of miRNA expression could allow it to act indirectly to repress target gene expression at the post-transcriptional level. Recently, several groups have unraveled important aspects of the connection between p53 and the miRNA network (Hermeking 2007).The conserved miR-34a-c family was found to be direct transcriptional targets of p53. MiRNA expression patterns were analyzed in wild-type p53 +/+ and p53 –/– mutant HCT-116 colon cancer cell lines after treatment with DNA damaging agents. Several miRNAs were induced in the wild type but not in the p53 –/– mutant cells, thus suggesting a p53-mediated expression. MiR-34a showed the strongest induction. Expression of miR-34a was sufficient to induce apoptosis through p53-dependent and independent mechanisms. MiR-34a-responsive genes are highly enriched for those that regulate cell-cycle progression, cellular proliferation, apoptosis, DNA repair and angiogenesis (Chang et al. 2007). By experimentally over-expressing miR-34a, p53 effects like cell-cycle arrest and apoptosis could be achieved. p53’s connection to the miR-34 family was successfully evaluated also on a model of lung carcinoma cells harboring regulated p53 alleles (Bommer et al. 2007) and p53 +/+ and p53 –/– mouse embryo fibroblasts (Corney et al. 2007). MiR-34a induction leads to dramatic reprogramming of gene expression. Among the down-regulated targets of the miR-34 family were well-characterized p53 targets like CDK4/6, cyclin E2, E2F5, BIRC3 and Bcl-2. Notably, these effects were nearly identical irrespective of whether miR-34-a, miR-34-b or miR-34-c was introduced. Others have identified SIRT1, a regulator of apoptosis in response to cellular stress, as an additional target of miR-34a. Interestingly, the suppression of SIRT1 by miR-34a resulted in apoptosis in wild-type colon cancer cells but not in p53 –/– mutants. This suggests a positive feedback loop between p53 and miR-34 (Yamakuchi et al. 2008).
Decreased levels of the miR-34 family have been found in many tumors, including CRC. MiR-34a expression was found to be down-regulated in 9 of 25 CRCs (Tazawa et al. 2007). Loss of 1p36, the genomic interval harboring miR-34a, is common in diverse human cancers (Chang et al. 2007) but one of the other mechanisms responsible for decrease of miR-34 family expression levels seems to be CpG island hypermethylation. MiR-34a promoter methylation was reported in 3 of 23 cases of colon cancer (Lodygin et al. 2008). MiR-34b/c were found to be epigenetically silenced in 9 of 9 cell lines examined and in 101 of 111 primary CRC tumors, but they were not in normal colonic epithelium. After treatment with demethylating agents, miR-34b/c expression was restored. That resulted in inhibition of tumor motility and metastasis formation (Toyota et al. 2008). The high frequency of their methylation in CRC and their contribution to the p53 network imply that miR-34a-c function as important tumor suppressors which can be lost during CRC development (Faber et al. 2009).
Like p53, members of the miR-34 family can be considered as tumor suppressors to date, making them potential candidates for causing cancer by way of their inactivation.
4.2.4 IGF Signaling
Insulin receptor substrate-1 (IRS-1) plays an important role in cell growth and cell proliferation. IRS-1, especially when activated by the type 1 insulin-like growth factor receptor (IGF-IR), sends an unambiguous mitogenic, anti-apoptotic, and anti-differentiation signal. IRS-1 levels are often increased in cases of human cancer and are low or even absent in differentiating cells (Pechlivanis et al. 2007). MiR-145 has been proposed as a tumor suppressor and it had been shown previously that miR-145 targets the 3′ untranslational region (3′UTR) of IRS-1 and dramatically inhibits the growth of colon cancer cells (Shi et al. 2007). More recently, IGF-IR was proven to be another direct target of miR-145. It was shown that an IRS-1 lacking its 3′UTR is no longer down-regulated by miR-145 and rescues colon cancer cells from growth inhibition induced by miR-145. An IGF-IR resistant to miR-145 (again by elimination of its 3′UTR) was not down-regulated by miR-145 but failed to rescue colon cancer cells from growth inhibition. These data indicate that miR-145 plays a significant role in IGF signaling in cancer pathogenesis (La Rocca et al. 2009).
4.2.5 E2F Family and Cell Cycle Regulation
The miR-17-92 cluster encodes six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1) located on chromosome 13q31.3. The human genomic locus encoding these miRNAs undergoes amplification in several types of lymphoma and solid tumors. The miR-17-92 cluster seems to be tightly linked to the functions of the E2F family of transcription factors, which are critical regulators of the cell cycle and apoptosis. E2F1, E2F2, and E2F3-activating E2Fs that induce expression of genes driving the progression from the G1 into the S phase – were among the first experimentally verified targets of the miR-17-92 cluster. Moreover, both E2F1 and E2F3 can directly activate transcription of these miRNAs, thereby establishing a negative feedback loop (Woods et al. 2007). The pro-tumorigenic activity of the miR-17-92 cluster also involves cell-non-autonomous functions that include induction of angiogenesis in solid tumors. Using a mouse model of colon cancer, Dews et al. (2006) demonstrated that the angiogenic activity of c-Myc is due at least in part to downstream activation of the miR-17-92 cluster. The anti-angiogenic factors thrombospondin-1 (TSP1) and connective tissue growth factor (CTGF) are negatively regulated by these miRNAs, which are potently induced by c-Myc in this model. Robust vascularization of tumors can be induced by expression of either c-Myc or the miR-17-92 cluster (Dews et al. 2006).
4.2.6 MET Signaling
Hu et al. (2010) provided evidence that miR-133b regulated tumor cell proliferation and apoptosis through modulation of the MET signaling pathway. In the CRC cell lines SW-620 and HT-29, ectopic expression of miR-133b potently affected tumor cell proliferation and apoptosis in vitro and in vivo by direct targeting of the receptor tyrosine kinase MET. Transfection of SW-620 and HT-29 cells with miR-133b significantly suppressed a luciferase-reporter containing the MET-3′UTR.
4.2.7 Cyclooxygenase-2
Cyclooxygenase-2 (COX-2) catalyzes the production of prostaglandin E2, one of the most important products of the arachidonate metabolism. Over-expressed COX-2 strongly contributes to the growth and invasiveness of tumor cells in patients with CRC (Strillacci et al. 2009). It has been demonstrated that COX-2 up-regulation depends upon various cellular pathways involving both transcriptional and post-transcriptional regulations. An inverse correlation was reported between COX-2 and miR-101 expression in CRC cell lines. It was demonstrated in vitro that the direct translational inhibition of COX-2 mRNA is mediated by miR-101. Moreover, this correlation was supported by data collected ex vivo, in which colon cancer tissues and liver metastases derived from CRC patients were analyzed. Impairment of miR-101 levels could represent one of the leading causes of COX-2 over-expression in CRC cells (Strillacci et al. 2009; Young 2010).
4.2.8 DNA Reparation
DNA reparation impairment has also been linked to CRC development and progression. A link between microenvironmental factors, microRNAs and DNA repair system has been highlighted by Crosby et al. (2009) in breast and cervical cancer cell lines. miR-210 and miR-373 expression can be induced by the hypoxia-inducible factor-1 alpha (HIF-1α). This transcription factor can bind to microRNA gene promoters and induce their expression. MiR-210 and miR-373 in turn down-regulate two major key players in DNA repair system RAD52 and RAD23B respectively implicated in homology-dependent repair (HDR) and nucleotide excision repair (NER). Although this interaction has not been shown in CRC yet, it might contribute to colon tumorigenesis.
4.2.9 Extracellular Matrix Breakdown and Epithelial-mesenchymal Transition
The extracellular matrix (ECM) and its remodeling play a crucial role in the development of blood supply and interaction with the mesenchymal stroma upon which tumor cells grow. ECM remodeling is one of the necessary conditions of tumor growth, survival, invasiveness, and metastasizing. The key enzymes, among the many involved in ECM breakdown, are proteinases, and among these are the urokinase plasminogen activator (uPA) cascade and the matrix metalloproteinases (MMPs) (Takayama et al. 2006). Substantial data indicate that miR-21 is significantly elevated in CRC and in many other tumors of various origins (Krichevsky and Gabriely 2009). Based upon a glioblastoma model, it was described that miR-21 regulates multiple genes associated with cellular motility and ECM remodeling. These included the RECK and TIMP3 genes which are suppressors of malignancy and inhibitors of MMPs (Gabriely et al. 2008). Specific inhibition of miR-21 with antisense oligonucleotides leads to elevated levels of RECK and TIMP3 and therefore reduces MMP activities. Although these observations originate from a glioblastoma model, up-regulation of miR-21 in CRC cells has been shown to increase their migratory and invasive abilities and miR-21 seems to act, in this case, in a similar manner (Gabriely et al. 2008). Furthermore, miR-21 was shown to act on PDCD4, a tumor suppressor gene that is an independent prognostic factor in resected CRC and a new negative regulator of intravasation through the invasion-related urokinase receptor gene (uPAR) (Asangani et al. 2008). Silencing of miR-21 by anti-miR-21 resulted in increased levels of PDCD4 in colorectal cell lines and decreased invasion in a chicken-embryo-metastasis assay. In addition, 22 resected human tumors showed higher miR-21 expression than did the corresponding normal mucosa and decreased amounts of PDCD4 protein while mRNA levels were unchanged. These results argue for miR-21’s having an important function in the pathogenesis of CRC, as it also shows an inverse correlation with survival.
EMT is the conversion of an epithelial cell into a mesenchymal cell. Morphologically, EMT is characterized by a decrease of E-cadherin, loss of cell adhesion, and increased cell motility leading to promotion of metastatic behavior of cancer cells (including CRC) (Natalwala et al. 2008). The transcriptional repressor zinc-finger E-box binding homeobox 1 (ZEB1) is a crucial inducer of EMT in various human tumors, and it recently was shown to promote invasion and metastasis of tumor cells. The functional links to EMT come from members of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429). ZEB1 directly suppresses transcription of miRNA-200 family members miR-141 and miR-200c, which strongly activate epithelial differentiation in pancreatic, colorectal and breast cancer cells (Burk et al. 2008). Notably, the EMT activators transforming growth factor β2 and ZEB1 are the predominant targets down-regulated by these miRNAs. These results indicate that ZEB1 triggers a miRNA-mediated feedforward loop that stabilizes EMT and promotes the invasion of cancer cells. Alternatively, depending on the environmental trigger, this loop might switch and induce epithelial differentiation, thereby explaining strong intratumoral heterogeneity. A recent study associated the expression of let-7 with two differentiation stages of a panel of cell lines (with epithelial and a mesenchymal gene signatures) and linked let-7 to EMT (Shell et al. 2007).
4.2.10 Alterations in MiRNA Processing Machinery
The idea of an altered miRNA processing was first postulated by Michael et al. (2003) who identified 28 microRNA sequences that included three completely new miRNAs (miR-320, miR-321, and miR-200c), seven mouse-specific miRNAs and two miRNAs (miR-143 and miR-145) that were consistently down-regulated in all cancer specimens by comparative analysis of CRC tissue and normal colonic mucosa. In the attempt to find an explanation for the deregulation of these mi-RNAs they analyzed the expression of the RISC complex associated genes, DICER and eIF2C2, by real-time PCR analysis in a subset of matched tissue-RNA samples. No discrepancies in gene expression between normal and tumoral tissues were found hypothesising that other factors might regulate microRNA expression in CRC (Valeri et al. 2009).
More recently frameshift mutations in TARBP2 were identified (TAR RNA-binding protein 2, an essential element of the DICER machinery) as very frequent in sporadic and hereditary CRCs associated with microsatellite instability. These mutations can cause decrease in TRBP protein expression, destabilization of DICER1 protein and defect in the processing of miRNAs, providing a possible explanation for miRNAs deregulation in colorectal cancers (Melo et al. 2009). Conversely SND1, another important component of the RISC complex has been reported as up-regulated gene in human colon cancers. SND1 mRNA was frequently up-regulated in human and mice cancers as well as in aberrant crypt foci and can down-regulate APC protein expression by post-transcriptional regulation. Based on these findings, authors speculated that SND1 may control gene expression of APC or other cancer-related genes through the regulation of miRNA-induced translational repression (Tsuchiya et al. 2007).
4.2.11 Others
In the study of Arndt et al. (2009), SW620 colon cancer cells were stably transduced with miR-143 or miR-145 expression vectors and analyzed in vitro for cell proliferation, cell differentiation and anchorage-independent growth. Signaling pathways associated with differentially expressed miRNAs were identified using a gene set enrichment analysis. Interestingly, miR-143 and miR-145 appeared to function in opposing manners to either inhibit or augment cell proliferation in a metastatic CRC model. The pathways targeted by miR-143 and miR-145 showed no significant overlap. Furthermore, gene expression analysis of metastatic versus non-metastatic isogenic cell lines indicated that miR-145 targets involved in cell cycle and neuregulin pathways were significantly down-regulated in the metastatic context.
Gregersen et al. (2010) employed a different microarray based approach to identify miR-145 targets. Based on seed site enrichment analyses and unbiased word analyses, a significant enrichment of miRNA binding sites in the 3′UTRs of transcripts down-regulated upon miRNA over-expression was found. Gene Ontology analysis showed an over-representation of genes involved in cell death, cellular growth and proliferation, cell cycle, gene expression and cancer. A number of the identified miR-145 targets have previously been implicated in cancer, YES and STAT1 were verified in vitro as direct targets.
4.3 Single Nucleotide Polymorphisms (SNPs) and MiRNAs: Risk Factors for CRC
The binding of miRNA to mRNA is critical for regulating the mRNA level and protein expression. This binding can be affected, however, by SNPs that can occur in the miRNA target site and can either abolish existing binding sites or create illegitimate binding sites. Therefore, SNPs within miRNA binding sites can have differing effects on gene and protein expression and represent another type of genetic variability that can influence the risk of certain human diseases, including CRC (Fig. 4.2). Various approaches have been used to predict and identify functional SNPs within miRNA binding sites and their biological relevance is beginning to be evaluated in large case–control studies (Chen et al. 2008). SNPs may occur also at the level of the pri-miRNA, pre-miRNA or the mature miRNA sequence (miR-polymorphisms). Such polymorphisms may be functional as to the biogenesis and action of the mature miRNA (Mishra and Bertino 2009).
Regarding CRC, out of eight candidates predicted by computer simulation, the two genes for CD86, a costimulatory ligand on lymphocytes, and for the insulin receptor carry an SNP that are significantly associated with the risk of sporadic CRC (odds ratios 2.74 and 1.94, respectively) (Landi et al. 2008). However, the biological relevance of these SNPs has not yet been confirmed by functional in vitro studies. Lee et al. (2010) analyzed the 40 SNPs of miRNA-related genes and their impact on the prognosis in the group 462 Korean CRC patients. None of tested SNPs was found to be an independent prognostic marker in CRC.
4.4 Plasma and Serum MiRNAs: Diagnosis and Monitoring of CRC
Circulating nucleic acids (CNAs) offer unique opportunities for an early diagnosis of CRC. Dysregulated expression of miRNAs in various tissues has been associated with a variety of human cancers. More recently, miRNAs’ occurrence in the serum and plasma of humans has been repeatedly observed. The levels of miRNAs in serum are more stable, reproducible, and consistent among individuals of the same species than are other CNAs (Chen et al. 2008). The detection of serum miRNAs have been tested in prostate cancer, ovarian cancer and CRC patients as possible early diagnostic biomarkers (Mitchell et al. 2008; Ng et al. 2009; Resnick et al. 2009).
In a study by Chen et al. (2008), CRC patients had a significantly different serum miRNA profile compared to healthy subjects (HS). In all cases, 69 miRNAs were detected in the CRC serum but not in HS. It is of interest to note that CRC patients shared a large number of serum miRNAs (e.g. miR-134, miR-146a, miR-221, miR-222, miR-23a, etc.) with lung cancer patients. Pearson correlation further indicated that the levels of miRNAs in serum from lung cancer patients and CRC patients were consistent, suggesting that there are some “common” tumor-relatedmiRNAs in serum (Chen et al. 2008). Differentially expressed miRNAs in the plasma of patients with CRC have been also reported (Ng et al. 2009). Expression pattern of 30 miRNAs (miR-17-3p, miR-92, miR-135b, miR-222, miR-95, etc.) in the plasma of patients with CRC were analyzed by real-time PCR expression profiling. Both miR-17-3p and miR-92 were significantly elevated (p < 0.0005). The plasma levels of these miRNAs were significantly reduced after surgery in 10 patients with CRC (p < 0.05). Further validation with an independent set of plasma samples (n = 180) indicated that miR-92 differentiates CRC not only from normal subjects but also from gastric cancer and inflammatory bowel disease. This marker yielded an ROC (receiver operating characteristic) curve area of 88.5%. In discriminating CRC from control subjects, the sensitivity was 89% and the specificity was 70%. MiR-92 has reasonable sensitivity for CRC detection and compares favorably with the fecal occult blood test (Ng et al. 2009). Huang et al. (2010) measured the levels of 12 miRNAs (miR-134, -146a, -17-3p, -181d, -191, -221, -222, -223, -25, -29a, -320a, and -92a) in plasma samples from patients with advanced colorectal neoplasia (carcinomas and advanced adenomas) and healthy controls using real-time PCR. Authors found that plasma miR-29a and miR-92a have significant diagnostic value for advanced neoplasia. miR-29a yielded an AUC (the areas under the ROC curve) of 0.844 and miR-92a yielded an AUC of 0.838 in discriminating CRC from controls. More importantly, these 2 miRNAs also could discriminate advanced adenomas from controls and yielded an AUC of 0.769 for miR-29a and 0.749 for miR-92a. Combined ROC analyses using these 2 miRNAs revealed an elevated AUC of 0.883 with 83.0% sensitivity and 84.7% specificity in discriminating CRC, and AUC of 0.773 with 73.0% sensitivity and 79.7% specificity in discriminating advanced adenomas. These data suggest that plasma miR-29a and miR-92a have strong potential as novel non-invasive biomarkers for early detection of CRC.
More recently, feasibility of fecal miRNAs as biomarkers for colorectal neoplasia screening was evaluated. miRNA expression profiles from stool of 29 patients showed higher expression of miR-21 and miR-106a in patients with adenomas and CRCs compared with individuals free of colorectal neoplasia (Link et al. 2010).
4.5 MiRNA Expression Profiles of CRC Tissue
Alterations in miRNA expression profiles have been successively detected in many types of human tumors (Garzon et al. 2006). The causes of the widespread differential expression of miRNA genes between malignant and normal cells can be explained by the gene location in cancer-associated regions, alterations in the miRNA processing machinery, and epigenetic mechanisms (Garzon et al. 2009). In reports on various cancer samples, generally lower miRNA levels were identified in tumors in comparison with normal tissue and, lower miRNA levels in poorly differentiated tumors compared to well-differentiated tumors in tissue samples (Lu et al. 2005) as well as in cell lines (Gaur et al. 2007). Studies focusing on miRNA expression profiling in CRC are summarized in Table 4.1, and some of these studies are described in detail below.
In 2003, Michael et al. (2003) published the first such study. Using cloning technology followed by northern blotting, he observed consistently reduced accumulation of the specific mature miR-143 and miR-145 in the adenomatous and carcinoma stages of colorectal neoplasia. The same blots, however, displayed consistent levels of the ~70-bp pre-miR-143 in each of the cell lines. The authors concluded that the levels of mature miR-143 in these cells were controlled post-transcriptionally. These data suggested that abnormal processing might affect miRNAs expression in colon cancer cells.
Bandres et al. (2006) examined by real-time PCR the expression of 156 mature miRNAs in colorectal tumors and adjacent non-neoplastic tissues from patients and CRC cell lines. This permitted them to identify a group of 13 miRNAs whose expression is significantly altered in this type of tumor. The most significantly deregulated miRNAs were miR-31, miR-96, miR-135b,miR-183 (up-regulated in tumors and CRC cell-lines), and miR-133b, miR-145 (down-regulated). In addition, the expression level of miR-31 was positively correlated with the stage of CRC tumor. These results, achieved through a standardized real-time PCR method, suggest that miRNA expression profile could have relevance to the biological and clinical behavior of colorectal neoplasia.
Velculescu’s group developed an experimental approach called miRNA serial analysis of gene expression (miRAGE) and used it to perform one of the largest experimental analyses of human miRNAs. Sequence analysis of 273,966 small RNA tags from human colorectal cells allowed them to identify 200 known mature mi-RNAs, 133 novel miRNA candidates, and 112 previously uncharacterized miRNA forms. To aid in evaluating the candidate miRNAs, they disrupted the Dicer locus in three human CRC cell lines and examined known and novel miRNAs in these cells. This study indicates that the human genome contains many more miRNAs than currently identified (Cummins et al. 2006).
From a large-scale analysis of miRNA expression profiles on 540 samples of solid cancers, including CRC, Volinia et al. (2006) identified a solid cancer miRNA signature composed by a large portion of over-expressed miRNAs. Among these miRNAs were some with well characterized cancer associations, such as miR-17-5p, miR-20a, miR-21, miR-92, miR-106a, and miR-155. A microarray-based approach for analysis of miRNA expression profiles in CRC was successfully applied also by Motoyama et al. (2009).
In another profiling study, Lanza et al. (2007) evaluated the expression of mi-RNAs and mRNAs in CRC samples characterized by microsatellite stability (MSS) or by high levels of microsatellite instability (MSI-H). Their analysis of miRNA expression profiles of MSI-H (n = 16) and MSS CRCs (n = 23) identified 14 differentially expressed miRNAs, while their analysis of messenger RNA expression profiles in these tumors identified 451 differentially expressed genes. Consequently, a smaller selected signature of best predictors of microsatellite status was generated: 27 genes, including 8 miRNAs, were identified as predictors. Further cluster analysis using just these 27 miRNAs and mRNAs also perfectly separated the two tumor classes. Cluster analyses run using either the mRNAs or the miRNAs independently did not perform as well in discriminating the tumor types. Therefore, the combined miRNA/mRNA fingerprint worked as the best discriminator for MSS versus MSI-H. In the study of Earle et al. (2010), relative expression levels of miR-92, -223, -155, -196a, -31, and -26b were significantly different among MSI subgroups (including low MSI and hereditary non-polyposis colorectal cancer (HNPCC) syndrome), and miR-31 and miR-223 were over-expressed in CRC of patients with HNPCC-associated cancer. To identify miRNAs that are differentially expressed in CRC and CRC subtypes, Sarver et al. (2009) carried out highly expression profiling of 735 miRNAs on samples obtained from a statistically powerful set of tumors (n = 80) and normal colon tissue (n = 28). Tumor specimens showed highly significant and large fold change differential expression of the levels of 39 miRNAs including miR-135b, miR-96, miR-182, miR-183, miR-1, and miR-133a, relative to normal colon tissue. Significant differences were observed in 6 miRNAs: decreased levels in MSS relative to MSI-H tumors included miR-552, miR-592, miR-181c, and miR-196. MiR-625 and miR-31 exhibited increased levels in MSI-H relative to MSS tumors.
Monzo et al. (2008) assessed the expression of mature miRNAs in human embryonic colon tissue, as well as in CRC and paired normal colon tissue. Overlapping miRNA expression was detected between embryonic colonic mucosa and CRC. The miR-17-92 cluster and its target, E2F1, exhibit a similar pattern of expression in human colon development and in colonic carcinogenesis – regulating cell proliferation in both cases. Authors of this study conclude that miRNA pathways play a major role in both embryonic development and neoplastic transformation of the colonic epithelium.
From a diagnostic point of view, miRNA expression profiles might also contribute significantly to the further determination of the tissue origin of the cancer of unknown primary sites. Cancer of unknown primary (CUP) is usually a very aggressive disease with a poor prognosis. Identifying of colorectal cancer among adenocarcinoma of unknown primary site may improve prognosis of these patients by giving them a chance for modern anti-cancer targeted therapy. Two recent studies examined metastases of unknown primary tumors with miRNA microarrays for their potential to identify the tissue of origin. After establishing a miRNA classifier (n = 68, 11 tumor types, 217 miRNAs), 12 out of 17 poorly differentiated tumors were accurately classified by miRNA profiling (Lu et al. 2005). A second publication reported an overall accuracy of 90% in classifying more than 400 malignant tumor samples of 22 tissue origins based on a set of 48 miRNAs (Rosenfeld et al. 2008).
A recent study on lymph node metastases of several malignant tumors, including CRC, identified three specific miRNAs (miR-148a, miR-34b/c, and miR-9), specifically down-regulated by CpG island hyper-methylation (Lujambio et al. 2008).
To offer new approach for preventing and controlling lymphatic metastasis in colon cancer, Huang et al. (2009) compared the miRNA expression profiles (723 human microRNA probes) of normal colonic epithelium from the two CRC patient groups; those with confirmed lymph node metastasis (n = 3), and those without detectable lymph node metastasis (n = 3). Two microRNA (hsa-miR-129 *, hsa-miR-137) were differentially expressed in the lymph node positive group compared with the lymph node negative group. After validation through real-time PCR method, hsa-miR-137 expression was significantly up-regulated nearly 6.6-fold in lymph node positive specimens (p = 0.036).
In general, there are several advantages of using miRNA expression profiling instead of its mRNA counterpart for biomarker identification and also for routine diagnostics. As a consequence of the fact that miRNAs target mRNAs with an imperfect sequence complementarity, a single miRNA can regulate the expression of more than 100 mRNAs simultaneously (Esquela-Kerscher and Slack 2006). This might explain why microarrays of 217 miRNAs have much higher information content than 16,000 mRNAs in distinguishing different tissues and tumors (Faber et al. 2009). It is relatively easier to discover reliable biomarkers from the approximately hundreds of miRNA candidates discovered to date than from over 40,000 genes. A further advantage is that, due to their small size and stem-loop structure, miRNAs are relatively more stable and less subjected to degradation during fixation and sample processing. One recent examination compared the miRNA-expression profiles from fresh frozen versus formalin-fixed paraffin-embedded (FFPE) CRC tissues. A good correlation coefficient of 0.86–0.89 was observed. Worthy of note is that differing formalin fixation times – inevitable in a routine pathology lab – did not significantly influence the expression of miRNAs in 40 CRC specimens (Xi et al. 2007). This can also be of benefit for large retrospective studies based on archived FFPE samples. Furthermore, miRNAs can be visualized at the cellular and subcellular levels by conventional as well as fluorescence in situ hybridization (Nuovo et al. 2008).
4.6 MiRNAs in CRC Prognosis and Prediction
Accumulating evidence shows that miRNA expression patterns are unique to certain cancers and have potential to be used as prognostic and predictive factors in clinical routine. Xi et al. (2007) performed Kaplan-Meier analysis for CRC patients with International Union Against Cancer (UICC) stages I-IV and found that tumors expressing high levels of miR-200c, recently connected to EMT, are correlated with poorer prognosis, regardless of tumor stage. These investigators also found that p53 mutation, commonly found in CRC, is strongly associated with greater than twofold miR-200c over-expression. Chen et al. (2010) identified siginificant decrese of miR-148a and miR-152 expression levels in CRC (both p < 0.001) tissue in comparison to their matched non-tumoral tissues. Authors further observed correlation of low expression levels of miR-152 and miR-148a with increased tumor size (p = 0.004 and 0.018, respectively) and advanced pT stage (p = 0.002 and 0.023, respectively).
MiR-21 is up-regulated in many solid tumors, including CRC (Krichevsky and Gabriely 2009). Recently, we have found that miR-21 over-expression shows a strong correlation with the established prognostic factors as nodal stage, metastatic disease and UICC stage (Slaby et al. 2007). Kulda et al. (2010) correlated miR-21 and miR-143 expression to disease-free interval (DFI) (p = 0.0026 and p = 0.0191, respectively). There was shorter DFI in patients with a higher expression of miR-21 and, surprisingly, also in patients with a higher expression of miR-143, which is a putative tumor suppressor. Using class comparison analysis, Shetter et al. (2008) later found that 37 miRNAs were differentially expressed in tumors of CRC patients. From this group, miR-20a, miR-21, miR-106a, miR-181b, and miR-203 were found by Cox regression analysis to be associated also with poor survival and were selected for validation. Validation was performed by measuring miRNAs’ expression using real-time PCR in tumor and paired non-tumor tissues in the validation cohort. In the validation set, only high expression of miR-21 was significantly associated with poor prognosis, and this association was independent of age, sex, and tumor location. Multivariate analysis further revealed that high miR-21 expression in tumors was associated with poor survival, independent of the tumor stage. In patients who received adjuvant therapy, high miR-21 expression indicated a poor response to therapy (Schetter et al. 2008).
Schepeler et al. (2008) found that miRNAs were associated with tumor microsatellite status in stage II colon cancer. The predictive molecular signature was composed of only four miRNAs (miR-142-3p, miR-212, miR-151, and miR-144). Furthermore, a biomarker based on miRNA expression profiles could predict recurrence of disease with an overall performance accuracy of 81%, thus, indicating a potential role for miRNAs in determining tumor aggressiveness. Kaplan-Meier survival curves showed that patients who had stage II CRC tumors with high expression of miR-320 or miR-498 had significantly shorter progression-free survival than did patients whose tumors showed low expression. These miRNAs were correlated with the probability of progression-free survival also by multivariate analysis. Although these results are promising, larger studies will be needed to prove whether miRNAsreally have significant potential to extend prognostic information based on the recent standard diagnostic procedures.
Another important question for management of CRC patients is the possibility of predicting therapy response. Nakajima et al. (2006) evaluated the significance of five mature miRNAs in tumors of CRC patients treated with 5-fluorouracil (5-FU)-based anti-metabolite S-1. They identified let-7 g and miR-181b as significant indicators for chemoresponse to S-1-based chemotherapy.
A study published by Rossi et al. (2007) reported a suggestive pattern of miRNAs rearrangement in HT-29 and HCT-116 human colon cancer cell lines after exposure to 5-FU, a classical anti-metabolite in broad clinical use. At clinically relevant concentrations, the drug up-regulated or down-regulated in vitro the expression of 19 and 3 miRNAs, respectively, by a factor of not less than two-fold. In some instances, 5-FU up-regulated miRNAs that are already over-expressed in tumor tissue, including, for example, miR-21 (Rossi et al. 2007).
In other instances, by contrast, the drug influenced the expression of miRNAs in a direction that is opposite to that induced by neoplastic transformation. A typical example is provided by miR-200b, which is up-regulated in various tumors but down-regulated by treatment with 5-FU. Interestingly, it is known that miR-200b targets mRNA that codes for a protein tyrosine phosphatase (PTPN12) which inactivates products of oncogenes, such as ABL, SRC, or KRAS (Schepeler et al. 2008).
Another study evaluated changes in miRNA expression profiles as a response to therapy, focusing on the effects of capecitabine chemoradiotherapy on rectal tumors in vivo (Svoboda et al. 2008). Tumor microexcisions were taken before starting a therapy and, again, after a 2-week therapy. The extent of tumor response to the therapy was investigated microscopically by an experienced pathologist according to Mandard’s tumor regression criteria. In this study, many miRNAs (miR-10a, miR-21, miR-145, miR-212, miR-339, miR-361) responded to capecitabine chemoradiotherapy in individual tumor samples. In most samples, however, only two miRNAs, miR-125b and miR-137, showed significant increase in expression levels after 2-week therapy.
Zhou et al. (2010) determined how 5-FU and oxaliplatin (L-OHP) modify the expression profiles (856 human miRNA probes) of miRNAs in HCT-8 and HCT-116 colon cancer cells. Fifty-six up- and 50 down-regulations of miRNA expression with statistical significance were identified in colon cancer cells following exposure to 5-FU or L-OHP compared to matched control cells. Expression levels of miR-197, miR-191, miR-92a, miR-93, miR-222, and miR-1826 were significantly down-regulated in both cell lines after the treatment of one drug or in one cell line following exposure to either drug.
HCT116 human colorectal cancer cells were used to investigate the biological and potential chemosensitizing role of miR-143 in the study of Borralho et al. (2009). Transient miR-143 over-expression resulted in an approximate 60% reduction in cell viability. In addition, stable miR-143 over-expressing cells were selected with G418 and exposed to 5-FU. Increased stable expression of miR-143 was associated with decreased viability and increased cell death after exposure to 5-FU. These changes were associated with increased nuclear fragmentation and caspase -3, -8 and -9 activities. In addition, extracellular-regulated protein kinase 5, nuclear factor-kappaB and Bcl-2 protein expression were down-regulated by miR-143, and further reduced by exposure to 5-FU.
MiR-215, through the suppression of denticleless protein homolog (DTL), a cell cycle-regulated nuclear and centrosome protein, induces decreased colon cancer cell proliferation by causing G2-arrest, thereby leading to an increase in their chemoresistance to the chemotherapeutic agents, methotrexate and Tomudex (Song et al. 2010).
Boni et al. (2010) investigated associations between 18 polymorphisms in both miRNA-containing genomic regions (primary and precursor miRNA) and in genes related to miRNA biogenesis with clinical outcome in 61 metastatic colorectal cancer (mCRC) patients treated with 5-FU and irinotecan (CPT-11). A significant association with tumor response and time to progression (TTP) was found for SNP rs7372209 in pri-miR26a-1 (p = 0.041 and p = 0.017, respectively). The genotypes CC and CT were favorable when compared with the TT variant genotype. SNP rs1834306, located in the pri-miR-100 gene, significantly correlated with a longer TTP (p = 0.04). In the miRNA-biogenesis pathway, a trend was identified between SNP rs11077 in the exportin-5 gene and disease control rate (p = 0.076).
There is increasing evidence that the let-7 miRNA exerts an effect as a tumor suppressor by targeting the KRAS mRNA. The let-7 complementary site (LCS6) T > G variant in the KRAS 3′UTR weakens let-7 binding. Graziano et al. (2010) analyzed whether the LCS6 variant may be clinically relevant to patients with metastatic colorectal cancer (MCRC) treated with anti-EGFR therapy. LCS6 genotypes and KRAS/BRAF mutations were determined in the tumor DNA of patients with MCRC who underwent salvage cetuximab-irinotecan therapy. There were 25% G-allele (T/G+G/G) carriers and 75% T/T genotype carriers. G-allele carriers were significantly more frequent in the KRAS mutation group than in patients with KRAS wild type (p = 0.004). In the patients without BRAF V600E mutation, overall survival (OS) and progression-free survival (PFS) times were compared between carriers of the LCS6 G-allele genotypes and carriers of the wild-type T/T genotype. LCS6 G-allele carriers showed worse OS (p = 0.001) and PFS (p = 0.004) than T/T genotype carriers (confirmed in the multivariate model including the KRAS status). In the exploratory analysis of the unresponsive patients with KRAS mutation, LCS6 G-allele carriers showed adverse OS and PFS times (Graziano et al. 2010).
Despite these results, more studies are needed that will examine the effects of chemotherapeutic agents on the miRNA expression profiles and their possible usage for predicting therapy response in CRC patients.
4.7 MiRNAs as Potential Therapeutic Targets in CRC
Since miRNAs constitute a robust network for gene regulation, they possess a great potential as both, a novel class of therapeutic targets and a powerful intervention tool (Fig. 4.3). The biosynthesis, maturation and activity of miRNAs can be manipulated with various oligonucleotides that encode the sequences complementary to mature miRNAs (Aslam et al. 2009). Over-expression of miRNAs can be induced either by using synthetic miRNA mimics or chemically modified oligonucleotides. Conversely, miRNAs can be silenced by antisense miRNA oligonucleotides (AMOs), locked nucleic acids (LNAs) and “antagomirs” (synthetic analogues of miRNAs). Cross-sensitivity with endogenous miRNAs and lack of specificity for cancer cells can cause non-specific side effects during miRNA modulation therapy. However, the use of an effective delivery system and less toxic synthetic anti-miRNA oligonucleotides may minimize such side effects (Krutzfeldt et al. 2005; Zhang and Farwell 2008). Gene therapies may be designed to treat CRC and to block the progression of precursor lesions by manipulating the tumor suppressive or oncogenic miRNAs. Such manipulation may control the tumor growth rate and have potential as a new therapy for both early and advanced cancers (Calin and Croce 2007; Tong and Nemunaitis 2008).
Studies have revealed that inhibition of miR-21 and miR-17-92 activity is associated with reduced tumor growth, invasion, angiogenesis and metastasis (Krichevsky and Gabriely 2009; Dews et al. 2006). Moreover, over-expression of miR-21 is associated with low sensitivity and a poor response to chemotherapy, and its inhibition may improve the response to chemotherapy. On the other hand, restoration of miR-145 expression has been associated with inhibition of tumor cells growth via down-regulation of IRS-1. Expression levels of miR-451, down-regulated in tumor tissues of CRC patients, were increased in vitro and caused reduced cell proliferation and increased sensitivity to radiotherapy (La Rocca et al. 2009). These miRNAs present examples of miRNAs validated as oncogenes or tumor suppressors in CRC and thus of potential candidates for miRNA-based targeted CRC therapy. Targeting such miRNAs may help to not only prevent the recurrence of disease in high-risk tumors in UICC stage II and control the growth of advanced metastatic tumors, but they also could provide another possibility for chemoresistant and radioresistant cancer patients. Although experimental miRNA therapy results look promising, only a limited number of studies have been conducted under in vivo conditions in animal models. There is still a long way to go to reach clinical testing of the first miRNA-based therapy for CRC in the future.
4.8 Perspectives and Challenges
The discovery of miRNAs has substantially changed the view on gene regulation, and new findings over the past few years have catapulted miRNAs to the center stage of cancer molecular biology. It is now evident that dysregulation of miRNAs is an important step in the development of many cancers, including CRC. A number of studies based on expression profiling have proven there are significant changes of miRNA expression levels in CRC tissue in comparison to colorectal epithelium, and these have identified groups of miRNAs enabling prognostic stratification of CRC patients and prediction of their responses to selected chemotherapeutic regimens and radiotherapy. To improve knowledge as to the roles of miRNAs in CRC pathogenetic pathways, functional effects of particular miRNAs have been successfully studied. The results of these studies suggest great potential for miRNAs as a novel class of therapeutic targets and as a powerful intervention tool in CRC. MiRNAs’ occurrence has been repeatedly observed also in serum and plasma, and miRNAs as novel minimally invasive biomarkers have indicated reasonable sensitivity for CRC detection and compare favorably with the fecal occult blood test. Figure 4.4 summarizes how miRNAs may enter the clinical management of CRC patients in the near future.
The potential usage of microRNAs in the clinical management of the colorectal cancer patients (Slaby et al. 2009)
References
Akao Y, Nakagawa Y, Naoe T. Let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–6.
Arndt GM, Dossey L, Cullen LM, et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer. 2009;9:374.
Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36.
Aslam MI, Taylor K, Pringle JH, et al. MicroRNAs are novel biomarkers of colorectal cancer. Br J Surg. 2009;96:702–10.
Bailey SG, Sanchez-Elsner T, Stephanou A, et al. Regulating the genome surveillance system: miRNAs and the p53 super family. Apoptosis. 2010;15:541–52.
Bandres E, Cubedo E, Agirre X, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29.
Bommer GT, Gerin I, Feng Y, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–307.
Boni V, Zarate R, Villa JC, et al. Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fluorouracil and irinotecan. Pharmacogenomics J. 2010. doi:10.1038/tpj.2010.58.
Borralho PM, Kren BT, Castro RE, et al. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009;276:6689–700.
Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9.
Calin GA, Croce CM. Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. J Clin Invest. 2007;117:2059–66.
Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52.
Chen K, Song F, Calin GA, et al. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29:1306–11.
Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006.
Chen X, Guo X, Zhang H, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–92.
Chen Y, Song Y, Wang Z, et al. Altered expression of miR-148a and miR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg. 2010;14:1170–9.
Cho WC. MicroRNAs in cancer – from research to therapy. Biochim Biophys Acta. 2010a;1805:209–17.
Cho WC. MicroRNAs: Potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010b;42:1273–81.
Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–74.
Corney DC, Flesken-Nikitin A, Godwin AK, et al. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8.
Crosby ME, Kulshreshtha R, Ivan M, et al. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–9.
Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–92.
Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–5.
Earle JS, Luthra R, Romans A, et al. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn. 2010;12:433–40.
Esquela-Kerscher A, Slack FJ. Oncomir – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69.
Faber C, Kirchner T, Hlubek F. The impact of microRNAs on colorectal cancer. Virchows Arch. 2009;454:359–67.
Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67.
Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80.
Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–79.
Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–7.
Gaur A, Jewell DA, Liang Y, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–68.
Graziano F, Canestrari E, Loupakis F, et al. Genetic modulation of the Let-7 microRNA binding to KRAS 3′-untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab-irinotecan. Pharmacogenomics J. 2010;10:458–64.
Gregersen LH, Jacobsen AB, Frankel LB, et al. MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS One. 2010;5:e8836.
Guo C, Sah JF, Beard L, et al. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–46.
He L, He X, Lowe SW, Hannon GJ. MicroRNAs join the p53 network – another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–22.
Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–8.
Hu G, Chen D, Li XM, et al. MiR-133b regulates the MET proto-oncogene and inhibits the growth of colorectal cancer cells in vitro and in vivo. Cancer Biol Ther. 2010;10:2.
Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–26.
Huang ZM, Yang J, Shen XY, et al. MicroRNA expression profile in non-cancerous colonic tissue associated with lymph node metastasis of colon cancer. J Dig Dis. 2009;10:188–94.
Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47.
Krichevsky AM, Gabriely G. MiR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53.
Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9.
Kulda V, Pesta M, Topolcan O, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet. 2010;200:154–60.
La Rocca G, Badin M, Shi B, et al. Mechanism of growth inhibition by microRNA 145: the role of the IGF-I receptor signaling pathway. J Cell Physiol. 2009;220:485–91.
Landi D, Gemignani F, Naccarati A, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–84.
Lanza G, Ferracin M, Gafa R, et al. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer. 2007;6:54.
Lee HC, Kim JG, Chae YS, et al. Prognostic impact of microRNA-related gene polymorphisms on survival of patients with colorectal cancer. J Cancer Res Clin Oncol. 2010;136:1073–8.
Link A, Balaguer F, Shen Y, et al. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766–74.
Lodygin D, Tarasov V, Epanchintsev A, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–600.
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8.
Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–61.
Melo SA, Ropero S, Moutinho C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–70.
Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58.
Michael MZ, O’Connor SM, van Holst Pellekaan NG, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91.
Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399–416.
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8.
Monzo M, Navarro A, Bandres E, et al. Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res. 2008;18:823–33.
Motoyama K, Inoue H, Takatsuno Y, et al. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069–75.
Nagel R, le Sage C, Diosdado B, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–802.
Nakajima G, Hayashi K, Xi Y, et al. Non-coding microRNAs hsa-let-7 g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genomics Proteomics. 2006;3:317–24.
Natalwala A, Spychal R, Tselepis C. Epithelial-mesenchymal transition mediated tumourigenesis in the gastrointestinal tract. World J Gastroenterol. 2008;14:3792–7.
Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–81.
Nuovo GJ. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods. 2008;44:39–46.
Pechlivanis S, Pardini B, Bermejo JL, et al. Insulin pathway related genes and risk of colorectal cancer: INSR promoter polymorphism shows a protective effect. Endocr Relat Cancer. 2007;14:733–40.
Resnick KE, Alder H, Hagan JP, et al. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–9.
Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–9.
Rossi L, Bonmassar E, Faraoni I. Modification of miR gene expression pattern in human colon cancer cells following exposure to 5-fluorouracil in vitro. Pharmacol Res. 2007;56:248–53.
Rossi S, Kopetz S, Davuluri R, et al. MicroRNAs, ultraconserved genes and colorectal cancers. Int J Biochem Cell Biol. 2010;42:1291–7.
Sarver AL, French AJ, Borralho PM, et al. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:401.
Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–24.
Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36.
Shell S, Park SM, Radjabi AR, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104:11400–5.
Shi B, Sepp-Lorenzino L, Prisco M, et al. MicroRNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem. 2007;282:32582–90.
Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402.
Slaby O, Svoboda M, Michalek J, et al. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;14;8:102.
Song B, Wang Y, Titmus Ma, et al. Molecular mechanism of chemoresistance by mir-215 in osteosarcoma and colon cancer cells. Mol Cancer. 2010;9:96.
Strillacci A, Griffoni C, Sansone P, et al. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res. 2009;315:1439–47.
Svoboda M, Izakovicova Holla L, Sefr R, et al. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int J Oncol. 2008;3:541–7.
Takayama T, Miyanishi K, Hayashi T, et al. Colorectal cancer: genetics of development and metastasis. J Gastroenterol. 2006;41:185–92.
Tazawa H, Tsuchiya N, Izumiya M, et al. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–7.
Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–55.
Toyota M, Suzuki H, Sasaki Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–32.
Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 2009;30:953–9.
Tsuchiya N, Ochiai M, Nakashima K, et al. SND1, a component of RNA-induced silencing complex, is up-regulated in human colon cancers and implicated in early stage colon carcinogenesis. Cancer Res. 2007;67:9568–76.
Valeri N, Croce CM, Fabbri M. Pathogenetic and clinical relevance of microRNAs in colorectal cancer. Cancer Genomics Proteomics. 2009;6:195–204.
Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61.
Wang X, Lam EK, Zhang J, et al. MicroRNA-122a functions as a novel tumor suppressor downstream of adenomatous polyposis coli in gastrointestinal cancers. Biochem Biophys Res Commun. 2009;387:376–80.
Wang YX, Zhang XY, Zhang BF, et al. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis. 2010;11:50–4.
Webster RJ, Giles KM, Price KJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–41.
Weiss GJ, Bemis LT, Nakajima E, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19:1053–9.
Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–4.
Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–74.
Yamakuchi M, Ferlito M, Lowenstein CJ. MiR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–6.
Young LE, Dixon DA. Posttranscriptional Regulation of Cyclooxygenase 2 Expression in Colorectal Cancer. Curr Colorectal Cancer Rep. 2010;6:60–7.
Zhang B, Farwell MA. MicroRNAs: a new emerging class of players for disease diagnostics and gene therapy. J Cell Mol Med. 2008;12:3–21.
Zhou J, Zhou Y, Yin B, et al. 5-Fluorouracil and oxaliplatin modify the expression profiles of microRNAs in human colon cancer cells in vitro. Oncol Rep. 2010;23:121–8.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer Netherlands
About this chapter
Cite this chapter
Slaby, O., Svoboda, M., Michalek, J., Vyzula, R. (2011). MicroRNAs in Colorectal Cancer. In: Cho, W. (eds) MicroRNAs in Cancer Translational Research. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-0298-1_4
Download citation
DOI: https://doi.org/10.1007/978-94-007-0298-1_4
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-0297-4
Online ISBN: 978-94-007-0298-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)