Abstract
One of the important problems in using gene expression profiles to forecast cancer is how to effectively select a few useful genes to build exact models from large amount of genes. Classification is also a major issue in data mining. The classification difficulties in medical area often classify medical dataset based on the outcomes of medical analysis or report of medical action by the medical practitioner. In this study, a prediction model is proposed for the classification of cancer based on gene expression profiles. Feature selection also plays a vital role in cancer classification. Feature selection techniques can be used to extract the marker genes to improve classification accuracy efficiently by removing the unwanted noisy and redundant genes. The proposed study discusses the bijective-soft-set-based classification method for gene expression data of three different cancers, which are breast cancer, lung cancer, and leukemia cancer. The proposed algorithm is also compared with fuzzy-soft-set-based classification algorithms, fuzzy KNN, and k-nearest neighbor approach. Comparative analysis of the proposed approach shows good accuracy over other methods.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others

Keywords
1 Introduction
Classification and feature reduction are wide areas of research in data mining. Many practical applications of classification involve a large volume of data and/or a large number of features/attributes [1]. Gene expression denotes the activation level of every gene within an organism at an exact point of time. The classification is the process of predicting the classes among the massive amount of dataset using some machine learning algorithms. The classification of different tumor types in gene expression data is of great importance in cancer diagnosis and drug discovery, but it is more complex because of its huge size [2]. There are a lot of methods obtainable to evaluate a gene expression profiles. A common characteristic of these techniques is selecting a subset of genes that is very informative for classification process and to reduce the dimensionality problem of profiles. In this study, entropy filter is used for selecting informative genes [2].
In bijective soft set, every element can be only mapped into one parameter and the union of partition by parameter set is universe [3]. In this study, improved bijective-soft-set-based classification is applied for generating decision rules from the reduced training dataset. The generated decision rules can be used for classification of test data given. In this study, the proposed bijective-soft-set-based approach is compared with fuzzy-soft-set-based classification algorithms, fuzzy KNN, and k-nearest neighbor approach [4]. In comparison with other methods, improved bijective-soft-set-based classification provides more accuracy.
The rest of the paper is structured as follows: Sect. 2 describes the fundamental concepts of soft set and bijective soft set. Section 3 describes overall structure of the proposed work, Sect. 4 discusses experimental analysis, and Sect. 5 describes the conclusion.
2 Basic Concepts of Set Theory and Bijective Soft Set Theory
In this section, we describe the basic notions of soft sets and bijective soft set. Let U be the initial universe of objects and E be a set of parameters in relation to objects in U. Parameters are often attributes, characteristics, or properties of objects [5].
Definition 1
A pair (F, A) is called a soft set over U, where F is a mapping given by F: A → P (U).
Definition 2
Let (F, B) be a soft set over a common universe U, where F is a mapping F: B → P (U) and B is non-empty parameter set [3].
Definition 3
Let U = {x 1, x 2 ,…, x n} be a common universe, X be a subset of U, and (F, E) be a bijective soft set over U. The operation of “(F, E) restricted AND X” denoted by \( (F,\,E)\; \tilde{\wedge}\,X \) is defined by U e ∈ E {F (e): F (e) ⊆ X} [3].
Definition 4
Let U = {x 1, x 2,…, x n} be a common universe, X be a subset of U, and (F, E) be a bijective soft set over U. The operation of “(F, E) relaxed AND X” denoted by \( (F,\,E)\,\tilde{ \wedge }\,X \) is defined by U e ∈ E {F (e): F (e) ∩ X ≠ ∅} [3].
3 Proposed Work
Based on the bijective soft set theory mentioned in the last section, we begin our experiment. The whole experimental process includes 3 steps: (1) discretization, (2) pretreatment, and (3) classification.
In the first step, datasets are discretized based on class–attribute contingency coefficient discretization algorithm because gene expression datasets include only continuous-valued attributes [6]. In the second step, informative genes are selected using entropy filtering. The efficiency of the genes is considered using entropy filter method. Entropy measures the uncertainty of a random variable. For the measurement of interdependency of two random genes X and Y, Shannon’s information theory is used [2]. Then, improved bijective-soft-set-based classification approach is applied for generating rules.
3.1 Improved Bijective-Soft-Set-Based on Classification: Proposed Approach
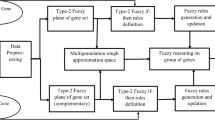
In the improved bijective-soft-set-based classification algorithm, two types of rules are generated. First type of rule is deterministic rule (certain rule) and are generated using AND and restricted AND operation [1]. Second type of rule is non-deterministic rule (possible rule) and are achieved by using AND and relaxed AND operations. For each non-deterministic rule, support is computed. Improved bijective-soft-set-based classification approach is presented in Fig. 1.
Table 1 represents a sample of the dataset as an example in order to extract the rules. Let A = {A1, A2} be the set of condition attributes, and D is the decision attribute. Decision attributes {B, D} stand for benign and malignant.
The proposed approach is explained with an example given in Table 1.
Step 1: Construct bijective soft set from conditional attributes [1].
Step 2: Construct bijective soft set from decision attribute.
B (Benign) = {X1, X2, X3, X4, X7, X9, X10} D (Malignant) = {X5, X6, X8}
Step 3: Apply AND operation on the bijective soft set (Fi, Ai).
(F1, A1) ∧ (F2, A2) = (H, C) = {{X1, X3, X9}, {X5, X6}, {X7, X8}, {X2, X4}}
Step 4: Generate deterministic rules by using U e ∈ E {F (e): F (e) ⊆ X}
(H, C) Restricted AND B (Benign) = {{X1, X3, X9}, {X2, X4}}
(H, C) Restricted AND D (Malignant) = {X5, X6}
(F, B) = {{X1, X3, X9}, {X2, X4}, {X5, X6}}
If A1 = 1 and A2 = 1 => d = Benign If A1 = 1 and A2 = 2 => d = Benign
If A2 = 2 and A2 = 1 => d = Malignant.
Step 5: Generate non-deterministic rules using U e ∈ E {F (e): F (e) ∩ X ≠ ∅}
If A1 = 1 and A2 = 1 => d = Benign If A1 = 1 and A2 = 2 => d = Benign
If A2 = 2 and A2 = 1 => d = Malignant If A1 = 2 and A2 = 2 => d = Benign
If A1 = 2 and A2 = 2 => d = Malignant.
Step 6: Support (Benign, Malignant) = \( \left( {\frac{1}{10 } \wedge \frac{2}{10}} \right) = 0.5. \)
4 Experimental Analysis
The dataset is collected from public microarray data repository [7]. In this study [8], the three cancer gene expression datasets selected for experiment are lung cancer dataset, leukemia dataset, and breast cancer dataset. The lung cancer dataset consists of 7,219 genes and 64 samples, and it belongs to the class ALL/AML. The leukemia cancer dataset consists of 7,219 genes and 96 samples, and it belongs to the class tumor/normal. The breast cancer dataset consists of 5,400 genes and 34 samples, and it belongs to the class benign/malignant.
4.1 Validation Measures
Validation is the key to making developments in data mining, and it is especially important when the area is still at the early stage of its development. Many validation methods are available. In this paper, we measure accuracy of the proposed algorithm based on precision, recall, and F-measure. The proposed algorithm’s accuracy is compared with three existing algorithms such as fuzzy-soft-set-based classification [4], KNN, and fuzzy KNN methods [4]. Figures 2, 3, and 4 show the comparative analysis of classification algorithms for gene expression datasets.
From the above results, it can be easily concluded that the IBISOCLASS algorithm is an effective one for classification of gene expression cancer dataset.
5 Conclusion
Classification is the key method in microarray technology. In this paper, classification technique is applied for predicting the cancer types among the genes in various cancer gene expression datasets such as leukemia cancer gene expression dataset, lung cancer gene expression dataset, and breast cancer gene expression dataset. In this study, improved bijective soft set approach is proposed for classification of gene expression data. The classification accuracy of the proposed approach is compared with the fuzzy soft set, fuzzy KNN, and KNN algorithm. The investigational analysis shows the effectiveness of the proposed improved bijective soft set approach over the other three methods. In future, it can be applied for other datasets also.
References
S. Udhaya Kumar, H. Hannah Inbarani, S. Senthil Kumar, “Bijective soft set based classification of Medical data”, International Conference on Pattern Recognition, Informatics and Medical Engineering, pp. 517–521, 2013.
Hamid Mahmoodian et al., “New Entropy-Based Method for Gene Selection”, IETE Journal of Research, vol. 55, no. 4, pp. 162–168, 2009.
K. Gong et al., “The Bijective soft set with its operations”, An International Journal on Computers & Mathematics with Applications, vol. 60, no. 8, pp. 2270–2278, 2010.
N. Kalaiselvi, H. Hannah Inbarani, “Fuzzy Soft Set Based Classification for Gene Expression Data”, International Journal of Scientific & Engineering Research, vol. 3, no. 10, 2012.
D. Molodtsov, “Soft set theory-first results”, An International Journal on Computers & Mathematics with Applications, vol. 37, no. 4–5, pp. 19–31, 1999.
Cheng-Jung Tsai, Chien-I Lee, Wei-Pang Yang, “A discretization algorithm based on Class-Attribute Contingency Coefficient”, An International Journal on Information Sciences, vol. 178, pp. 714–73, 2008.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer India
About this paper
Cite this paper
Udhaya Kumar, S., Hannah Inbarani, H., Senthil Kumar, S. (2014). Improved Bijective-Soft-Set-Based Classification for Gene Expression Data. In: Krishnan, G., Anitha, R., Lekshmi, R., Kumar, M., Bonato, A., Graña, M. (eds) Computational Intelligence, Cyber Security and Computational Models. Advances in Intelligent Systems and Computing, vol 246. Springer, New Delhi. https://doi.org/10.1007/978-81-322-1680-3_14
Download citation
DOI: https://doi.org/10.1007/978-81-322-1680-3_14
Published:
Publisher Name: Springer, New Delhi
Print ISBN: 978-81-322-1679-7
Online ISBN: 978-81-322-1680-3
eBook Packages: EngineeringEngineering (R0)