Abstract
The use of traditional microbiological culturing methods for the study of microbes had limited success since it has been estimated that 99% of microbes cannot be cultivated easily. Over the past decade, “metagenomics,” which is the culture-independent genomic analysis of microbes, has been developed to overcome these difficulties. Metagenomic analysis involves the basic steps like (1) the selection of an environmental niche, (2) the isolation of genetic material directly from an environmental sample, (3) manipulation of the genetic material, (4) library construction, and (5) the analysis of genetic material in the metagenomic library. The screening of clones can be done for phylogenetic markers or for other conserved genes by hybridization or multiplex PCR or for expression of specific traits, such as enzyme activity or antibiotic production, or they can be sequenced randomly. This chapter gives an overview of metagenomics including its success as well as future biotechnological applications in pharmaceuticals, bioactive molecules, biocatalysts, biomaterials, and others. There can be little doubt that the field of metagenomics gene discovery offers enormous scope and potential for both fundamental microbiology and biotechnological development.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Microbes, being ubiquitous in nature, are essential for every part of human life. It is through the capacity of these tiny minuscules that our planet is replenished with the key elements of life, such as carbon, nitrogen, oxygen, phosphorous, and sulfur in accessible forms. All plants and animals have closely associated microbial communities that provide them necessary nutrients, growth factors, and vitamins. The billions of benign microbes that live in the human gut help us to digest food, break down toxins, and fight off disease-causing counterparts. We also depend on microbes to clean up pollutants in the environment, such as oil and chemical spills. Microbial communities not only play key role in maintaining environmental quality and health but also participate in the upkeep of individual plants and animals. They can also live in extreme environments, at temperatures, pressure, and pH levels in which no other life forms can dare to tread.
Think about the countless jobs these tiny things do for us, starting from making antibiotics, many drugs in clinical use, enzymes, and other chemicals for industrial use to remediate pollutants in soil and water, to enhance crop productivity, to produce biofuels, to ferment Variety of foods, and to provide unique signatures that form the basis of microbial detection in disease diagnosis and forensic analysis. This is an endless list, and knowing them well not only includes all the processes involved and the genes responsible for all kinds of work they do but also tapping the vast genetic diversity existing in the microbial world. It is, therefore, no surprise that more than 900 genomes have already been sequenced.
We currently have little information (<1% of all bacterial species) on the vast majority of microorganisms present in Earth’s different environments, mainly due to our inability to culture them in the laboratory. Historically, our inability to culture microorganisms is due to lack of knowledge of their physiology and environmental cues that may help in designing suitable culture medium. However, new cultivation techniques are beginning to address this problem (Handelsman 2004). Nonetheless, most of our knowledge has been gleaned from the relatively small number of presently culturable representatives. So miniscule is this representation that it gives no clues as to what constitutes a microbial world and what all can it do.
Going back to nature, therefore, what do we really want to know in microbial ecology and evolution? The fundamental questions include what types of species are present (phylogenetic questions), how many of each type are present at a given time and location (genomic questions), what are they doing there (metabolic and functional questions), and what resources are they using (biogeochemical questions)? The realization that most microorganisms cannot be grown readily in pure culture forced microbiologists to question their belief that the microbial world had been conquered.
Microbiology has experienced a number of transformations during its history of three and a half century. Each of these stages has altered microbiologists’ view of microorganisms and how to study them (Handelsman 2004). The roots of microbiology are firmly associated with the invention of microscope. The first record of a human being seeing a bacterial cell dates back to 1663, when Antonie van Leeuwenhoek watched bacteria that he recovered from his own teeth through his homemade microscope. He was a keen observer and an outstanding combiner of ground glass pieces to obtain a magnified view of a sample. His observations and detailed illustrations of microbial life prompted many other observers (both scientists and nonscientists) to take an interest in the microscopic world. His colorful descriptions of bacteria made their study compelling; in his descriptions of the many shapes of the bacteria he sampled from his teeth, he marveled that one “shot through the water like a pike does through water,” firmly establishing that these tiny objects were indeed alive. For the next 200 years, refinement in microscopy enabled microbiologists to view heterotrophs, autotrophs, and obligate parasites alike with better details. Robert Koch’s postulates and his own innovation in developing culture media were instrumental in making another shift. From 1880s forward, the microbiological world was virtually restricted to the culture tubes of a microbiology laboratory.
Microbiologists were attracted to the power and precision of studies of bacteria in pure culture, and as a result, most of the knowledge that fills modern microbiology textbooks today is derived from organisms maintained in pure culture. Because culturing provided the platform for building the depth and details of modern microbiological knowledge, for a long time microbiologists ignored the challenge to identify and characterize uncultured organisms. They focused instead on the rich source of diversity found in the readily culturable model organisms, and this contributed to the explosion of knowledge in microbial physiology and genetics in the 1960s to mid-1980s. Meanwhile, the study of uncultured microorganisms remained in the hands of a few persistent enthusiasts who began to accumulate hints that flitted at the edge of the microbiological consciousness, suggesting that culturing did not capture the full spectrum of microbial diversity. Many of the organisms could not be cultured on agar medium because their temperature requirements exceeded the melting point of the agar. Or that we did not, and still do not, understand their growth requirements. Therefore, elucidating the physiological function of microorganisms without culturing them required ingenuity. One of the indicators that cultured microorganisms did not represent much of the microbial world was the often observed “great plate count anomaly” – the discrepancy between the sizes of populations estimated by dilution plating and by microscopy. This discrepancy is particularly dramatic in some aquatic environments, in which plate counts and viable cells estimated by acridine orange staining can differ by four to six orders of magnitude, and in soil, in which 0.1–1% of bacteria are readily culturable on common media under standard conditions. It is now widely accepted that the application of standard microbiological methods for the recovery of microorganisms from the environment has had limited success in providing access to the true extent of microbial biodiversity. It follows that much of the extant microbial genetic diversity (collectively termed the metagenome) remains unexplored and unexploited, an issue of considerable relevance to a wider understanding of microbial communities. Incessant quest of man for newer and newer chemicals, drugs, and other resources from microbes that may have important bearing to the biotechnology industry has provided further impetus to this line of study. The recent development of technologies designed to access this wealth of genetic information through environmental nucleic acid extraction has provided a means of avoiding the limitations of culture-dependent genetic exploitation.
The visualization of microbial world was changed radically in 1985 by Carl Woese, whose work reflected that rRNA gene provides evolutionary chronometers (Woese 1987). A new branch of microbial ecology was created by Pace and his colleagues (Lane 1985; Stahl et al. 1985) by using direct analysis of 5S and 16S rRNA gene sequences from different environments. The analysis of these sequences was used to describe the diversity of microorganisms without culturing (Pace et al. 1986). The early studies relied on sequencing of reverse transcription-generated cDNA copies or direct sequencing of RNA. The development of PCR technology was the next technical breakthrough as by designing the appropriate primers virtually any gene and almost the entire gene could be amplified (Giovannoni et al. 1990). The new technique accelerated the discovery of diverse taxa as habitats across the earth could be surveyed (Barns et al. 1994; Eden et al. 1991). The application of PCR technology provided a view of microbial diversity that was not distorted by the culturing bias and revealed that the uncultured majority is unbelievably diverse and contains members that diverge deeply from the readily culturable minority. 16S rRNA gene sequences also provided an aid to culturing efforts in addition to providing a universal culture-independent means to assess the diversity. Culturing efforts have intensified recently due to nucleic acid probes labeled with fluorescent tags providing such an assay, facilitating quantitative assessment of enrichment and growth. Successes have included pure cultures of members of the SAR11 clade, now termed the genus Pelagibacter (Cho and Giovannoni 2004; Connon and Giovannoni 2002), which represents more than one-third of the prokaryotic cell types in the surface of the ocean but was known only by its 16S rRNA signature until 2002 (Morris et al. 2002). The Acidobacteria phylum (Janssen et al. 2002) is the corollary to SAR11 in terrestrial environments. Acidobacteria are abundant in soil, typically representing 20–30% of the 16S rRNA sequences amplified by PCR from soil DNA, but until recently only three members had been cultured (Barns et al. 1999). Given that many organisms will not be coaxed readily into pure culture, a critical advance is to extend the understanding of the uncultured world beyond cataloging 16S rRNA gene sequences, and microbiologists have striven to devise methods to analyze the physiology and ecology of these diverse, uncultured, hitherto unknown organisms.
Metagenomics
Among the methods designed to gain access to the physiology and genetics of uncultured microorganisms, metagenomics, or environmental genomics, the genome analysis of a population has emerged as a powerful centerpiece. Direct isolation of genomic DNA from an environment circumvents culturing the organisms under study, followed by cloning of genomic fragments into a culturable host that captures it for further study and preservation. With the feasibility of such a technique, numerous advances have been derived from sequence-based and functional analysis in samples from water and soil from diverse habitat and those associated with eukaryotic hosts.
Definition of Metagenomics
What is metagenomics? A review paper published in 2004 defines “metagenomics” as functional and sequence-based analysis of the collective microbial genomes contained in an environmental sample (Handelsman 2004). The report from the American Institute of Microbiology (2002) defines metagenomics as that “entails large-scale sequencing of pooled, community genomic material, with either random or targeted approaches, assembly of sequences into unique genomes or genome clusters, determination of variation in community gene and genome content or expression over space and time, and inference of global community activities, function, differentiation, and evolution from community genome data.” Probably the oldest paper that used the term “metagenome” was published in 2000 (Woese 1987). However, the concept that organisms could be identified without cultivation by retrieving and sequencing them directly from nature is much older. Metagenomic approaches to capture microbial diversity in natural habitats have been employed by many researchers for years. The terms used to describe such methods include environmental DNA libraries, zoo libraries, soil DNA libraries, recombinant environmental libraries, whole genome treasures, community (environmental) genome analysis, whole genome shotgun sequencing, random community genomics, and probably others. Among them, metagenomics seems to be the most commonly used term to describe such studies and was used for the title of the first International Conference titled Metagenomics 2003 organized by Dr. Christa Schleper in Darmstadt, Germany. Metagenomics combines the power of genomics, bioinformatics, and systems biology. Operationally, it is novel in that it involves study of the genomes of many organisms simultaneously.
Metagenomics is employed as a means of systematically investigating, classifying, and manipulating the entire genetic material isolated from environmental samples. This is a multistep process that relies on the efficiency of five main steps. The procedure consists of (a) the selection of an environmental niche, (b) the isolation of genetic material directly from an environmental sample, (c) manipulation of the genetic material, (d) library construction, and (e) the analysis of genetic material in the metagenomic library.
A sample is first collected that represents the environment under investigation because the biological diversity will be different in different environments. The second step of the procedure is the isolation of the DNA. As the samples may contain many different types of microorganisms, the cells are broken open using chemical methods such as alkaline conditions or physical methods such as sonication. Once the DNA from the cells is free, it must be separated from the rest of the materials in the sample. This is accomplished by taking advantage of the physical and chemical properties of DNA. Some methods of DNA isolation used include density centrifugation, affinity binding, and solubility/precipitation. Commercial kits are now available and are properly used for isolation of DNA from mixed samples (Lloyd-Jones and Hunter 2001).
Once the DNA is collected, it is manipulated so that it can be introduced in a chosen model organism. Genomic DNA is relatively large, so it is cut up into smaller fragments using enzymes called restriction endonucleases. These are special enzymes that cut DNA at a particular sequence of base pairs. Depending upon the enzyme used, this results in the smaller, linear fragments of DNA carrying either staggered or flush ends. The fragments are then combined (ligated) with suitable vectors. Vectors are small units of DNA that can be transformed into cells where they can replicate and produce the proteins encoded on the introduced DNA using the machinery that the cells use to express normal genes. The vectors also contain a selectable marker. Selectable markers provide a growth advantage that the model organism would not normally have otherwise (such as resistance to a particular antibiotic) and are used to identify which cell contains vectors (transformed). The ones which do not contain vector (untransformed) are selected out.
The next step is to introduce the vectors with the metagenomic DNA fragments into the model organism, to generate metagenomic library. This allows the DNA from organisms that would not grow under laboratory conditions to be replicated, expressed, and studied. DNA inserted in the vector is transformed into cells of a model organism, typically Escherichia coli. Though the first choice always falls on E. coli, it is becoming clear that this bacterium cannot express all the genes. A search for an alternative host has to be kept in mind and should become a part of all such protocols. Transformation is the physical insertion of foreign DNA into a cell, followed by stable expression of proteins. It can be done by chemical, electrical, or biological methods. The method of transformation is determined based on the type of sample used and the required efficiency of the reaction. The metagenomic DNA in the vectors represents the entire DNA in the same sample initially, but the vectors are designed such that only one kind of DNA fragment from the sample will be maintained in each individual cell. The transformed cells are then grown on selective media so that only the cells carrying vectors will survive. Each group of cells that grows in a unit is called a colony. Each colony consists of many cloned cells that originated from one single cell. The population of cells containing all of the metagenomic DNA samples in vectors constitutes what is called metagenomic library. Each colony can be used to create a stock of cells for future study of a single fragment of the DNA from the environmental sample.
The clones can be then screened for expression of specific traits, such as enzyme activity or antibiotic production (function-based approach), or they can be sequenced randomly (sequence-based approach). The clones can also be screened for phylogenetic markers or “anchors,” such as 16S rRNA and recA, or for other conserved genes by hybridization or multiplex PCR. This helps in taxonomic delineation of the source of the DNA (uncultured microbe). Each approach has strengths and limitations; together these approaches have enriched our understanding of the uncultured world, providing insight into groups of prokaryotes that are otherwise entirely unknown.
Metagenomic DNA Libraries
The basic steps of DNA library construction include generation of suitably sized DNA fragments, cloning of fragments into an appropriate vector, and screening for the gene of interest (Fig. 10.1). DNA fragmentation is a significant problem in constructing metagenomic libraries. Mainly vigorous extraction methods from environmental samples often result in excessive DNA shearing particularly when a higher yield is desired. An alternative approach uses blunt-end or T–A ligation to clone randomly sheared metagenomic fragments (Wilkinson et al. 2002).
Cosmid and bacterial artificial chromosome (BAC) libraries have been widely used for the construction of metagenomic libraries because of their ability to carry large DNA fragments (Beja 2004). Cloning such fragments of metagenomic DNA allows entire functional operons to be targeted with the possibility of recovering entire metabolic pathways. This approach has successfully been applied for the isolation of several multigenic pathways such as that responsible for the synthesis of the antibiotic violacein (Brady et al. 2001). Cosmid-sized (35–45 kbp) inserts in E. coli can also be stably maintained using fosmid vectors (Beja 2004).
The limitation of E. coli as a host for comprehensive mining of metagenomic samples is highlighted by the low number of positive clones obtained during a single round of screening (typically less than 0.01%). This suggests that without sample enrichment, the discovery of specific genes in a complex metagenome is technically challenging.
The assumption that expression in an E. coli host will not impose a further bias is largely untested. Although the E. coli transcriptional machinery is known to be relatively promiscuous in recognizing foreign expression signals, a bias in favor of Firmicutes genes has been established. The further development of host screening systems is therefore a fruitful approach for the more effective future exploitation of metagenomes.
Screening of Metagenomic Clones
Two strategies are generally used to screen and identify novel genes from metagenomic libraries: sequence-based analysis and function-based analysis (Table 10.1). Both sequence- and function-based screenings have individual advantages and disadvantages, and they have been applied successfully to discover genes from metagenome (Table 10.2).
Sequence-Based Analysis
Sequence-based analysis can involve complete sequencing of cloned DNAs. As described earlier, clones containing phylogenetic anchors indicate the probable taxonomic group and identify the source of the DNA fragment. Alternatively, random sequencing can be conducted, and its function could be deduced by comparing it with the entries available in databases. Once a gene of interest is identified, phylogenetic anchors can be sought in the flanking DNA to provide a link of phylogeny with the functional gene. Identification of phylogenetic markers is a powerful approach guided by sequence analysis. It was first proposed by the DeLong group, which produced the first genomic sequence linked to a 16S rRNA gene of an uncultured archaeon (Stein et al. 1996). Subsequently, they identified an insert from seawater bacteria containing a 16S rRNA gene that affiliated with the Proteobacteria. The sequence of flanking DNA revealed a bacteriorhodopsin-like gene. Its gene product was shown to be an authentic photoreceptor, leading to the insight that bacteriorhodopsin genes are not limited to Archaea but are in fact abundant among the Proteobacteria of the ocean (Black et al. 1995; Bohlool and Brock 1974).
Sequencing random clones is an alternative to phylogenetic marker-driven approach, which has produced dramatic insights, especially when conducted on a massive scale. Sequence-based analysis can infer the distribution and redundancy of functions in a community, linkage of traits, genomic organization and detect horizontal gene transfer. The recent monumental sequencing efforts, which include reconstruction of the genomes of uncultured organisms in a community in the Sargasso Sea (Venter et al. 2004) and acid mine drainage (Tyson et al. 2004), illustrate the power of large-scale sequencing efforts to enrich our understanding of uncultured communities. These studies have made new linkages between phylogeny and function, indicated the surprising abundance of certain types of genes, and reconstructed the genomes of organisms that have not yet been cultured.
The power of this approach is likely to increase, as the collection of phylogenetic markers is growing. With the diversity of such markers, it became possible to assign more and more fragments of anonymous DNA to the organisms from which they could have likely been derived. There is limited utility of use of phylogenetic markers either as the initial identifiers of DNA fragments under study or as indicators of taxonomic affiliation for DNA fragments carrying genes of interest because their function is limited. Hence, the small number of available markers is a deterrent to provide reliable placement of the DNA source in the “Tree of Life” (Henne et al. 1999).
Function-Based Analysis
In function-based screening, clones expressing desired traits are selected from libraries, and molecular and biochemical aspects of active clones are analyzed. Identification of clones that express a function is a powerful yet challenging approach to metagenomic analysis. Faithful transcription and translation of the gene or genes of interest and secretion of the gene product is required for its success, if the screen or assay requires it to be extracellular. Functional analysis has identified novel antibiotics (Courtois et al. 2003; Gillespie et al. 2002), antibiotic resistance genes (Diaz-Torres et al. 2003; Riesenfeld et al. 2004), Na+(Li+)/H+ transporters (Majernik et al. 2001), and degradative enzymes (Healy et al. 1995; Henne et al. 1999, 2000a, b), to name a few. The power of the approach is that it does not require the gene(s) of interest be recognizable by sequence analysis, making it the only approach to metagenomics that has the potential to identify entirely new classes of genes for new or known functions. However, function-based screening has several limitations. This method requires expression of the function of interest in the host cell (e.g., E. coli), as well as clustering of all the genes required for the said function. Heterologous expression still remains a barrier in extracting the maximum information from functional metagenomics analyses.
When the functions of interest do not provide the basis for selection, high-throughput screens can substitute them. For example, active clones display a characteristic and easily distinguishable appearance on certain indicator media, even when plated at high density. Henne et al. (1999) detected clones that utilize 4-hydroxybutyrate in libraries of DNA derived from agricultural or river valley soil with the indicator dye tetrazolium chloride (Henne et al. 1999). Very rare lipolytic clones in the same libraries were detected by production of clear halos on media containing rhodamine and either triolein or tributyrin Henne et al. (1999). Catabolic enzyme genes can also be screened by substrate-induced gene expression (SIGEX).
High-throughput screening can also be done to identify compounds that induce the expression of genes under the control of a quorum-sensing promoter. This is a very powerful approach as the screen is intracellular, thus detecting that metagenomics DNA which is in the same cell as the sensor for quorum-sensing induction (Handelsman 2004). One of the very good examples of such a sensor comprises luxR promoter, which is induced by acylated homoserine lactones, linked to gfp. Promoter luxR resides on a plasmid in an E. coli strain that cannot induce quorum sensing. However, if an inducer of the luxR-mediated transcription of gfp is expressed from metagenomics DNA, the cell fluoresces and can be captured by fluorescence microscopy.
This sensor system can also detect inhibitors of quorum sensing, if acylated homoserine lactones are added to medium and fluorescence-activated cell sorting is set to collect the nonfluorescent cells. Arrays of genes have been identified from the metagenomics libraries of mid gut of the gypsy moth and microbiota of the soil.
The discovery of new biological motifs is dependent in part on functional analysis of metagenomic clones. Assignment of functions to numerous “hypothetical proteins” in the databases has been done through functional screens of metagenomic libraries. To identify and overcome the barriers to heterologous gene expression and to detect rare clones efficiently in the immense libraries that represent all of the genomes in complex environments, further innovations in the techniques will be required. An emerging and powerful direction for metagenomic analysis is the use of functional anchors, which are the functional analogs of conventional phylogenetic anchors. Functional anchors define the functions that can be assessed rapidly in all of the clones in a library. When a collection of clones with a common function is assembled, they can be sequenced to identify phylogenetic anchors and genomic structure in the flanking DNA. Such an analysis can provide a slice of the metagenome that cuts across clones with a different selective tool, determining the diversity of genomes containing a particular function that can be expressed in the host carrying the library. Technological developments that promote functional expression and screening are bound to advance this new frontier of functional genomics (Handelsman 2004). Although function-driven screens usually result in identification of full-length genes (and therefore functional gene products), one limitation of this approach is its reliance on the expression of the cloned gene(s) and the functioning of the encoded protein in a foreign host.
Metagenomic studies have also been applied to environmental transcriptomes, where direct retrieval and analysis of microbial transcripts is done. In this approach, environmental mRNA is isolated. These mRNAs were then reverse transcribed, amplified with random primers, cloned, and functionally analyzed. This is a means of exploring functional gene expression within natural microbial communities without bias towards known sequences and provides a new approach for obtaining community specific variants of key functional genes (Pace et al. 1986).
Phage-display expression libraries provide a means for isolating DNA sequences by affinity selection of the surface-displayed expression product. This method is efficient and amenable to high-throughput screening, offering the potential to enrich even rare DNA sequences in the metagenome. Phage display was pioneered by George Smith in 1985, and it leads to full realization of the value of protein–ligand interaction. Phage display is used for many purposes. As a natural selection procedure, it is useful for generating targets for drug discovery (Benhar 2001; Trepel et al. 2002; Gnanasekar et al. 2004), epitope mapping (Matthews et al. 2002), and for screening antibodies (Prinz et al. 2004). Antibodies were one of the first proteins to be displayed on a phage surface (McCafferty et al. 1990), and the isolation of monoclonal antibodies has been one of the most successful applications of phage display to date (Hoogenboom et al. 1998).
Bacteriophages are estimated to total 1,031 virus particles (Brussow and Hendrix 2002). Due to the bacteria-killing activity of some phages, and to the diminishing power of antibiotics to treat disease, phage therapy is becoming commercially popular (Alisky et al. 1998; Miedzybrodzki et al. 2007; Capparelli et al. 2007; Easton 2009), and several important studies have been carried out on the possibility of developing phage as an alternative to antibiotics (Weber-Dabrowska et al. 2000; Wagenaar et al. 2005). Bacteriophages are used for phage display due to their natural ability to infect bacterial cells and because they can incorporate foreign DNA into their circular genome and transport them into a bacterial cell during infection (Smith and Petrenko 1997). However, phage display is limited by the expression capacity of the bacteriophage, a protein size with an upper limit of around 50 kDa (Crameri and Suter 1993). Filamentous phage display allows assembly in, and secretion from, an infected bacterium without compromising the host cell membrane (Mullen et al. 2006). E. coli cells infected with such bacteriophage become a factory for phage production, as the host machinery is commandeered to generate phage virions.
SIGEX
In 2005, Uchiyama et al. introduced a third type of function-based screen, which was termed as SIGEX. It has been developed for isolating novel catabolic genes from environmental metagenomes, particularly genes that are difficult to obtain using conventional gene cloning methods. This high-throughput screening approach employs an operon-trap gfp expression vector in combination with fluorescence-activated cell sorting. In SIGEX, restriction enzyme-digested metagenome fragments are ligated into an operon-trap vector (e.g., p18GFP), and a library is constructed and grown in a liquid culture by transforming a cloning host (e.g., E. coli). The library is subjected to a substrate-dependent gene-induction assay, and positive cells are selected by detecting activity of a co-expressed marker (e.g., GFP) encoded in the vector. High-throughput screening is possible if FACS is used to select GFP-expressing cells (Fig. 10.2).
In this way, Uchiyama and Miyazaki isolated aromatic hydrocarbon-induced genes from a metagenomic library derived from groundwater (Uchiyama et al. 2005).
Limitations of this method are (1) only genes homologous to known genes can be obtained, (2) genes obtained may be partial, (3) many enzymes are difficult to be expressed in a heterologous host as an active form, (4) catabolic genes that are distant from a relevant transcriptional regulator cannot be obtained, and (5) it is sensitive to the orientation of genes with desired traits (Yun and Ryu 2005).
METREX
A similar type of screen, designated metabolite-regulated expression (METREX), has been published by Williamson et al. (2005). The goal of this study was to design and evaluate a rapid screen to identify metagenomic clones that produce biologically active small molecules. To identify clones of interest, biosensor detecting small, diffusible signal molecules that induce quorum sensing is placed inside the same cell as the vector harboring a metagenomic DNA fragment. If the clone produces a quorum-sensing inducer, the cell produces GFP and can be identified by fluorescence microscopy or captured by fluorescence activated cell sorting. METREX detected quorum-sensing inducers among metagenomic clones that a traditional overlay screen would not. One inducing clone carrying a LuxI homologue has been identified. This way, later, Guan et al. (2007) identified a new structural class of quorum-sensing inducers from the mid-gut bacteria of gypsy moth larvae by employing this method. A monooxygenase homolog which produced small molecules that induced the activities of LuxR from Vibrio fischeri and CviR from Chromobacterium violaceum has been detected (Williamson et al. 2005; Guan et al. 2007).
PIGEX
In 2010, Uchiyama and Miyazaki introduced another screening based on inducer gene expression known as product-induced gene expression. It is a reporter assay-based screening method for enzymes, which was used to screen metagenomic library containing large number of clones. In this case, enzyme activities are detected by expression of gfp, which is triggered by product formation. In their study, transcriptional activator BenR was replaced upstream of gfp. E. coli cells harboring the benR-gfp cassette would fluoresce in the presence of a benzoate precursor compound if they expressed an enzyme capable of actively transforming the precursor into benzoate. This reporter assay system would allow the identification of desired enzymatic activities by linking product formation to reporter gene expression. Using this system, amidases were targeted which can convert benzamide to benzoate. Ninety-six thousand clones were screened, and 11 amidase genes were recovered from 143 fluorescent wells, 8 of which were homologous to known bacterial amidase genes, while 3 were novel genes (Uchiyama and Miyazaki 2010).
Sample Enrichment
In a metagenomic screening process (e.g., expression screening of metagenomic libraries), the target gene(s) represent a small proportion of the total nucleic acid fraction. Pre-enrichment of the sample thus provides an attractive means of enhancing the screening hit rate. The discovery of target genes can be significantly improved by applying one of several enrichment options, ranging from whole-cell enrichment to the selection and enrichment of target genes and genomes (Miller et al. 1999).
Culture enrichment on a selective medium favors the growth of target microorganisms. The inherent selection pressure can be based on nutritional, physical, or chemical criteria, although substrate utilization is most commonly employed. For example, a fourfold enrichment of cellulase genes in a small insert expression library was obtained by culture enrichment on carboxymethyl cellulose (Miller et al. 1999). Although culture enrichment will inevitably result in the loss of a large proportion of the microbial diversity by selecting fast-growing culturable species, this can be partially minimized by reducing the selection pressure to a mild level after a short period of stringent treatment.
Nucleic Acid Extraction and Enrichment Technology
Numerous community nucleic acid extraction methods have been developed. Mainly two principal strategies for the recovery of metagenomes are (a) cell recovery and (b) direct lysis. Extraction of total metagenomic DNA is essentially a compromise between the vigorous extraction required for the representation of all microbial genomes and the minimization of DNA shearing and the co-extraction of inhibiting contaminants as isolation of individual cell is rather a difficult process. Chemical lysis is a gentler method in comparison to mechanical bead beating, recovering higher molecular weight DNA. Chemical lysis can also select for certain taxa by exploiting their unique biochemical characteristics.
Total DNA extraction does not typically contain an even representation of the population’s genome within a given environmental sample. The dominant organism overshadows the rare organisms. This could lead to bias towards conclusion and downstream manipulations such as PCR. This can be overcome by means of experimental normalization. Separation of genomes can be done by caesium chloride gradient centrifugation in the presence of intercalating agent, such as bisbenzimide, for the buoyant density separation. The separation of genome is based on their %G + C content. Equal amount of each band on the gradient is combined to represent a normalized metagenome. Normalization can also be achieved by denaturing the fragmented genomic DNA first, then reannealing them under very stringent conditions (68°C for 12–36 h). The concept being, abundant ssDNA will anneal more rapidly as their number is higher than the rare dsDNA species. The ssDNA strands will be separated from dsDNA, resulting in enrichment of rarer sequences within the environmental sample.
Genome Enrichment Strategies
Many strategies are being employed for genome enrichment. One strategy is to target the active component of microbial populations. Such a strategy is aimed to tell us which species are functionally active in specific processes (Miller et al. 1999).
Stable-Isotope Probing (SIP)
Genome enrichment strategies can be used to target the active components of microbial populations. Stable-isotope probing (SIP) techniques involve the use of a stable-isotope-labeled substrate and density gradient centrifugal separation of the “heavier” DNA or RNA. After growing a mix of different microbial species on a simple 13C-labeled substrate like 13C-methanol, the 13C DNA produced by methanol-utilizing species can be clearly separated from the 12C DNA originating from species unable to utilize methanol. After DNA extraction from the growth medium, the newly formed (“heavy”) 13C DNA can be separated from the (“light”) 12C DNA by density-gradient centrifugation. The 13C DNA can then be identified by comparing with DNA libraries and subsequently linked to the active microbial species. This method is called stable-isotope probing (DNA-SIP). Actively growing microorganisms can also be labeled with 5-bromo-2-deoxyuridine (BrdU), and the labeled DNA or RNA is separated by immunocapture or density gradient centrifugation (Urbach et al. 1999).
Suppressive Subtractive Hybridization (SSH)
This technique identifies the differences between different DNAs derived from microorganisms. Adaptors are ligated to the DNA populations, and subtractive hybridization is carried out to select for DNA fragments unique to each DNA sample. It is completely PCR based and eliminates the step of single-stranded tester cDNA purification by streptavidin–biotin or hydroxyapatite. For cDNA subtraction, the tester pool is divided in two fractions, and a different adaptor is ligated to each fraction. An excess of driver cDNA, without linkers (adapters), is denatured and hybridized with each tester (with linkers) cDNA pools (first hybridization). Both samples are mixed together with addition of more single-stranded driver (second hybridization). The resulting pool is a mixture of single stranded, double stranded with only one linker, double stranded like the original pools, and double stranded with both linkers corresponding to the tester-specific fragments. Filling the ends of the linkers allows creating templates to be amplified by PCR. Conception of the adaptors is such that the cDNA possessing the same kind of adaptor on both sides will form a hairpin preventing amplification. Only the ones possessing both linkers will be amplified exponentially. The resulting PCR product is enriched in tester-specific cDNAs. The products are cloned and characterized to confirm their specificity by cDNA microarray. This is a powerful tool for genome enrichment, but the complexity of metagenomes makes this detection difficult. Using multiple rounds of subtractive hybridization can increase the sensitivity of the process (Green et al. 2001).
Gene Enrichment Strategies
To selectively enrich for a specific target gene within a metagenome, a more practical approach would be to use one of several differential expression technologies that rely on the isolation of mRNA to target transcriptional differences in gene expression. For example, differential expression analysis (DEA) is a very effective tool for gene enrichment (Ochman et al. 1993).
Differential Expression Analysis (DEA)
In this approach, the expression of genes upregulated for the specific activity can be identified. DEA targets transcriptional differences in gene expression. Many variations in the basic concept exist today which include selective amplification via biotin and restriction-mediated enrichment (SABRE), integrated procedure for gene identi-fication (IPGI), serial analysis of gene expression (SAGE), tandem arrayed ligation of expressed sequence tags (TALEST), and total gene expression analysis (TOGA). These techniques have been effectively applied for eukaryotic gene discovery, but none have been applied in a metagenomic context. Their high sensitivity and selectivity should enable small differences in expression of single copy genes to be detected (Futamata et al. 2001).
Gene Targeting
A number of PCR-based approaches designed to recover the flanking regions of a DNA fragment once its sequence is known have been reported (Futamata et al. 2001). Although suitable for use at a single-genome level, these methods are technically more difficult to apply at the metagenomic level due to the increased complexity of a multigenomic DNA sample. A desire to simplify this process led us to look at the development of other novel approaches.
One potentially powerful approach is based on in vitro hybridization of a genomic DNA sample with the target gene fragment acting as a probe. Genomic DNA is fragmented, and priming sites are introduced by ligation of adapters. The gene-specific PCR product is then used as a driver to selectively hybridize to full-length gene fragments in the DNA sample. These partially double-stranded full-length gene fragments can then be selectively separated from the single-stranded background (genomic DNA). To remove any residual background, the adapters are removed; because the full-length gene fragments are only partially double stranded, the priming sites will remain intact as the restriction enzyme can only act on double-stranded DNA within the priming site. The full-length gene can then be amplified. This method is particularly powerful for multigenomic cloning as the use of degenerate gene-specific primers on a metagenomic sample typically yields a population of target gene fragments. Genes coding for catabolic enzymes such as methane monooxygenase, ammonia monooxygenase, catechol dioxygenase, and phenol hydroxylase have been retrieved from the environment in order to gain insight into the genetic diversity of catabolic populations. It is currently expected that such genetic information could aid in understanding and advancing bioremediation (Daniel 2005).
However, as a tool for biocatalyst discovery, gene-specific PCR has two major drawbacks. First, the design of primers is dependent on existing sequence information and skews the search in favor of known sequence types. Functionally similar genes resulting from convergent evolution are not likely to be detected by a single gene-family-specific set of PCR primers. Second, only a fragment of a structural gene will typically be amplified by gene-specific PCR, requiring additional steps to access the full-length genes. Amplicons can be labeled as probes to identify the putative full-length gene(s) in conventional metagenomic libraries.
Environment Niche Selection
The microbial diversity of both cultured and uncultured microorganisms is a direct reflection of the environment from where they are derived. Though highly variable, two major communities taken into consideration are soil and marine ones. To this is added another community that exists in association with or parasitic to eukaryotic organisms.
Metagenomics of Soil
Phylogenetic surveys of soil ecosystems have shown that the number of prokaryotic species found in a single sample exceeds that of known cultured members. Soil metagenomics, which comprises isolation of soil DNA and the production and screening of clone libraries, can provide a cultivation-independent assessment of the largely untapped genetic reservoir of soil microbial communities. This approach has already led to the identification of novel biomolecules. However, owing to the complexity and heterogeneity of the biotic and abiotic components of soil ecosystems, the construction and screening of soil-based libraries is difficult and challenging. This review describes how to construct complex libraries from soil samples and how to use these libraries to unravel functions of the resident microbial communities (Dunbar et al. 1999).
The recovery of microbial soil DNA that represents the resident microbial community and is suitable for cloning or PCR is still an important challenge, considering the diversity of microbial species (both cultured and uncultured), the large populations of soil microorganisms, and the complex soil matrix, which contains many compounds (such as humic acids) that bind to DNA and interfere with the enzymatic modification of DNA. By using universal primers for bacteria and archaea, phylogenetic surveys can be carried out by PCR amplification of 16S rRNA genes from soil DNA. These results have allowed cataloging and comparison of the microbial diversity in different soil habitats and the comparative analysis of changes in community structure owing to altered environmental factors (Ovreas 2000; Dunbar et al. 2002; Zhou et al. 2002; Yeager et al. 2004; Henne et al. 1999).
Construction of Soil DNA Libraries
Same steps are involved in constructing soil-based libraries, as the cloning of genomic DNA of individual microorganisms, that is, fragmentation of the soil DNA by restriction-enzyme digestion or mechanical shearing, insertion of DNA fragments into an appropriate vector system, and transformation of the recombinant vectors into a suitable host.
Construction of libraries from soil DNA and screening of these libraries by functional and sequence-based approaches was the major breakthrough in soil metagenomics. This technology paved the way for elucidating the functions of organisms in soil communities, for genomic analyses of uncultured soil microorganisms, and for the recovery of entirely novel natural products from soil microbial communities. In landmark studies, novel genes that encoded useful enzymes and antibiotics were recovered by direct cloning of soil DNA into plasmid, cosmid, or BAC vectors and screening of the generated libraries (Brady and Clardy 2000; Rondon et al. 2000; Ogram et al. 1987). The genes were identified using functional screens, and some having little homology to known genes were identified. This illustrates the enormous potential of the analysis of soil-based metagenomic libraries.
Several factors are important for the success of projects to generate and screen soil derived metagenomic libraries. For example, composition of the soil sample, collection and storage of the soil sample, the DNA extraction method used for high quality DNA recovery, representation of the isolated DNA from the microbial community present in the original sample, the host vector systems used for cloning, maintenance and screening and the screening strategy, all may affect the final outcome.
Many soil DNA extraction protocols have been published, and commercial soil DNA extraction kits are available (Lloyd-Jones and Hunter 2001). Two main methods are known for the DNA extraction from soil: direct lysis of cells contained in the sample matrix followed by separation of DNA from the matrix and cell debris, pioneered by Ogram et al. (Gabor et al. 2003), or separation of the cells from the soil matrix followed by cell lysis. The crude DNA recovered by both methods is purified by standard procedures. The amounts of DNA isolated from different soil types using a selection of protocols range from less than 1 μg to approximately 500 μg of DNA per gram of soil (Brady and Clardy 2000). More DNA is recovered using the direct lysis approaches, perhaps because of the loss of biomass during separation. For example, Gabor et al. (Delong 2005) recorded a 10- to 100-fold reduction in the DNA yield using the cell separation approach compared with the direct lysis approach.
Metagenomics of Marine Microbial Community
Marine microbial communities were among the first microbial communities to be studied using cultivation-independent genomic approaches. Ocean-going genomic studies are now providing a more comprehensive description of the organisms and processes that shape microbial community structure, function, and dynamics in the sea. Through the insight of microbial community genomics, a more comprehensive view of uncultivated microbial species, genes and biochemical pathways, distributions, and naturally occurring genomic variability is being brought into sharper focus. Besides providing new perspectives on oceanic microbial communities, these new studies are now poised to reveal the fundamental principles that drive microbial ecological and evolutionary processes (Béjà et al. 2000).
Marine Microbial Case Studies
Several studies have used either large-insert DNA cloning techniques or whole genome shotgun (WGS) approaches or both to characterize marine microbial assemblages. The outcomes of these studies include the discovery of unsuspected mechanisms of light-driven energy generation in the ocean (Béja et al. 2002; Preston et al. 1996), a massive survey of the gene complement of Sar-gasso Sea microorganisms, and the characterization of metabolic pathways of methane-oxidizing archaea in deep-sea sediments (Hallam et al. 2004; Falkowski and de Vargas 2004; Kruger et al. 2003; Teeling et al. 2004).
Photobiology of Marine Picoplankton
Early forays into environmental genomics demonstrated the feasibility of obtaining informative genomic “snapshots” from uncultivated marine microorganisms (Beja et al. 2000; Nelson et al. 1999). Several surprising discoveries have come to light through recent surveys of genome fragments from bacterioplankton that were archived in BAC libraries. To identify genome fragments containing phylogenetic markers (for instance, rRNAs) and sequence flanking the genomic regions, a type of phylogenetically anchored chromosome walking (Kawarabayasi et al. 1999; Stein et al. 1996) has been one of the important approaches. A 130-kb BAC clone was isolated from an uncultivated SAR86 bacterium (Zhou et al. 2002) (an abundant component of α-proteobacteria in ocean surface waters) using this method. Sequencing of the 130-kb fragment revealed a new class of genes of the rhodopsin family (named proteorhodopsin) that had never before been observed in bacteria as a whole or in the ocean community. The new genes have similarities to the known genes called rhodopsins that capture light energy from the sun and couple this with carbon cycling in the ocean through non-chlorophyll based pathways. When the bacterial proteorhodopsin was expressed in E. coli, it functioned as a light-driven proton pump (Zhou et al. 2002). So this genomic survey of uncultivated marine bacteria led directly to the discovery of a new type of light-driven energy generation in oceanic bacteria. Later studies confirmed the presence of retinal-bound proteorhodopsin in the ocean and showed that optimized spectral “tuning” of bacterial rhodopsins matches depth-specific light availability. Shotgun sequencing from the Sargasso Sea has now verified both the abundance and diversity of this new class of photoproteins. The emerging understanding of proteorhodopsin taxonomic and environmental distributions is providing new insights into gene and genome evolution in microbial populations (Tamas et al. 2002).
Sargasso Sea Metagenomics
The Sargasso Sea is a part of the North Atlantic Ocean, lying roughly between the West Indies and the Azores. Here, the heart of the Bermuda Triangle is covered by the strongest and most notorious sea on the planet – the Sargasso Sea so named because there is a kind of seaweed, which lazily floats over its entire expanse, called Sargassum. Environmental investigations in the nutrient-poor waters near Bermuda in the Sargasso Sea led to the discovery of 1,800 new species of bacteria and more than 1.2 million new genes. Scientists used a whole-genome shotgun sequencing technique to clone random DNA fragments from the many microbes present in the sample, generating a treasure-house of new information.
The Sargasso Sea is a complex and physically sprawling ecosystem. The phylogeny of the community members of this diversity has not been exhaustively surveyed, and the inputs and outputs are more difficult to quantify. Craig Venter, who pioneered the Human Genome Project, led a group of scientists who embarked on the largest metagenomics project to date, in which they sequenced over 1 billion bp and claim to have discovered 1.2 million new genes (Venter et al. 2004). They placed 794,061 genes in a conserved hypothetical protein group, which contains genes to which functions could not be confidently assigned. The next most abundant group contained 69,718 genes apparently involved in energy transduction. Among these were 782 rhodopsin-like photoreceptors, increasing the number of sequenced proteorhodopsin genes by almost 10-fold. Linkage of the rhodopsin genes to genes that provide phylogenetic affiliations, such as genes encoding subunits of RNA polymerase, indicated that the proteorhodopsins were distributed among taxa that were not previously known to contain light-harvesting functions, including the Bacteroides phylum (Venter et al. 2004).
An intriguing initial observation is that many of the genomes in the Sargasso Sea contain genes with similarity to those involved in phosphonate uptake or utilization of polyphosphates and pyrophosphates, which are present in this extremely phosphate-limited ecosystem. The phosphorus cycle is not well understood, and this collection of genomes provides a new route for discovery of the mechanisms of phosphorus acquisition and transformation. The resulting data represent the largest genomic data set for any community on earth and offer a first glimpse into the broad ensemble of adaptations underlying diversity in the oceans. Because microbes generally are not preserved in the fossil record, genomic studies provide the key to understanding how their biochemical pathways evolved (Vezzi et al. 2005).
Future studies will allow more insights into how these molecules function as well as opportunities for mining and screening the data for specific applications. The vast data set provides a foundation for many new studies by other researchers. Analyses using iron-sulfur proteins as benchmarks led one group, for example, to conclude that these data reflect diversity equal to that in all the currently available databases, suggesting that microbial diversity thus far has been vastly underestimated (Beja et al. 2000).
Practical Approach
Large-Insert Bacterial Artificial Chromosome and Fosmid Libraries
Several strategies for cultivation-independent genomic survey of marine microbial communities have been used. More recently, fosmids and bacterial artificial chromosomes (BACs) have been applied in genomic analyses of naturally occurring marine microorganisms (Béja et al. 2002; Preston et al. 1996). These vectors are particularly useful for stable, high-fidelity propagation of large DNA inserts. DNA fragments of up to 200 kb can be stably cloned in these vectors; therefore, one clone could represent 5–10% of the entire genome of a small bacterium. BAC clones prepared from microbial assemblage DNA can be easily screened to identify and characterize the cloned gene fragments for functions or to evaluate phylogeny. The first example of characterization of a microorganism using this approach examined an abundant but uncultivated group of planktonic marine archaea (Stein et al. 1996). Several studies have expanded the characterization of uncultivated archaeal species using this general approach. BAC libraries are repositories of genomic material and can also serve as a valuable reference resource for further sequencing and in vitro biochemical experimentation.
Small-Insert Whole-Genome Shotgun Libraries
Another approach for cultivation-independent microbial genome characterization is a variant of whole-genome shotgun (WGS) sequencing. For pure bacterial cultures, the WGS approach has been important for obtaining complete genome sequences, including those of several marine bacteria and archaea. WGS sequencing has also been used to sequence microbial symbionts and, in one case, an extremely simple microbial biofilm assemblage (Chen et al. 2003). WGS sequencing relies on the preparation and end sequencing of small-DNA-insert libraries and subsequent sequence assembly in silico. The high-throughput nature of this approach makes it extremely attractive. Variations on this theme, using linker ligation and subsequent amplification techniques, have also been used to generate shotgun libraries from naturally occurring viral populations.
Till now, it seems that WGS approaches alone cannot adequately deconvolute whole genome sequences from complex microbial assemblages. As with the human genome sequencing effort, the most complete and reliable datasets will probably result from a combination of sequencing and analysis strategies. These will also probably include front-end cell purification strategies to reduce inherent complexity, followed by combined WGS and large-insert sequencing strategies. In combination, these approaches could enhance the accuracy, coverage, and reliability of genomics-based efforts to understand complex microbial communities. Nevertheless, WGS sequencing of microbial communities represents a powerful, even if expensive, approach for high-volume, single-pass gene survey and sampling (Delong 2005).
Viral Metagenomics
Viruses, most of which infect microorganisms, are the most abundant biological entities on the planet. Identifying and measuring the community dynamics of viruses in the environment is complicated more so, because less than 1% of microbial hosts have been cultivated. Also, there is no single gene that is common to all viral genomes, so total uncultured viral diversity cannot be monitored using approaches analogous to ribosomal DNA profiling. Metagenomic analyses of uncultured viral communities circumvent these limitations and can provide insights into the composition and structure of environmental viral communities.
Viral metagenomes mostly comprise novel sequences. Viral metagenomics, which also focuses on shotgun sequencing of metagenomes, gives insight into the vast and previously untapped diversity of viral communities in, for example, near-shore marine water samples (Breitbart et al. 2004), marine sediment sample (Breitbart et al. 2003), human fecal sample (Cann et al. 2005), and other fecal sample (Pedulla et al. 2003). When the marine sequences were first published, approximately 65% of them had no significant similarity to any sequence in the GenBank nonredundant database. Analyses 2 years later revealed that most of the viral sequences are still unique, despite the fact that the GenBank database has since more than doubled in size. Likewise, in the equine fecal metagenome, 68% of the sequences have no similarity to any sequence in GenBank (Pedulla et al. 2003). Genomic analyses of cultured phages also show that most of the open reading frames (ORFs) are novel (Wommack et al. 1999). By contrast, only about 10% of the sequences from environmental microbial metagenomes (Homann et al. 2004) and cultured microbial genomes (Tyson et al. 2004) are novel when analyzed in similar ways. These observations indicate that while much of the global microbial metagenome is being sampled, the global viral metagenome is still relatively uncharacterized. That there is an even greater amount of biodiversity than that attributed to prokaryotic communities allows further hypotheses to be developed about the role of viral communities. Daubin and Ochman have gone on to hypothesize that the unique genes in microbial genomes in fact were acquired from the phage genomic pool (Wommack et al. 1999).
Isolation of viral community DNA representative for metagenomic analyses is complicated by the presence of free and cellular DNA. If the free DNA is not removed, the viral DNA signal will be lost. Similarly, at ∼50 kb long, the average viral genome is about 50 times smaller than the average microbial genome (2.5 Mb), so any cellular contamination will overwhelm the viral signal (Homann et al. 2004). By an estimate, 200 L of seawater or 1 kg of solid material is a typical starting sample consisting of fecal, soil, and sediment samples suspended in osmotically neutral solutions before filtration. To separate the intact viral particles from the microorganisms and free DNA, a combination of differential filtration with tangential flow filters (TFF), DNase treatment, and density centrifugation in cesium chloride (CsCl) is used. Viruses sensitive to CsCl will disintegrate in this step, and very large or very small viruses will be lost in the filtration step. As assessed by pulse-field gel electrophoresis (Stahl et al. 1985) and epifluorescence microscopy, this protocol seems to capture most of the viral community. Once intact virions have been isolated, the viral DNA is extracted and cloned. Cloning of viral metagenomes representative is challenging, due to low DNA concentrations (∼10–17 ng DNA per virion), modified DNA bases, for example, 5-(4-aminobutylaminomethyl) uracil and 5-methyl cytosine, and the presence of lethal viral genes such as holins and lysozymes. In order to circumvent these problems, it is necessary to concentrate virions from several hundred liters, in most water samples, to obtain enough DNA for cloning. The linker-amplified shotgun library (LASL) technique includes a PCR amplification step, which makes it possible to clone small amounts of DNA (1–100 ng). The PCR step also converts modified DNA into unmodified DNA. A shearing step disrupts lethal virus genes by shearing DNA into small fragments (∼2 kb) and provides the random fragments necessary for cloning. It is possible to make representative metagenomic libraries, using this protocol, that contain viral fragments that are proportional to their concentrations in the original sample. LASLs typically contain millions of random clones. RNA and single-stranded DNA (ssDNA) viruses, however, cannot be cloned using this approach. Preliminary studies with random-primed reverse transcriptase and random-primed strand-displacement DNA polymerases indicate that these viral groups could be analyzed using metagenomic approaches (Homann et al. 2004).
Host-Associated Bacteria: Genomic Insights into Pathogenesis and Symbiosis
Pathogenesis
The amenability of host-associated microbes to physical separation makes them well suited to this approach, which is in contrast to organisms that reside in complex environmental communities. The first complete genome of an uncultured bacterium, the syphilis spirochaete, Treponema pallidum, was published in 1998 – a landmark in genome sequencing. Although the bacterial origin of syphilis was recognized a century ago, the infectious agent could not be isolated in continuous culture. The DNA that was used for sequencing the intracellular pathogen was obtained from the testes of infected rabbits by a series of lysis and centrifugation steps that eventually resulted in an essentially pure bacterial preparation. Sequence analysis immediately identified potential contributors to virulence and aided the development of DNA-based diagnostics (Piel 2004).
A year and a half of painstaking growth in coculture with human fibroblasts was necessary to obtain sufficient DNA to sequence the genome of the Whipple disease bacterium, Tropheryma whipplei. The sequence revealed deficiencies that indicated an explanation for the failure to propagate in culture. Based on these genomic insights, Renesto et al. (2003) used a standard tissue-culture medium, supplemented with amino acids that were implicated by the sequence analysis, to successfully cultivate T. whipplei in the absence of host cells, shortening their doubling time by an order of magnitude. This is one of many cases in which DNA sequence information has been used to improve culture techniques, diagnostics, and therapies for fastidious organisms (Wommack et al. 1999).
Symbiosis
Many bacterial symbionts that have highly specialized and ancient relationships with their hosts do not grow readily in culture. Many of them live in specialized structures, often in pure or highly enriched state, in host tissues, making them ideal candidates for metagenomic analysis because the bacteria can be separated readily from host tissue and other microorganisms. This type of analysis has been conducted with Cenarchaeum symbiosum, a symbiont of a marine sponge, a Pseudomonas-like bacterium that is a symbiont of Paederus beetles, Buchnera aphidicola, an obligate symbiont of aphids, the Actinobacterium, Tropheryma whipplei, the causal agent of the rare chronic infection of the intestinal wall, and the Proteo-bacterium symbiont of the deep-sea tube worm Riftia pachyptila. These systems provide good models for metagenomic analysis of more complex communities and thus warrant further attention in this review, although the term metagenomics typically connotes the study of multispecies communities (Handelsman 2004).
Tube Worm Symbiosis: Proteobacterium
Riftia pachyptila, the deep-sea tubeworm, lives 2,600 m below the ocean surface, near the thermal vents that are rich in sulfide and reach temperatures near 400 °C. The tube worm does not have a mouth or digestive tract, and therefore it is entirely dependent on its symbiotic bacteria, which provide the worm with food. The bacteria live in the trophosome, a specialized feeding sac inside the worm (Piel 2004). The bacteria and trophosome constitute more than half of the animal’s body mass. The bacteria oxidize hydrogen sulfide, thereby producing the energy required to fix carbon from CO2, providing sugars and amino acids (predominantly as glutamate) that nourish the worm (Piel 2004). The worm contributes to the symbiosis by collecting hydrogen sulfide, oxygen, and carbon dioxide and transporting them to the bacteria.
The bacterium is a member of the γ-Proteobacteria, as identified by 16S rRNA gene sequence. The bacteria have not been grown in pure culture in laboratory media, but they provide an excellent substrate for metagenomics because they reach high population density in the trophosome and exist there as a single species. Hughes et al. (1997) isolated DNA from the bacterial symbiont and constructed fosmid libraries that were used to understand the physiology of the bacteria. Robinson et al. (1998) identified a gene with similarity to ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) from the same fosmid library. All the residues associated with the active site are conserved in the protein sequence deduced from the DNA sequence, and it has highest similarity with the RubisCO from Rhodospirillum rubrum. The characterization of this gene lends further support to the premise that the chemoautotrophic bacterial symbiont in R. pachyptila fixes carbon for its host. The libraries were also screened for two-component regulators with a labeled histidine kinase gene as a probe. They identified a two-component system whose components complemented an envZ and a phoR creC double mutant, respectively.
The discovery of a functional envZ homologue indicates that the symbiont carries a response regulator that is typical of γ-Proteobacteria, although the signals eliciting responses from these proteins have not yet been functionally identified (Handelsman 2004).
Gut Microbiome
Our body surfaces are home to microbial communities whose aggregate membership outnumbers our human somatic and germ cells by at least an order of magnitude. The majority of microbes that reside in the gut, have a profound influence on human physiology and nutrition, and are crucial for human life. Furthermore, the gut microbes contribute to energy harvest from food, and changes of gut microbiome may be associated with bowel disease or obesity. They synthesize essential amino acids and vitamins and process components of otherwise indigestible contributions to our diet such as plant polysaccharides (Qin et al. 2010).
To understand and exploit the impact of the gut microbes on human health and well-being, it is necessary to decipher the content, diversity, and functioning of the microbial gut community. 16S ribosomal RNA gene sequence-based methods revealed that two bacterial divisions, Bacteroidetes and the Firmicutes, constitute over 90% of the known phylogenetic categories and dominate the distal gut microbiota. Metagenomic sequencing represents a powerful alternative to rRNA sequencing for analyzing complex microbial communities (Gill et al. 2006).
Illumina-based metagenomic sequencing has been used by Qin et al. (2010), where assembling and characterization of 3.3 million nonredundant microbial genes, derived from 576.7 gigabases of sequence, have been done from fecal samples of 124 European individuals. The gene set, ∼150 times larger than the human gene complement, contains an overwhelming majority of the prevalent microbial genes of the cohort and includes a large proportion of the prevalent human intestinal microbial genes. The genes are largely shared among individuals of the cohort. Over 99% of the genes are bacterial, indicating that the entire cohort harbors between 1,000 and 1,500 prevalent bacterial species and each individual at least 160 such species, which are also largely shared. It has been found that gut microbiome has signifi-cantly enriched metabolism of glycans, amino acids, and xenobiotics; methanogenesis; and 2-methyl-erythritol 4-phosphate pathway-mediated biosynthesis of vitamins and isoprenoids.
The application of metagenomics to the medical field has led to a highly productive integration of clinical, experimental, and environmental microbiology. The functional roles played by human microbiota are closely looked into either through animal models or studies of human populations. Of particular interest is the fact that several human diseases have been linked to alterations in the composition and dynamics of human microbiota. The inputs from human microbiome and these based on human gene expression and variability and their application are a subject of great scientific challenge and interest (Frank et al. 2011).
Biogeochemical Cycles
Metagenomics provides an important insight into the community-wide assessment of metabolic and biogeochemical function. Analysis of specific functions across all members of a community can generate integrated models about how organisms share the workload of maintaining the nutrient and energy budgets of the community in an environment. The models can then be tested with genetic and biochemical approaches. An example of such an analysis is the nearly complete sequencing of the metagenome of a community in acid drainage of the Richmond mine. This mine is known to be representing one of the most extreme environments on Earth. The microbial community forms a pink biofilm that floats on the surface of the mine water. The drainage water below the biofilm has a pH between 0 and 1 and high levels of Fe, Zn, Cu, and As. The temperature around the biofilm water is 42 °C and is microaerophilic, having no source of carbon or nitrogen other than the gaseous forms in the air. Few of the most prominent bacterial members of the community are Leptospirillum, Sulfobacillus, and sometimes Acidimicrobium, one archaeal species, Ferro-plasma acidarmanus, and other members of its group, the Thermoplasmatales. The mine is rich in sulfide minerals, including pyrite (FeS2), which is dissolved as a result of oxidation, and is catalyzed by microbial activity. Tyson et al. (2004) were able to clone total DNA, because of its simple community structure, and sequenced most of the community with high coverage. The G + C content of the genomes of the dominant taxa in the mine differs substantially, thus providing the good indicator of its source. Sequence alignment of 16S rRNA and tRNA synthetase genes confirmed the organismal origins of the clones. Nearly complete genomes of Leptospirillum group II and Ferroplasma type II were reconstructed, and substantial sequence information for the other community members could be obtained (Tyson et al. 2004).
The metagenomic sequence substantiated a number of significant hypotheses. First, it appears that Leptospirillum group III contains genes with similarity to those known to be involved in nitrogen fixation, suggesting that it provides the community with fixed nitrogen. This was a surprise because the previous supposition was that a numerically dominant member of the community, such as Leptospirillum group II, would be responsible for nitrogen fixation. However, no genes for nitrogen fixation were found in the Leptospirillum group II genome, leading to the suggestion that the group III organism is a keystone species that has a low numerical representation but provides a service that is essential to community function. Ferroplasma type I and II genomes contain no genes associated with nitrogen fixation but contain many transporters that indicate that they likely import amino acids and other nitrogenous compounds from the environment.
Energy appears to be generated from iron oxidation by both Ferroplasma and Leptospirillum spp. The genomes of both groups contain electron transport chains, but they differ significantly. The genomes of Leptospirillum group II and III contain putative cytochromes that typically have a high affinity for oxygen. The cytochromes may play a role in energy transduction as well as in maintaining low oxygen tension, thereby protecting the oxygen-sensitive nitrogenase complex. All of the genomes in the acid mine drainage are rich in genes associated with removing potentially toxic elements from the cell. Proton efflux systems are likely to be responsible for maintaining the nearly neutral intracellular pH, and metal resistance determinants pump metals out of the cells, maintaining nontoxic environment in the interior of the cells (Tyson et al. 2004).
The acid mine drainage community provides a model for the analysis of other communities. Determining the origin of DNA fragments and assigning functions may be more difficult for communities that are phylogenetically or physiologically more complex and variable, but the approach will be generally useful for all communities (Tyson et al. 2004).
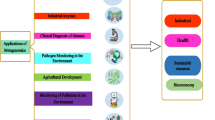
Metagenomics and Industrial Application
Besides the vast academic output, one of the major interests in metagenome analysis is its immense economic potential. Different industries have different motivations to explore the enormous resource that constitute the uncultivated microbial diversity. Currently, there is a global political drive to promote white (industrial) biotechnology as a central feature of the sustainable economic future of modern industrialized societies. This requires the development of novel enzymes, processes, products, and applications. Metagenomics promises to provide new molecules with diverse functions, but ultimately, expression systems are required for any new enzymes and bioactive molecules to become an economic success.
Metagenomics (Barns et al. 1999) has the potential to substantially impact industrial production. The dimensions of the enormous biological and molecular diversity, as shown by Torsvik and Venter (Lorenz and Eck 2005) and their coworkers, are truly astonishing. A pristine soil sample might contain in the order of 104 different bacterial species. More than one million novel open reading frames, many of which may encode putative enzymes, could be identified in a single effort such that sampled marine prokaryotic plankton retrieved from the Sargasso Sea.
Different industries are interested in exploiting the resource of uncultivated microorganisms that has been identified through large-scale environmental genomics for several reasons detailed below.
The Ideal Biocatalyst
For any industrial application, enzymes need to function sufficiently well according to several application-specific performance parameters. With the exception of yeasts and filamentous fungi, access to novel enzymes and biocatalysts has largely been limited by the comparatively small number of cultivable bacteria.
Novelty
For industries that produce bulk commodities such as high-performance detergents, a single enzyme backbone with superior functionality that has an entirely new sequence would be useful to avoid infringing competitors’ intellectual property rights.
Maximum Diversity
The pharmaceutical and supporting fine-chemicals industries often seek entire sets of multiple, diverse biocatalysts to build in-house toolboxes for biotransformations (Lorenz and Eck 2005). These toolboxes need to be made rapidly accessible to meet the strict timelines of a biosynthetic-feasibility evaluation in competition with traditional synthetic chemistry.
Elusive Metabolites
Many pharmacologically active secondary metabolites are produced by bacteria that live in complex consortia or by bacteria that inhabit niches that are difficult to reconstitute in vitro. So although there are reports on how to circumvent this general problem of microbial cultivation either by mimicking natural habitats (Lorenz and Eck 2005) or by allowing for interspecies communication after single cell microencapsulation (Kaeberlein et al. 2002), the cloning and heterologous expression of biosynthetic genes that encode secondary metabolites (usually present as gene clusters) is the most straightforward and reproducible method of accessing their biosynthetic potential.
White Biotechnology
“Industrial” or white biotechnology is currently a buzzword in the European biobusiness community. The term was coined in 2003 by the European Association for Bioindustries (EuropaBio), based on a case study report, and it denotes all industrially harnessed bio-based processes that are not covered by the red biotechnology (medical) or green biotechnology (plant) labels. White biotechnology has its roots in ancient human history, and its products are increasingly a part of everyday life, from vitamins, medicines, biofuel, and bioplastics to enzymes in detergents or dairy and bakery products. It is the belief of industrial promoters, analysts, and policy makers that white biotechnology has the potential to impact industrial production processes on a global scale. The main long-term applications of white biotechnology will be focused on replacing fossil fuels with renewable resources (biomass conversion), replacing conventional processes with bioprocesses (including metabolic engineering), and creating new high-value bioproducts, including nutraceuticals, performance chemicals, and bioactives. While the possibilities are immense, some success stories are already being cited (Zengler et al. 2002).
Screening Enzymes for Industrial Use
Diversa, the largest and most prominent specialist biotech company for the commercialization of metagenome technologies, has described several approaches to access “uncultivable” microorganisms. Applying a classical growth-selection-based expression strategy, diverse environmental libraries were constructed in E. coli using phage λ or Bac vectors. After growth in media containing nitriles as the sole nitrogen source, more than 100 new and diverse nitrilase genes were recovered (Zengler et al. 2002). The resulting enzyme library is marketed to serve the fine-chemical and pharmaceutical industries (Robertson et al. 2004).
In addition to new technologies to amplify DNA from limited resources using random primers and strand-displacing DNA polymerase from phage Φ29 (DeSantis et al. 2002), a strategy promoted by Lucigen (Middleton, Wisconsin, USA), it is clear that current mass-sequencing efforts in several laboratories will facilitate the in silico identification of open reading frames that might encode potentially useful enzymes (Zhou et al. 2002).
Once new genes are cloned and screened for activity, the main stumbling block is the expression of pure protein in sufficient amounts at reasonable costs. A cheap and efficient enzyme, usually produced in efficient expression systems like bacilli or filamentous fungi, is a key factor for success, particularly when the enzyme functions as part of the final (bulk) product such as in detergents. In the fine-chemical industry, there might be a similar consideration for bulk product synthesis. Particularly in the pharmaceutical industry, the time taken for a target compound to come to the market is decisive, and in these applications, it might be even more important for a company to have a large collection of biochemically diverse catalysts, even if these molecules are not expressed in large amounts.
There is ample demand for novel enzymes and biocatalysts, and metagenomics is currently thought to be one of the most likely technologies to provide the candidate molecules required (Detter et al. 2002). The diversity of potential substrates for enzymatic transformations in the fine-chemical industry and the short time that is usually available to propose viable synthetic routes (particularly for the pharmaceutical industry) make it useful to produce pre-characterized enzyme libraries using generic substrates, before screening for a specific enzyme that is required for biotransformation of a particular substrate of interest (Lorenz et al. 2002).
Since the inception of two pioneering commercial metagenomics ventures in the late 1990s (Recombinant Biocatalysis Ltd of La Jolla and TerraGen Discovery Inc. of Vancouver), these technologies have been taken up by several of the biotechnology giants and have been the focal area of several start-up companies. Recombinant Biocatalysis Ltd, now Diversa Corporation, is the acknowledged leader in the field with impressive lists of libraries derived from global biotopes and of cloned enzymes in a range of enzyme classes. Several other smaller biotechnology companies appear to be competing in the same market sector, and others are obviously knocking at the door. The relatively small size of the industrial enzyme market compared with the pharmaceuticals market suggests that a switch in product focus might not be unexpected. Although the authors are unaware of any successfully commercialized therapeutics derived from metagenomic screening programs, the normal timelines for the identification, development, evaluation, and approval of products for the pharmaceuticals market are longer than the existence of metagenomics as a research field (Vakhlu et al. 2008).
Next-Generation Sequencing Approaches to Metagenomics
Next-generation sequencing approaches enable us to gather many more times sequence data than was possible a few years ago. The first next-generation high-throughput sequencing technology, the 454 GS20 pyrosequencing platform, which was developed by Roche, became available in 2005. The GS20 platform has now been replaced by GSFLX platform. Illumina released Solexa GA in early 2007, and more recently, SOLiD and Heliscope were released by Applied Biosystems and Helicos. Rapid advances in sequencing technology are fueling a vast increase in the number and scope of metagenomics projects. Most metagenome sequencing projects so far have been based on Sanger or Roche-454 sequencing, as only these technologies provide long enough reads, while Illumina sequencing has not usually been considered suitable for metagenomic studies due to a short read length of only 35 bp. However, now that reads of length 75 bp can be sequenced in pairs, Illumina sequencing has become a viable option for metagenome studies as well. A new software MEGAN has been evolved for metagenome analysis that allows one to process sequencing reads in pairs and makes assignments of such reads based on the combined bit-scores of their matches to reference sequences. By using next-generation sequence data in metagenomics experiments, a wide range of new analyses are possible. Metagenomic study has an increasingly powerful partner in the next-generation sequence technology, and this partnership is likely to get more productive as softwares and hardwares mature (MacLean et al. 2009; Mitra et al. 2010).
Metatranscriptomics
As metagenomic DNA-based analyses cannot differentiate between expressed and non-expressed genes, it is unable to reflect the actual metabolic activity. To identify RNA-based regulation and expressed biological signatures in complex ecosystems, sequencing and characterization of metatranscriptomics have been employed. There are several difficulties associated with the processing of environmental RNA sample mainly due to the recovery and enrichment of high quality mRNA by the removal of other RNA species. Short half-lives of mRNA and low yield of cDNA include technical challenges in the isolation procedure. Initially, metatranscriptomics had been limited to the microarray and high-density array technological analysis of mRNA derived cDNA cloning. Detection sensitivity of microarray is not equal for all imprinted sequences as it can give information about those sequences for which it was designed and the result is also dependent on chosen hybridization condition. Transcript cloning avoids some of the problems, but it introduces other biases associated with cloning system and library construction. Substantial progress for the efficient analysis of more complex expression profiles has become available with the development of next-generation sequencing technologies like Roche’s 454, Illumina’s SOLEXA, and ABI’S SOLiD (Schloss and Handelsman 2003; Warnecke and Hess 2009).
In 2006, Leininger and colleagues were the first employing pyrosequencing to unravel active genes of soil microbial communities. This study has revealed that archaeal transcripts of key enzyme for ammonia oxidation were several magnitudes higher in soil than the bacterial version of it. In 2008, Friaz-Lopez et al. produced ≥50 Mbp by 454 pyrosequencing – still using first generation of this technology (MacLean et al. 2009). Gilbert et al. followed shortly afterwards with ≥300 Mbp of sequence data using second generation called GS-FLX (Gilbert et al. 2008). Other metatranscriptomic studies employing direct sequencing of cDNA have targeted the ocean surface water from North Pacific subtropic gyre, a phytoplankton bloom in the Western English Channel, coastal waters of a fjord close to Bergen, Norway, etc. All these studies demonstrated successful application of high-throughput sequencing technologies to exploit unknown transcripts that have been isolated directly from complex environment (Simon and Daniel 2011).
Metaproteomics
With the availability of metagenomic sequences, it is now possible to apply postgenomic techniques – particularly proteomics – to complex microbial communities as well. In 2004, Wilmes and Bond coined the term “metaproteomics” as a shotgun for large-scale characterization of the entire protein complement of environmental microbiota at a given time point. Protein expression is a reflection of specific microbial communities. Elucidation of metaproteomic expression is supposed to be central to functional studies of microbial consortia. In this study, an outer membrane protein and an acetyl coenzyme A acyl transferase were produced by a microbial community derived from activated sludge. These are highly expressed and putatively originated from an unculturable polyphosphate-accumulating Rhodocyclus strain dominating in the activated sludge (Mitra et al. 2010; Simon and Daniel 2011).
The landmark metaproteomic investigation successfully combined “shotgun” MS proteomics with the community genome analyses. This study analyzed the protein complement of a low-complexity natural biofilm, growing inside the Richmond Mine at Iron Mountain, CA (USA), having very low pH (0.8), a temperature of 42°C, and high level of heavy metals (Detter et al. 2002). It was found that sampled biofilm was dominated by Leptospirillum group II along with the presence of Leptospirillum group III, Sulfobacillus, and Archaea related to Ferroplasma acidarmanus. Using the genome dataset, a total of 12,148 protein sequences were constructed. Challenges for metaproteomic analysis include uneven species distribution, large heterogeneity within microbial community, and broad range of protein expression levels within microorganisms. Despite these hurdles, metaproteomics will provide a new dimension of environmental catalysis (Simon and Daniel 2011; Leininger et al. 2006).
Metagenomics is a burgeoning field with new challenges encountered at every step in each instance. The gamut of challenges runs from inefficiencies in sampling, DNA extraction methods, and construction of libraries to inadequacies in data analysis and visualization tools. Added to this are limited computational power and data storage constraints due to the huge amounts of genomic data flooding in from initiatives worldwide. Some of these intricacies will have to be kept in mind, while garnering the full advantages of the metagenomic analyses, both from academic and application point of view (Leininger et al. 2006).
Low Abundance Species Overlooked
The high complexity environment of the Sargasso Sea comprising ∼1,800 different species was daunting in terms of metagenome assembly and analysis. Many current assembly software are befuddled by the large numbers of complex, polymorphic metagenomic data, as are the annotation software, which are designed for use on “closed” (completely assembled) microbial genomes. Assembly is also hampered by shallow sequence coverage resulting from failures to sample uniformly, particularly in high-complexity environments where relative abundance of individual species varies. Most of the sequences obtained may be from the most predominant species in the environment, while sequences from low-abundance species may go undetected. These low-abundance species may well play a critical role in the ecophysiology of the habitat (Leininger et al. 2006; Wilmes and Bond 2004).
Lack of Reference Genomes
Sometimes, assembly can be assisted by the availability of a preexisting reference genome that can serve as a blueprint for piecing environmental genomic data together. Of course, such reference genomes are presently only available for a subset of cultured species, so assembling genomes of more divergent or novel species is not always an easy task. Finally, intraspecies heterogeneity or polymorphisms, or high levels of sequence conservation between phylogenetically unrelated genomes, all can confound the assembly software and result in false or chimeric assemblies (http://web.camera.calit2.net/cameraweb).
Standardizing Metadata
Metadata refers to the temporal, spatial, and physicochemical data associated with the sampling site from which organisms were derived for the metagenomics study. Typical examples are time, date, latitude, longitude, temperature, pH, salinity, etc. The purpose of making such metadata available is to enable correlation of deciphered ecology with the environmental conditions that may favor one population structure over another. Presently, there are no established standards for submission of metadata, and a Genomics Standards Consortium is involved in soliciting opinions from the research community to define a minimal set of metadata required for every genomic and metagenomic project (http://web.camera.calit2.net/cameraweb).
Conclusions
It is perhaps too early to state that metagenomic gene discovery is a technology that has “come of age.” New approaches and technological innovations are pouring in on a regular basis and many of the technical difficulties still waiting to be fully resolved. However, there can be little doubt that the field of metagenomics gene discovery offers enormous scope and potential for both fundamental microbiology and biotechnological development.
The genomes of the total microbiota found in nature contain huge untapped genetic information, which is accessible by metagenomic approaches. Yet the surface of this resource has been barely scratched as far as microbial genomes are concerned. The awareness of the real scope of microbial genome diversity and growing interest in biotechnological application of microbial products as pharmaceuticals, bioactive catalysts, biomaterials, and so forth must prompt the development of new research techniques for the direct and indirect acquisition of these genomes. Although there is unarguably a great need for future leaps in techniques for isolating and culturing novel microorganisms, the recent development of metagenomics, a field that effectively circumvents the microbial isolation and culturing, has been a major breakthrough.
References
Alisky J, Iczkowski K, Rapoport A, Troitsky N (1998) Bacteriophages show promise as antimicrobial agents. J Infect 36:5–15
Arima K, Wooly J (2008) Metagenomics. In: Xu Y, Gogarten JP (eds) Computational methods for understanding Bacterial and Archeal Genomes. Imperial College Press, London, pp 345–466
Barns SM, Fundyga RE, Jeffries MW, Pace NR (1994) Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA 91:1609–1613
Barns SM, Takala SL, Kuske CR (1999) Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl Environ Microbiol 65:1731–1737
Beja O (2004) To BAC or not to BAC: marine ecogenomics. Curr Opin Biotechnol 15:187–190
Beja O et al (2000) Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902–1906
Béjà O et al (2000) Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol 2:516–529
Béja O et al (2002) Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630–633
Benhar I, Eshkenazi I, Neufeld T, Opatowsky J, Shaky S, Rishpon J (2001) Recombinant single chain antibodies in bioelectrochemical sensors. Talanta 55:899–907
Black C, Fyfe JAM, Davies JK (1995) A promoter associated with the neisserial repeat can be used to transcribe the uvrB gene from Neisseria gonorrhoeae. J Bacteriol 177:1952–1958
Bohlool BB, Brock TD (1974) Immunofluorescence approach to the study of the ecology of Thermoplasma acidophilum in coal refuse material. Appl Microbiol 28:11–16
Brady SF, Clardy J (2000) Long-chain N-acyl amino acid antibiotics isolated from heterologously expressed environmental DNA. J Am Chem Soc 122:12903–12904
Brady SF et al (2001) Cloning and heterologous expression of a natural product biosynthetic gene cluster from cDNA. Org Lett 3:1981–1984
Breitbart M et al (2003) Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 185:6220–6223
Breitbart M et al (2004) Diversity and population structure of a nearshore marine sediment viral community. Proc Biol Sci 271:565–574
Brussow H, Hendrix RW (2002) Phage genomics: small is beautiful. Cell 108:13–16
CAMERA (Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis) (2010) v1.3.2.32site. http://web.camera.calit2.net/cameraweb
Cann A, Fandrich S, Heaphy S (2005) Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes 30:151–156
Capparelli R, Parlato M, Borriello G, Salvatore P, Iannelli D (2007) Experimental phage therapy against Staphylococcus aureus in mice. Antimicrob Agents Chemother 51:2765–2773
Chen CY et al (2003) Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res 13:2577–2587
Cho JC, Giovannoni SJ (2004) Cultivation and growth characteristics of diverse group of oligotrophic marine gammaproteobacteria. Appl Environ Microbiol 70:432–440
Connon SA, Giovannoni SJ (2002) High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol 68:3878–3885
Courtois S, Cappellano CM, Ball M, Francou FX, Normand P, Helynck G, Martinez A, Kolvek SJ, Hopke J, Osburne MS, August PR, Nalin R, Guerineau M, Jeannin P, Simonet P, Pernodet JL (2003) Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl Environ Microbiol 69:49–55
Crameri R, Suter M (1993) Display of biologically active proteins on the surface of filamentous phages: a cDNA cloning system for the selection of functional gene products linked to the genetic information responsible for their production. Gene 137:69–75
Daniel R (2005) Metagenomics of soil. Nat Rev Microbiol 3:470–478
Delong EF (2005) Microbial community genomics in the ocean. Nat Rev Microbiol 3(6):459–469
DeSantis G et al (2002) An enzyme library approach to biocatalysis: development of nitrilases for enantioselective production of carboxylic acid derivatives. J Am Chem Soc 124:9024–9025
Detter JC et al (2002) Isothermal strand-displacement amplification applications for high-throughput genomics. Genomics 80:691–698
Diaz-Torres ML, McNab R, Spratt DA, Villedieu A, Hunt N, Wilson M, Mullany P (2003) Novel tetracycline resistance determinant from the oral metagenome. Antimicrob Agents Chemother 47:1430–1432
Dunbar J, Takala S, Barns SM, Davis JA, Kuske CR (1999) Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microbiol 65:1662–1669
Dunbar J, Barns SM, Ticknor LO, Kuske CR (2002) Empirical and theoretical bacterial diversity in four Arizona soils. Appl Environ Microbiol 68:3035–3045
Easton S (2009) Functional and metagenomic analysis of the human tongue dorsum using phage display: submitted for the degree of Doctorate of Philosophy. http://discovery.ucl.ac.uk/18512/
Eden PA, Schmidt TM, Blakemore RP, Pace NR (1991) Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int J Syst Bacteriol 41:324–325
Falkowski PG, de Vargas C (2004) Shotgun sequencing in the sea: a blast from the past? Science 304:58–60
Frank DN, Zhu W, Sartor RB, Li E (2011) Investigating the biological and clinical significance of human disbioses. Trends Microbiol 19:427–434
Futamata H et al (2001) Group-specific monitoring of phenol hydroxylase genes for a functional assessment of phenol-stimulated trichloroethylene bioremediation. Appl Environ Microbiol 67:4671–4677
Gabor EM, de Vries EJ, Janssen DB (2003) Efficient recovery of environmental DNA for expression cloning by indirect methods. FEMS Microbiol Ecol 44:153–163
Gilbert JA, Field D, Huang Y, Edwards R, Li W et al (2008) Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One 3:e3042
Gill RS et al (2006) Metagenomic analysis of the human distal gut microbiome. Science 312(5778):1355–1359
Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR, Rondon MR, Clardy J, Goodman RM, Handelsman J (2002) Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl Environ Microbiol 68:4301–4306
Giovannoni SJ, Britschgi TB, Moyer CL, Field KG (1990) Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60–63
Gnanasekar M, Rao KV, He YX, Mishra PK, Nutman TB, Kaliraj P, Ramaswamy K (2004) Novel phage display-based subtractive screening to identify vaccine candidates of Brugia malayi. Infect Immun 72:4707–4715
Green CD et al (2001) Open systems: panoramic views of gene expression. J Immunol Methods 250:67–79
Guan C, Ju J, Bradley BR et al (2007) Signal mimics derived from a metagenomic analysis of the gypsy moth gut microbiota. Appl Environ Microbiol 73(11):3669–3676
Hallam SJ et al (2004) Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457–1462
Handelsman J (2004) Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685
Healy FG, Ray RM, Aldrich HC, Wilkie AC, Ingram LO, Shanmugam KT (1995) Direct isolation of functional genes encoding cellulases from the microbial consortia in a thermophilic, anaerobic digester maintained on lignocellulose. Appl Microbiol Biotechnol 43:667–674
Henne A, Daniel R, Schmitz RA, Gottschalk G (1999) Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl Environ Microbiol 65:3901–3907
Henne A, Schmitz RA, Bomeke M, Gottschalk G, Daniel R (2000a) Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl Environ Microbiol 66:3113–3116
Henne A, Schmitz RA, Bomeke M, Gottschalk G, Danie R (2000b) Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl Environ Microbiol 66:3113–3116
Hoogenboom HR, De Bruine AP, Hufton SE, Hoet RM, Arends JW, Roovers RC (1998) Antibody phage display technology and its applications. Immunotechnology 4:1–20
Homann MJ et al (2004) Rapid identification of enantioselective ketone reductions using targeted microbial libraries. Tetrahedron 60:789–797
Hughes, DS, Felbeck H, Stein LJ (1997) A histidine protein kinasehomolog from the endosymbiont of the hydrothermal vent tubeworm Riftia pachyptila. Appl Environ Microbiol 63:3494–3498
Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M (2002) Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions acidobacteria, actinobacteria, proteobacteria, and verrucomicrobia. Appl Environ Microbiol 68:2391–2396
Kaeberlein T, Lewis K, Epstein SS (2002) Isolating ‘uncultivable’ microorganisms in pure culture in a simulated natural environment. Science 296:1127–1129
Kawarabayasi Y et al (1999) Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res 6(83–101):145–152
Kruger M et al (2003) A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426:878–881
Lane J (1985) Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA 82:6955–6959
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol WG, Prosser IJ, Schuster CS, Schleper C (2006) Archea predominate among ammonia oxidizing prokaryotes in soils. Environ Microbiol 442:806–809
Lloyd-Jones G, Hunter DWF (2001) Comparison of rapid DNA extraction methods applied to contrasting New Zealand soils. Soil Biol Biochem 33:2053–2059
Lorenz P, Eck J (2005) Metagenomics and industrial applications. Nat Rev Microbiol 3:510–516
Lorenz P, Liebeton K, Niehaus F, Eck J (2002) Screening for novel enzymes for biocatalytic processes: accessing the metagenome as a resource of novel functional sequence space. Curr Opin Biotechnol 13:572–577
MacLean D, Jones GDJ, Studholme JD (2009) Application of ‘next generation’ sequencing technologies to microbial genetics. Nat Rev Microbiol 7:287–296
Majernik A, Gottschalk G, Daniel R (2001) Screening of environmental DNA libraries for the presence of genes conferring Na + (Li+)/H + antiporter activity on Escherichia coli: characterization of the recovered genes and the corresponding gene products. J Bacteriol 183:6645–6653
Matthews LJ, Davis R, Smith GP (2002) Immunogenically fit subunit vaccine components via epitope discovery from natural peptide libraries. J Immunol 169:837–846
McCafferty J, Griffiths AD, Winter G, Chiswell DJ (1990) Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348:552–554
Miedzybrodzki R, Fortuna W, Weber-Dabrowska B, Gorski A (2007) Phage therapy of staphylococcal infections (including MRSA) may be less expensive than antibiotic treatment. Postepy Hig Med Dosw (Online) 61:461–465
Miller DN et al (1999) Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol 65:4715–4724
Mitra S, Schubach M, Hudson HD (2010) Short clones or long clones? A simulation study on the use of paired reads in metagenomics. BMC Bioinformatics 11(Suppl 1):S12.1–S12.11
Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ (2002) SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810
Mullen LM, Nair SP, Ward JM, Rycroft AN, Henderson B (2006) Phage display in the study of infectious diseases. Trends Microbiol 14:141–147
Nelson KE et al (1999) Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323–329
Ochman H, Jose Ayala F, Hartl DL (1993) Use of polymerase chain reaction to amplify segments outside boundaries of known sequences. Methods Enzymol 218:309–332
Ogram A, Sayler GS, Barkay T (1987) The extraction and purification of microbial DNA from sediments. J Microbiol Methods 7:57–66
Ovreas L (2000) Population and community level approaches for analysing microbial diversity in natural environments. Ecol Lett 3:236–251
Pace NR, Stahl DA, Lane DJ, Olsen GJ (1986) The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol 9:1–55
Pedulla ML et al (2003) Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–182
Piel J (2004) Metabolites from symbiotic bacteria. Nat Prod Rep 21:519–538
Preston CM, Wu KY, Molinski TF, DeLong EF (1996) A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA 93:6241–6246
Prinz DM, Smithson SL, Westerink MA (2004) Two different methods result in the selection of peptides that induce a protective antibody response to Neisseria meningitidis serogroup CJ. Immunol Methods 285:1–14
Qin J et al (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi:10.1038
Renesto P et al (2003) Genome-based design of a cell-free culture medium for Tropheryma whipplei. Lancet 362:447–449
Riesenfeld CS, Goodman RM, Handelsman J (2004) Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol 6:981–989
Robertson DE et al (2004) Exploring nitrilase sequence space for enantioselective catalysis. Appl Environ Microbiol 70:2429–2436
Robinson JJ, Stein JL, Cavanaugh CM (1998) Cloning and sequencing of a form II ribulose-1,5-biphosphate carboxylase/oxygenasefrom the bacterial symbiont of the hydrothermal vent tubeworm Riftia pachyptila. J Bacteriol 180:1596–1599
Rondon MR et al (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66:2541–2547
Schloss PD, Handelsman J (2003) Biotechnological prospects from metagenomics. Curr Opin Biotechnol 14:303–310
Simon C, Daniel R (2011) Metagenomic analyses: past and future trends. Appl Environ Microbiol 77:1153–1161
Smith GP, Petrenko VA (1997) Phage display. Chem Rev 97:391–410
Stahl DA, Lane DJ, Olsen GJ, Pace NR (1985) Characterization of a Yellowstone hot spring microbial community by 5S rRNA sequences. Appl Environ Microbiol 49:1379–1384
Stein JL, Marsh TL, Wu KY, Shizuya H, DeLong EF (1996) Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment front a planktonic marine archaeon. J Bacteriol 178:591–599
Tamas I et al (2002) 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376–2379
Teeling H, Meyerdierks A, Bauer M, Amann R, Glockner FO (2004) Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ Microbiol 6:938–947
Trepel M, Arap W, Pasqualini R (2002) In vivo phage display and vascular heterogeneity: implications for targeted medicine. Curr Opin Chem Biol 6:399–404
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF (2004) Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43
Uchiyama T, Abe T, Ikemura T, Wantanabe K (2005) Substrate-induced gene expression screening of environmental metagenome libraries for isolation of catabolicgenes. Nat Biotechnol 23:88–93
Uchiyama T, Miyazaki K (2010) Product-induced gene expression (PIGEX): a product-responsive reporter assay for enzyme screening of metagenomic libraries. Appl Environ Microbiol 76(21):7029–7035
Urbach E et al (1999) Immunochemical detection and isolation of DNA from metabolically active bacteria. Appl Environ Microbiol 65:1207–1213
Vakhlu J, Sundan KA, Johri NB (2008) Metagenomics: Future of microbial gene mining. Indian J Microbiol 48:202–215
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74
Vezzi A et al (2005) Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459–1461
Wagenaar JA, Van Bergen MA, Mueller MA, Wassenaar TM, Carlton RM (2005) Phage therapy reduces Campylobacter jejuni colonization in broilers. Vet Microbiol 109:275–283
Warnecke F, Hess M (2009) A perspective: metatranscriptomics as a tool for the discovery of novel biocatalysts. J Biotechnol 142:91–95
Weber-Dabrowska B, Mulczyk M, Gorski A (2000) Bacteriophage therapy of bacterial infections: an update of our institute’s experience. Arch Immunol Ther Exp (Warsz) 48:547–551
Wilkinson DE et al (2002) Efficient molecular cloning of environmental DNA from geothermal sediments. Biotechnol Lett 24:155–161
Williamson LL, Bradley RB, Schloss PD et al (2005) Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl Environ Microbiol 71(10):6335–6344
Wilmes P, Bond LP (2004) The application of two dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ Microbiol 6:911–920
Woese CR (1987) Bacterial evolution. Microbiol Rev 51:221–271
Wommack KE, Ravel J, Hill RT, Chun J, Colwell RR (1999) Population dynamics of Chesapeake Bay virioplankton: total-community analysis by pulsed field gel electrophoresis. Appl Environ Microbiol 65:231–240
Yeager CM et al (2004) Diazotrophic community structure and function in two successional stages of biological soil crusts from the Colorado plateau and Chihuahuan desert. Appl Environ Microbiol 70:973–983
Yun J, Ryu S (2005) Screening for novel enzymes from metagenome and SIGEX as a way to improve it. Microb Cell Fact 4:1–5
Zengler K et al (2002) Cultivating the uncultured. Proc Natl Acad Sci USA 99:15681–15686
Zhou J et al (2002) Spatial and resource factors influencing high microbial diversity in soil. Appl Environ Microbiol 68:326–334
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer India
About this chapter
Cite this chapter
Srivastava, S., Ghosh, N., Pal, G. (2013). Metagenomics: Mining Environmental Genomes. In: Kuhad, R., Singh, A. (eds) Biotechnology for Environmental Management and Resource Recovery. Springer, India. https://doi.org/10.1007/978-81-322-0876-1_10
Download citation
DOI: https://doi.org/10.1007/978-81-322-0876-1_10
Published:
Publisher Name: Springer, India
Print ISBN: 978-81-322-0875-4
Online ISBN: 978-81-322-0876-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)