Abstract
The prevalence of breast cancer within industrialized populations and the accessibility of tissue for research have meant that genomic, transcriptomic, and proteomic studies have often focused on breast cancer. This has led to major advances in the understanding of this disease and to findings that have been translated to many other types of cancer. Like other common cancers, breast cancer shows a lipogenic phenotype, meaning that significant quantities of lipids are synthesized and stored within breast cancer cells. As the importance of this lipogenic phenotype is becoming better appreciated, studies are beginning to focus upon how lipogenesis is regulated in breast cancer and the critical genes and pathways involved. Lipidomic studies have also begun to characterize lipid profiles in breast cancer cells and tissues and to study the biological consequences of these altered profiles. This chapter will provide an overview of lipid biology in human breast cancer, focusing upon our current understanding of breast cancer lipogenesis, how this contributes to tumor formation and progression, what is understood of its molecular basis, and how the techniques of lipidomics are beginning to be applied to this disease.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Breast cancer is the most common cancer affecting women and a major cause of death from cancer. It is thus both an important clinical problem and a tractable disease to explore. Primary tumors are almost always surgically excised, leading to availability of primary material for study in different tissue forms. Furthermore, many breast cancer cell lines have been derived, are readily cultured in vitro, and have been characterized in extensive molecular detail [1, 2]. The combined availability of both primary tissue and cell lines has meant that breast cancer researchers have acted as “early adopters” of profiling technologies, and breast cancer has often served as a test case for new technology implementation. Some of the earliest gene expression profiling studies were conducted in breast cancer [3, 4], which then paved the way for analyses of less common cancer types. More recently, next-generation sequencing analyses of very large breast cancer cohorts [5, 6] have permitted a level of molecular characterization of the breast cancer genome that would have been difficult to foresee even 10 years ago. The significance of these approaches, both to our understanding of breast cancer as a disease and to our ability to interrogate and understand other cancer types and biological systems, cannot be overstated.
Like other common cancers, breast cancer shows a lipogenic phenotype, meaning that significant quantities of lipids are synthesized and stored within breast cancer cells. Lipids are of unique importance to mammary gland biology, as lipids are a major and important constituent of milk and drive the rapid postnatal growth and development of mammalian infants. As the significance of the lipogenic phenotype within cancer cells is becoming more broadly appreciated, studies are increasingly focusing upon how lipogenesis is regulated in breast cancer and the critical genes and pathways involved. Lipidomic studies have also begun to characterize lipid profiles in breast cancer cells and tissues and to study the biological consequences of these altered profiles. The present chapter will therefore provide an overview of lipid biology in human breast cancer, focusing upon our current understanding of breast cancer lipogenesis, how this contributes to tumor formation and progression and what is understood of its molecular basis. We will also describe dietary influences on the risk of developing breast cancer, and overweight and obesity as causes of breast cancer, before discussing lipidomics methods and how these are beginning to be applied to the study of breast cancer.
Lipid Requirements of Mammalian Cells
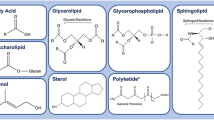
Actively proliferating cells must generate biomass in order to build new cells and hence require a variety of lipids to build new membranes, lipid cofactors, and lipid-modified proteins [7, 8]. Examples of the structures of some of the major classes of biological lipids are shown in Fig. 11.1. Lipids build the extensive networks of intracellular and pericellular membranes that define and partition cellular organelles and their functions. In mammalian cells, most membrane lipids consist of glycerol-phospholipids (PLs), such as phosphatidylcholine, phosphatidylserine, phosphatidylinositol, phosphatidylethanolamine, sterols (mainly cholesterol), and sphingolipids (mainly sphingomyelin) [9]. Fatty acids in general, and the 16-carbon saturated fatty acid palmitate in particular, can also be used for other functions. For example, palmitate and other fatty acids are added by enzymatic processes to increase the hydrophobic nature of proteins to facilitate membrane-associated signalling [10, 11]. Among the proteins that are modified by palmitate are Ras, Wnt, hedgehog, and small protein GTPases [10, 11], each of which is implicated in a variety of cancers.
Living cells acquire fatty acids for their metabolic demands from two major sources, exogenous dietary and de novo endogenous synthesis (Fig. 11.2). Proliferative embryonic cells actively use de novo synthesized fatty acids, whereas most adult normal cells (with the exception of the liver and lactating mammary gland) preferentially use exogenous fatty acids. Fatty acids are present in the diet as triglycerides, and following dietary intake, these are packaged in the intestinal epithelium into chylomicrons (Fig. 11.3). These chylomicrons are then secreted into the lymphatic system and enter the circulation via the thoracic duct. Lipolysis of these particles, initially in tissues such as the heart and lungs by the enzyme lipoprotein lipase (LPL), produces free fatty acids (FFA) that quickly associate with serum albumin. Remnant particles return to the liver where their triglycerides are assembled with apolipoprotein B 100 (apoB) for secretion as very low-density lipoprotein (VLDL) particles. In the circulation, lipolysis of VLDL produces LDL, which in turn is taken up by peripheral tissues, whereas FFA uptake is mediated by fatty acid translocase/CD36 (Figs. 11.2 and 11.3).
Summary of cellular lipogenesis using endogenous and exogenous substrates. Free fatty acid (FFA) is obtained from two pathways, de novo synthesis and FFA uptake. Fatty acid is synthesized from acetyl-CoA by multiple enzymes including acetyl-CoA carboxylase (ACACA) and fatty acid synthase (FASN). FFA uptake is mediated by fatty acid translocase/CD36. FFA is used for either energy production via β-oxidation or complex lipid synthesis such as monoglyceride (MG), diglyceride (DG), and phospholipids (PL), primarily phosphatidylcholine (PC) and phosphatidylethanolamine (PE). To avoid lipotoxicity, excess FFA must be converted to triglyceride (TG), which is then incorporated into lipid droplets. Cholesterol (Chol) is derived from both acetyl-CoA and uptake through the LDL receptor (LDL-R). Cholesterol forms part of the plasma membrane, but excess cholesterol needs to be esterified into cholesterol ester (CE), which is then incorporated into lipid droplets (shaded circles). When cells require energy generation from reserved lipid stores, these can be released through lipolysis. This process is regulated by adipose triglyceride lipase (ATGL) and the PAT protein perilipin (PLIN)
Uptake of dietary lipids. Following dietary intake, lipids are packaged in the intestinal epithelium into chylomicrons (CM), which enter the circulation. Lipoprotein lipase (LPL) converts CM into chylomicron remnants (CMr), releasing free fatty acid (FFA). CMr are cleared by the liver where their triglycerides are assembled with apolipoprotein B 100 (apoB) by microsomal triglyceride transfer protein (MTP) for secretion as very low-density lipoproteins (VLDL) particles. In the circulation, lipolysis of VLDL by LPL produces FFA and low-density lipoproteins (LDL), which in turn are taken up by peripheral tissues via fatty acid translocase/CD36 and the LDL receptor (LDL-R), respectively. LDL is rich in FFA and cholesterol (Chol); hence, an uptake by LDL-R increases both cellular FFA and cholesterol. Additionally, FFA can be obtained from lipolysis of adipocyte lipid stores, facilitated by adipose triglyceride lipase (ATGL), which are then mobilized to target cells for uptake by CD36
In tissues capable of de novo lipogenesis, FFA are also synthesized from the precursor acetyl-CoA by multiple enzymes including acetyl-CoA carboxylase (ACACA) and fatty acid synthase (FASN) (Fig. 11.2). Cholesterol can also be synthesized from acetyl-CoA, as well as taken up through the LDL receptor (LDL-R). FFA are used either for energy production via β-oxidation or for the synthesis of complex lipids such as monoglycerides, diglycerides, and phospholipids (Fig. 11.2). As FFA are toxic to cells, excess FFA must be converted to triglyceride, which is then incorporated into lipid storage organelles, known as lipid droplets (Fig. 11.2). Similarly, excess cholesterol must be esterified into cholesterol ester (CE), which is also incorporated into lipid droplets. Lipid droplets commonly consist of a core of neutral lipids surrounded by phospholipid monolayer and associated proteins and vary greatly in size [8, 12]. Small lipid droplets represent reservoirs that can be rapidly accessed. Conversely, it is more efficient to store lipid in large versus small lipid droplets, and hence, adipose cells contain a single unilocular lipid droplet for maximum storage efficiency [12]. When cells require energy generation from reserved lipid stores, these can be released through lipolysis. This process is regulated by adipose triglyceride lipase (ATGL) and the PAT protein perilipin (PLIN) (Fig. 11.2).
Lipids in Normal Breast Biology
Mammalian infants are typically born after long gestation periods, yet remain highly reliant on their mothers after birth. As lipids represent the most dense source of energy, lipids in milk are vital to drive rapid neonatal growth and are particularly required for postnatal brain development. Milk lipid composition is the most variable attribute of milk and is affected by animal genetics, physiology, and the environment [13]. Lipid-rich milk therefore promotes neonate growth, progressively reducing neonate dependency and promoting survival. Breast-feeding human infants is known to avert serious health problems in neonates, children, and adults, leading to huge savings in medical costs [14]. The regulation of milk fat composition in ruminants is also of major economic significance, both in terms of livestock breeding and in improving the quality of milk available to consumers [13].
The mammary gland largely develops postnatally, undergoing proliferation under the influence of ovarian hormones during puberty, but then remaining largely quiescent until pregnancy. During this time, the epithelial ductal tree expands into the mammary fat pad and undergoes further branching. Alveolae develop and begin to produce milk during the late stage of pregnancy. Lipids are both taken up from the circulation and synthesized within breast epithelial cells, packaged into lipid droplets, and then released into the alveolar lumen as bilayer membrane-coated structures called milk fat globules [15]. Despite the medical and economic importance of milk in human and other species, milk composition and its regulation remain incompletely understood [14].
Lipids in Breast Cancer
As breast epithelial cells take up, synthesize, and secrete lipids during late pregnancy and lactation, it is not surprising that breast cancer is one of many cancers characterized by a lipogenic phenotype. Similar to embryonic cells, the breast and other types of cancer cells endogenously synthesize 95 % of fatty acids, despite the abundance of extracellular fatty acids available to them [16–18]. Cancer cells are highly dependent on de novo lipogenesis for their proliferation, and the lipogenic pathway is activated at a relatively early stage in various types of tumors [19]. The majority of newly synthesized fatty acids in cancer cells are converted predominantly to phospholipids and then incorporated into membrane lipids by proliferating cancer cells. It has been recently suggested that activation of de novo lipid synthesis in cancer cells leads to increased incorporation of saturated fatty acids into cell membranes, which in turn protects cells from both endogenous and exogenous damage [20]. Altered membrane properties occurring in response to de novo lipogenesis may also influence the uptake and activity of chemotherapeutic drugs in cancer cells [20].
Early Studies Demonstrating a Lipogenic Phenotype in Breast Cancer
Efforts to study biochemical alteration of breast cancer were initiated over 40 years ago. In 1966, Rees et al. investigated the lipid composition of mammary glands and mammary carcinomas from rats in various hormonal states using thin-layer chromatography (TLC) and gas–liquid chromatography [21]. Although they could identify triglyceride and phospholipid profiles in the tissues investigated, quantification of lipid species was limited to percentages of total lipids [21]. The limitations of TLC also challenged Hilf et al. in 1970 when comparing lipids in human breast cancer and normal breast tissue [22]. Although they were able to identify differences in cholesterol, FFA, triglycerides, and cholesterol esters in infiltrating ductal carcinomas compared to normal breast tissue, it was unclear which species of lipids were uniquely altered [22]. Nevertheless, they found that cholesterol, FFA, and cholesterol esters were increased in breast cancer, while triglyceride levels were decreased [22]. Sakai et al. reported similar findings, with additional data on the fatty acid composition of phospholipids and triglycerides [23]. The fatty acid compositions of phospholipids were significantly different between human breast cancer and noncancerous excised breast tissues [23]. Specifically, the proportion of monounsaturated (oleate 18:1) and polyunsaturated (docohexanoate 22:6n-3) fatty acids in the major phospholipids was significantly higher in cancer compared to noncancerous tissues [23].
Molecular Basis of Lipogenesis in Breast Cancer Cells
Lipids can either be obtained through the diet or synthesized within cells (Fig. 11.2). The processes of lipid uptake, synthesis, and subsequent metabolism are regulated by numerous transporters and enzymes (Fig. 11.2), the discussion of most of which is beyond the scope of this review. Since the expression of a vast number of proteins is deregulated in cancer cells through genetic, transcriptional, and posttranscriptional mechanisms, it would not be unexpected if some regulators of lipid uptake, synthesis, and metabolism would be thus affected, if only by chance. However, if these deregulated processes provide an advantage to the cancer cell, they will be selected for within the highly competitive environment of cancer tissue. It is now recognized that metabolic deregulation is a hallmark of cancer [24] and that changes in the expression and function of key lipogenic enzymes is actively selected for during tumorigenesis.
In cancer cells, increased glucose uptake results in increased conversion of pyruvate to acetyl-CoA in the mitochondria. Acetyl-CoA is then incorporated into the tricarboxylic acid cycle, which produces citrate in the presence of ATP. Accumulated citrate is exported to the cytoplasm where it is converted by ATP-citrate lyase (ACLY) to generate cytosolic acetyl-CoA, the precursor for FFA synthesis (Fig. 11.2). Acetyl-CoA is then carboxylated by acetyl-CoA carboxylase (ACACA) to synthesize malonyl-CoA, which is then converted to palmitate by fatty acid synthase (FASN) [19]. The unbiased analysis of large numbers of genes and proteins through genomics or proteomics approaches, respectively, has made it increasingly apparent that ACACA, ACLY, and FASN play key roles in tumor progression (Fig. 11.2). Of these three proteins, the expression of FASN and its role in mediating tumor growth has been most heavily investigated, as will be discussed in the following section.
Fatty Acid Synthase (FASN)
Increased FASN expression, relative to normal tissue, has been documented in tumors of the prostate, breast, colon, ovary, endometrium, bladder, and lung [25]. Additionally, FASN overexpression has been noted in melanoma, retinoblastoma, and soft tissue sarcoma [25–27]. FASN overexpression is primarily regulated at the transcriptional level in tumors following oncogene activation, tumor suppressor loss, or growth factor stimulation [28]. FASN levels can also be modulated by posttranslational modification and gene duplication [29, 30]. The expression levels of FASN are highest in metastatic tumors, correlate with decreased survival, and are predictive of poor outcome and disease recurrence in several tumor types [31–34]. These data suggest that FASN not only provides a metabolic advantage that may drive tumor cell survival and proliferation but may also promote a more aggressive tumor phenotype.
In normal physiology, fatty acid synthesis is crucial for development, as mice with the homozygous deletion of Fasn display an embryonic lethal phenotype [35]. On the other hand, with the exception of the liver, adipose tissue, and lactating mammary gland, FASN is expressed at low or undetectable levels in most normal adult tissues [25]. Therefore, unlike in cancer cells, fatty acid synthesis does not seem to be required for normal adult tissue maintenance. Accordingly, mice harboring liver-specific deletions of Fasn display normal liver function and no obvious phenotype, as long as they are maintained on a normal diet [36].
Coincident with the differences in FASN expression between normal and tumor tissues, there also seem to be mechanistic differences in how fatty acids are used in normal and tumor cells. In the liver and adipose tissue, fatty acids are synthesized in response to excess caloric intake. These fatty acids primarily partition toward triglyceride synthesis for fat storage. In contrast, tumor FASN-derived fatty acids preferentially partition into phospholipids that segregate into the plasma membrane or lipid rafts [37]. Additionally, it has been hypothesized that FASN also contributes to the redox status of tumor cells through oxidation of NADPH during the fatty acid synthesis cycle [38]. When all factors are taken into account, it is likely that FASN and fatty acid synthesis provide substrates to affect multiple cellular functions which support a proliferative phenotype.
ERBB2 Signalling and Lipogenesis
ERBB2 (HER2/neu) is a member of the epidermal growth factor receptor (EGFR) family of receptor tyrosine kinases that regulates biological functions ranging from cellular proliferation to transformation, differentiation, motility, and apoptosis. ERBB2 expression levels must be tightly controlled to ensure normal cellular function [39]. In vitro and in vivo studies clearly demonstrate that deregulated ERBB2 expression and activity play a pivotal role in oncogenic transformation, tumorigenesis, and metastasis [40–44]. In breast cancer, amplification of the ERBB2 gene is associated with poor prognosis, shorter relapse time, and low survival rate [40–44].
Aberrant expression of ERBB2 can trigger the activation of multiple downstream signalling pathways, including the phosphatidylinositol 3′-kinase (PI3K)/PTEN/AKT pathway and the Ras/Raf/mitogen-activated protein kinase (MAPK) pathway. These pathways induce cell proliferation and differentiation, decrease apoptosis, and/or enhance tumor cell motility and angiogenesis. Despite the recognized association of ERBB2 and these signalling pathways, less has been known about the specific effectors regulated by ERBB2 that ultimately contribute to its oncogenic effects. The use of transcriptomic analyses to identify genes that were differentially expressed in response to exogenous ERBB2 expression in breast epithelial cells identified increased FASN transcript and protein levels [45]. Similarly, in a panel of human breast cancer cell lines endogenously expressing different levels of ERBB2 and FASN, high levels of both FASN protein expression and FASN enzymatic activity were found to positively correlate with both ERBB2 amplification and ERBB2 protein overexpression [46]. A proteomic study further revealed that proteins involved in glycolysis and de novo lipogenesis pathways were highly expressed in ERBB2-positive breast carcinomas [47], supporting the notion that ERBB2-driven oncogenesis depends upon the lipogenic phenotype [19]. Additionally, mouse NIH-3T3 fibroblasts and human breast epithelial MCF10A cells engineered to overexpress ERBB2 exhibited a significant upregulation of FASN transcript and protein levels [48]. Increased FASN protein levels were also reported to be significantly higher in ERBB2-positive invasive breast tumors examined in tissue microarray format [49].
Control of endogenous FASN levels occurs through modulation of the expression and/or maturation status of the transcription factor sterol regulatory element-binding protein-1c (SREBP-1c). In ERBB2-overexpressing tumor cells, SREBP-1c expression and activation is driven by constitutive activation of the P13K/AKT and/or MAPK/ERK1/2 pathways [19]. Supporting this notion, pharmacological inhibitors of PI3K and MAPK downregulate SREBP-1c and decrease FASN transcription, ultimately reducing lipogenesis in ERBB2-overexpressing cancer cells [50]. FASN overexpression by ERBB2-mediated oncogenic stimuli can also be abrogated by deletion of the major SREBP-binding site from the FASN promoter [51].
An alternative mechanism for ERBB2-FASN induction has also been proposed by Yoon et al [52]. They reported that the induction of FASN in ERBB2-overexpressing breast cancer cells was neither accompanied by changes in FASN transcript levels nor was mediated by the activation of SREBP-1c. Rather, the 5′- and 3′-untranslated regions of FASN mRNAs appeared to be involved in selective FASN translational induction that was mediated by the mammalian target of rapamycin (m-TOR)-regulated signal transduction. In this translational mechanism of FASN regulation, the activation of mTOR significantly increased the synthetic rate of FASN, whereas ERBB2-induced upregulation of FASN protein expression was inhibited by both the PI3K inhibitor LY294002 and the mTOR inhibitor rapamycin [52]. These observations suggest that ERBB2-driven FASN overexpression can be regulated at multiple levels.
Gene Amplification of Lipogenic Genes
Gene amplification is a frequently employed mechanism which increases the expression of targeted genes. Classical cytogenetics approaches first identified genomic regions which were subjected to increased copy number, and then the advent of comparative genomic hybridization (CGH) considerably facilitated genomic copy number studies [53]. Array-based CGH and copy number analyses using single-nucleotide polymorphism profiling have largely superseded classical CGH, and next-generation sequencing is playing an increasingly significant role in identifying, quantifying, and physically mapping copy number changes in cancer and other cell types [54, 55]. Whereas many genes may be affected by copy number changes, only a proportion of these are likely to contribute to the cancer phenotype and represent gene amplification targets.
Genomic profiling and other approaches have shown that genes encoding key enzymes within the lipogenic pathway are increased in copy number and/or overexpressed in breast cancer. As described above, the oncogene ERBB2 located at chromosome 17q (35.1 MB) is amplified in approximately 15 % of breast cancer cases [56] and increases lipogenesis within cancer cells, at least in part by regulating FASN expression and function (see the section “ERBB2 Signalling and Lipogenes”).
It is striking that genes coding for three key enzymes of the fatty acid biosynthetic pathway also reside on human chromosome 17q, namely, FASN (77.6 MB), ACACA (32.7 MB), and ACLY (37.3 MB) [51, 57–59]. A number of these and other lipogenic genes cluster at chromosome 17q12-q21 within 5 MB of each other (Fig. 11.4) and could be commonly affected by copy number increases [60]. In contrast, FASN lies toward the telomeric end of chromosome 17q and does not form part of the lipogenic gene cluster around ERBB2 (see Fig. 11.4). To date, only one study has evaluated the correlation of FASN expression with gene copy number alterations in cancer cells. Using fluorescence in situ hybridization analysis in paraffin-embedded tissue microarrays, a significant increase in FASN copy number was found in a proportion of prostate adenocarcinomas and metastases, which was associated with increased FASN protein detection [30]. It is as yet unclear whether increased FASN copy number plays a significant role in driving increased FASN levels in breast cancer cells.
Positions of chromosome 17q genes with known roles in lipogenesis (shown using hg 18 chromosome 17 coordinates, in MB), with the corresponding cytogenetic bands indicated on the lower ideogram. Approximate positions of genes are shown using vertical arrows: ACACA, 32.7 MB; ACLY, 37.3 MB; MED1 (previously known as PPAR γ-binding protein), 34.8 MB; STARD3 and ERBB2, 35.1 MB; N1RD1, 35.5 MB; and FASN, 77.6 MB. All genes except FASN map within 4.6 MB and could be commonly affected by genomic events leading to increased copy number in breast and other cancers
Experimental evidence has begun to support the concept that increased copy number at the ERBB2 amplicon allows cancer cells to produce high levels of intracellular lipid, while concomitantly promoting the conversion of FFA to triglycerides to avoid lipotoxicity [61, 62]. It has been proposed that the co-amplification of other lipogenic genes with ERBB2 further increases the reliance of such tumors on lipogenesis [62]. Two genes that have been identified to be important for ERBB2-positive breast cancer cell survival, but not that of other breast cancer cells or normal mammary epithelial cells, are mediator complex subunit 1 (MED1, previously known as peroxisome proliferator-activated receptor (PPAR) γ-binding protein or PBP) and the nuclear receptor NR1D1 (nuclear receptor subfamily 1, group D, member 1), a PPARγ target protein [62]. The MED1 and NR1D1 genes within the ERBB2 amplicon (see Fig. 11.4) not only positively affect transcriptional rates of the lipogenic genes FASN, ACLY, and ACACA but also further regulate lipid storage during adipocyte differentiation [63–68]. More recent experimental evidence supports the notion that co-amplification of MED1 and NR1D1 synergistically enhances FFA to triglyceride conversion in ERBB2-positive cells in order to avoid lipotoxicity [39, 62].
Lipogenic amplification target genes have also been identified at other genomic loci beyond chromosome 17q. For example, the “Spot 14” (S14 or THRSP) gene encodes a nuclear protein that is associated with fatty acid synthesis and is located at chromosome 11q13 [69]. High Spot 14 levels as detected by immunohistochemistry were significantly associated with tumor recurrence in breast cancer, but were not associated with either hormone receptor or ERBB2 status in the cohort examined [70].
Overweight and Obesity as Causes of Breast Cancer
Since the 1980s, the percentages of overweight and obese adults and children have risen markedly in the Western world, leading to an impending global health crisis of unprecedented proportion. This has been attributed to a combination of ready access to calorie-rich foods and reduced rates of activity. Overweight and obesity also represent a major environmental cause of cancer [71], which may overtake tobacco use as the leading such cause of cancer as smoking rates decline. The manner in which obesity predisposes individuals to cancer is still a subject of debate, and the causal role that obesity plays is likely to be different in the case of different cancer types. It may be difficult to separate, for example, the effects of obesity from the effects of lack of exercise, or from increased or reduced intakes of particular dietary components, which may have effects beyond contributing to the overweight or obese state.
Increased circulating levels of estrogen serve to drive the proliferation of estrogen receptor-positive breast cancers, and to date, the significance of obesity in relation to breast cancer incidence and risk has been proposed to lie primarily in adipose tissue representing the major site for estrogen synthesis in postmenopausal women [72]. In this tissue, estrogen is synthesized from androgens by aromatase, which is a major drug target in postmenopausal women with estrogen receptor-positive disease. Women with large breasts were reported to have a higher incidence of breast cancer relative to women with average-sized breasts, which could reflect amounts of increased glandular tissue from which tumors can derive and/or higher local estrogen levels generated from increased adipose tissue [73]. Obesity is also known to result in adipose tissue becoming increasingly dysfunctional, leading to the secretion of a variety of factors termed adipokines, which may promote tumor initiation or progression [74]. However, it is possible that a high-fat diet leading to obesity may also promote breast cancer through other mechanisms, as will be discussed below.
Diet and Breast Cancer
A large body of evidence substantiates an important role for de novo lipogenesis in cancer. Given the fact that breast cancer derives from cells with the ability to both synthesize lipid and take up lipid from the circulation, it is important to consider possibly dietary influences on breast cancer risk and development. To date, the role of dietary saturated fat in contributing to breast cancer risk is somewhat controversial. Positive associations between saturated fat or animal fat consumption and cancer have been reported in cohort studies [75] and in studies investigating cancer incidence in 20 countries [76]. Dietary intake of palmitic acid has also been significantly associated with increased breast cancer risk [77]. In general, inaccuracies in reporting dietary intake and difficulties in conducting mechanistic studies on human populations have hampered investigations on the role of dietary saturated fat in cancer development. However, measuring fatty acid composition of adipose tissue using lipidomics techniques may provide a composite measure of dietary fat intake over several years, due to the low turnover rate of stored lipids within adipose tissue [78].
Uptake of Dietary Lipids by Breast Cancer Cells: Part of the Picture?
While there are difficulties in conducting epidemiological dietary studies, some molecular studies have indicated the possible involvement of fatty acid uptake (in particular, saturated fatty acid uptake) in fuelling cancer cells. Studies have shown that LDL receptors are upregulated in tumor cells [79]; therefore, the LDL receptor-mediated pathway is a possible route for fatty acid delivery to peripheral tissues, especially tumor cells. Kuemmerle et al. also reported that cancer cells could also uptake released fatty acid from the lipolysis process through fatty acid translocase CD36 [80]. Immunohistochemical analysis confirmed the presence of LPL and CD36 in breast liposarcoma and prostate cancer tissues [80].
Excessive intake of dietary lipids is a well-known cause for obesity, which is in turn a risk factor for breast cancer [81, 82]. In the mammary gland, a large percentage of the cells are adipocyte or adipocyte precursor cells [83]. The abdominal fatty tissue known as the omentum has been described as a preferred metastasis location for ovarian cancer. Nieman et al. reported that adipocyte-ovarian cancer coculture led to the direct transfer of lipids from adipocytes to ovarian cancer cells and promoted in vitro and in vivo tumor growth [84]. Furthermore, coculture induced lipolysis in adipocytes and β-oxidation in cancer cells, suggesting that lipids stored in adipocytes can act as an energy source for the cancer cells [84]. Considering that the breast is an organ rich in adipose tissue, the transfer of fatty acids between breast cancer cells and breast adipocytes could also occur.
Therapeutic Targeting of Lipogenesis in Breast Cancer
A number of approaches have either been tested or may be applied to target lipogenesis in breast cancer. However, despite overwhelming evidence of the importance of lipogenesis in cancer, and in breast cancer in particular, progress in targeting this pathway has been described as modest at best [85]. Limiting factors have been described as the previous lack of crystallographic structures for relevant targets that has impeded drug design and in establishing structure–antitumor relationships [85]. The most heavily investigated target to date is FASN. Numerous FASN inhibitors have been reported and tested in the context of breast cancer, but their application has been limited in some cases by anorexic side effects [86]. Researchers are continuing to develop alternative inhibitors without these side effects [87]. Other key metabolic enzymes that could represent therapeutic targets in cancer cells are ACACA and ACLY. These targets are of great interest for the treatment of diabetes and obesity but have been explored to a limited extent in the context of cancer [85].
Due to the health and economic impact of the obesity pandemic, novel therapies are being aggressively developed against a variety of targets [88]. With rapidly improving knowledge of the significance of altered lipid synthesis and possibly uptake by cancer cells, such agents show increasing possibility of being adopted for cancer use. Redeploying approved drugs has advantages over the development of novel agents, in that there are preexisting pharmacokinetic, toxicity, and side effects data. For example, agents targeting fatty acid-binding proteins are being developed in the context of insulin resistance and other conditions [89] but could conceptually be applied to cancers where fatty acid-binding proteins are known to be overexpressed. The PAT protein family, which regulates lipid storage in lipid droplets, is also viewed as potential drug targets in the treatment of obesity [88] and are expressed in some lipogenic cancers [90]. The eventual targeting of lipid droplet-associated proteins could be applied to treat lipogenic cancers characterized by increased expression of these targets, where the overexpression of lipogenic genes may represent predictive biomarkers.
Characterizing Lipogenesis in Breast Cancer Cells
Lipid Detection Methods
Thin-layer chromatography (TLC), gas chromatography (GC), and high-performance liquid chromatography (HPLC) have been used in lipid research for many years. In 1966, Rees et al. studied the influence of hormonal status on lipid composition of rat mammary carcinomas, mammary glands, and related tissue [21]. In this study, they used TLC and GC to identify the levels of glycerolipids, sterols, and phospholipids relative to the percentage of total lipids. Following its emergence, TLC became widely accepted as a conventional analysis method for lipids in the 1960s [91, 92], with the advantages of being fast, simple, and inexpensive. However, the major limitation of TLC is its restricted resolution, which significantly hinders its application.
Since most lipids are not volatile and some lipids are easily degraded under high temperature, GC is not a very widely used method in lipidomics, due to the complexity of derivatization required before separation [93]. The derivatization may eliminate much structural information about lipid molecular species, especially polar lipids. Therefore, when using GC to analyze different categories of lipids, complex pre-separation is absolutely necessary [94]. These problems result in the much less frequent application of GC than liquid chromatography. Nevertheless, GC technology is appropriate for the analysis of fatty acids, because the resolution capacity of GC is much higher than that of liquid chromatography. The separation of cis/trans isomers, which is rarely achieved with other lipid detection methods, can be achieved using conventional GC–MS methods.
HPLC is the most widely used separation technique in lipidomics. In contrast to other separation techniques, HPLC has good reproducibility and high resolution and can separate almost all lipid molecular species. HPLC systems are relatively isolated from the environment, limiting the contact between samples and air and thus avoiding self-oxidation and degradation of lipids. In recent years, lipid separation by liquid chromatography and detection by mass spectrometry has become one of the core techniques for the growing field of lipidomics (see the section “Lipidomic Approaches”).
Other methods to detect lipids in biological systems include nuclear magnetic resonance (NMR) spectroscopy and biochemical approaches. NMR spectroscopy is an excellent tool to study molecular structures of purified lipids (1H-NMR and 13C-NMR) and for investigating the structure and dynamics of lipid membranes (1H, 2H, and 31P high-resolution and solid-state NMR) [95]. For the analysis of phospholipid mixtures, 31P-NMR is by far the most appropriate approach. The linear response and relatively high speed of 31P-NMR allow for accurate and selective analysis with high sample throughput [96]. One disadvantage is that NMR techniques have only moderate sensitivity compared with mass spectrometry. Many lipids can also be detected using biochemical approaches (e.g., optical/colorimetric assays). This type of measurement is highly quantitative, but often experimentally challenging in that optimization of conditions can require significant effort.
Lipidomic Approaches
The term “lipidome” describes the complete lipid profile within a cell, tissue, or organism and is a subset of the “metabolome,” which also includes the three other major classes of biological molecules, namely amino acids, sugars, and nucleic acids [97]. Efforts to characterize lipids in cells are relatively recent and have been driven by some spectacular advances in mass spectrometry instrumentation and applications. The dramatic increase in lipidomic research over the past decade has been triggered by impressive developments in analytical technologies, initiated by the application of electrospray ionization mass spectrometry (ESI-MS) to the characterization of membrane phospholipids [98–101]. Technical developments include very high sensitivity and specificity mass and chromatographic resolutions and the increased availability of authentic synthetic lipid standards. These, coupled with impressive developments in data and bioinformatics analysis, have facilitated the detailed molecular analysis of a wide diversity of lipids, ranging from phospholipids and triglycerides to sterols and glycolipids.
Lipidomic analysis by ESI-MS can be categorized in two broad groups, either coupled to liquid chromatography (LC-MS) or shotgun lipidomics (Fig. 11.5), in which the specificity of analysis of different lipid classes in a directly infused sample is provided by diagnostic tandem MS/MS scans. Shotgun lipidomics is an excellent technique for identifying the major pools of phospholipids. Using this method, lipid class (head group) identification is accomplished using precursor ion scans (PIS) and neutral loss scans (NLS) in positive-ion modes and/or negative-ion modes. PIS and NLS are full scan methods mainly offered by triple quadrupole or quadropole time of flight (Q-TOF) MS devices [102]. The fatty acid content of individual lipids is then identified by PIS analysis in negative-ion mode. For example, phosphatidylinositol species PI 38:4 (PI 18:0/20:4) would be identified by a precursor ion scan of 241 m/z in negative-ion mode and associated fatty acid side chains would be identified as 283 m/z (C18:0) and 303 m/z (C20:4) [103]. Overall, shotgun lipidomics analyses are prone to ion suppression of detection of minor components by molecules that become preferentially ionized but are rapid and accurate for quantification using a limited number of internal standards.
For the analysis of lipid classes using LC-MS, ion suppression is less of an issue and this approach enables resolution of isobaric molecular species of identical molecular mass, but different molecular structures. LC-MS in lipidomics is characterized by an additional layer of separation preceding m/z analysis. A chromatographic separation step substantially increases the number of detectable lipids due to reduced suppression effects in the ion source [104, 105]. In this manner, the identification of very low abundance lipids is possible without any manual intervention in the analysis process [106]. Normal phase and reversed phase, as two different modes of HPLC, have both been used for different purposes in lipidomics analysis. The normal-phase method is used to separate different classes of lipids based on polar head groups and the reverse-phase method is often used to separate different molecular species in one class based on their different fatty acyl chains [93]. In addition to m/z values, LC-MS also offers retention time values for identification purposes. LC-MS is, however, more time-consuming, and as different periods of gradient liquid chromatography elution will have different ionization capacity, multiple standards are required for accurate quantification.
Similar to other “omics” technologies, lipidomics generates large sets of data. The diversity of lipid chemical structures presents a challenge both from experimental and informatics standpoints. So far, although there is a general consensus in the lipidomics community to adopt the lipid classification introduced by Lipid Maps [97], there is no similar consistency for data analysis programs to interrogate lipidomics mass spectrometry results. The need for a robust, scalable bioinformatics infrastructure is high at a number of different levels: (a) establishment of a globally accepted classification system; (b) creation of databases of lipid structures, lipid-related genes and proteins; (c) efficient analysis of experimental data; (d) efficient management of metadata and protocols; (e) integration of experimental data and existing knowledge into metabolic and signalling pathways; and (f) development of informatics software for efficient searching, display, and analysis of lipidomics data [97]. These requirements need to be addressed by collaborative efforts between researchers working in biology, chemistry, and bioinformatics.
Technical Developments in Lipidomics Relevant to Breast Cancer Research
In 2008, Haynes et al. described a method for quantitation of subpicomole amounts of long-chain and very-long-chain fatty acyl-CoA by reverse-phase liquid chromatography combined with electrospray ionization tandem mass spectrometry in positive-ion mode with odd-chain length fatty acyl-CoAs as internal standards [107]. RAW264.7 macrophage cells and human breast cancer MCF7 cells were used as examples in this optimization, and their analysis revealed large differences in fatty acyl amounts and subspecies distributions [107]. The amounts of very-long-chain fatty acyl (>C20) and long-chain fatty acyl (<C20) were similar in cancer cells, whereas in noncancerous cells, the majority of fatty acyls were long chain [107]. Further lipidomics studies in breast cancer cell lines were performed using positive and negative modes on electrospray linear ion trap and electrospray triple quadrupole mass spectrometry [108, 109]. These instruments combine sensitivity, specificity, selectivity, and speed for accurate analysis of phospholipids [110]. Comparing three different breast cell lines (nonmalignant mammary epithelial MCF10A cells, nonmetastatic breast cancer T-47D cells, and metastatic breast cancer MDA-MB-231 cells), they reported that phosphatidylcholines and phosphatidylinositols were decreased in nonmalignant cells relative to cancer cells [109]. Furthermore, the MDA-MB-231 cell line possessed the highest levels of phosphatidic acids, phosphatidylcholines, and phosphatidylinositols [109]. Advanced mass spectrometry has also been applied to characterize lipid profiles directly in breast cancer patients, with palmitic acid, stearic acid, linoleic acid, and total fatty acid being emphasized as having the greatest potential to act as biomarkers of breast cancer [111].
Integration of Lipidomics, Genomics, and Proteomics in Breast Cancer Research
The application of genomics, transcriptomics, and proteomics to breast cancer has generated huge amounts of information regarding the molecular changes that occur in breast cancer tissues and cell lines. In comparison to genomics, transcriptomics, and proteomics, lipidomics is a relatively new approach [99]. As such, the number of publications including the term “lipidomics” (1,475 publications, October, 2012, identified using the full-text search function of Highwire) is far exceeded by those including the term “genomics” (>154,000) or “proteomics” (>66,000) (Fig. 11.6). Nonetheless, the overall number of publications including both the terms “cancer” and “lipidomics” is rising rapidly (Fig. 11.7).
Venn diagram showing the numbers of articles including the search terms “genomics” (n = 154,904), “proteomics” (n = 66,439), and/or “lipidomics” (n = 1,475) or any combination thereof. Relatively high proportions of lipidomics articles included either “genomics” (n = 256, 17.4 %), “proteomics” (n = 336, 22.8 %), or both terms (n = 155, 10.5 %), reflecting the status of lipidomics being a relatively new field, which is open to integration with other “omics” disciplines. Article numbers were generated using the full-text search function of the Highwire literature search engine on 12 October 2012. The Venn diagram is shown for illustration and is not drawn to scale
Cumulative numbers of scientific publications that include the terms “lipidomics” and “cancer,” published by the end of each calendar year from 2001 (zero articles) to 2012 (470 articles in total). Article numbers were generated using the full-text search function of the Highwire literature search engine on 12 October 2012
Many studies have performed genomic and transcriptomic analyses of breast cancer, in order to identify genes with significant relationships between gene copy number and transcript level and hence genes which may represent amplification targets or tumor suppressor genes [112–114]. Predictions from transcriptomic studies are then frequently validated using protein detection techniques, to identify genes that are reproducibly differentially expressed at both the transcript and protein levels [115]. These integrative approaches have highlighted ways in which lipid metabolism and profiles are altered in tumors. For example, associations between gene copy number and expression identified both ACACA (chromosome 17, 32.7 MB) and NR1D1 (chromosome 17, 35.5 MB) (Fig. 11.4) as being potentially druggable amplification targets in breast cancer [116].
In contrast, very few studies have attempted to integrate lipidomic and other -omic profiles in any biological context [117]. However, the ability of lipidomics to illuminate molecular mechanisms of disease when combined with transcriptomics data has been recently demonstrated in the context of breast cancer. Lipidomics analysis of a large cohort of human breast tissues revealed increased incorporation of de novo synthesized fatty acids into membrane phospholipids in tumors versus normal breast tissues [118]. In silico transcriptomics data [119] were then interrogated to identify candidate proteins possibly underpinning these changes. Candidate proteins were investigated using immunohistochemistry, revealing that breast cancers with high levels of de novo synthesized fatty acids also demonstrated high FASN and ACACA levels in cancer cells in situ [118]. A similar approach was employed by Brockmöller et al. to investigate the expression of glycerol-3-phosphate acyltransferase (GPAM) in breast cancer tissue and to describe associations between GPAM immunohistochemical staining and metabolomic profiles [120].
While few breast cancer studies have integrated lipidomics with other high-throughput approaches, the importance of integrating lipidomics with other “omics” technologies is clearly understood by the research community. This is indicated by the fact that of all publications including the term “lipidomics” (n = 1,475), 17 and 23 % also mentioned “genomics” or “proteomics,” respectively, and 11 % included all three terms (Fig. 11.6). The slightly higher co-use of the terms “lipidomics” and “proteomics” could reflect the fact that proteomics and lipidomics employ similar platforms, and it has been proposed that many proteomics groups could undertake lipidomics projects [99]. Thus, in the short term, we might expect more frequent integration of proteomics and lipidomics approaches in cancer.
Conclusion and Future Perspective
Breast cancer is both an important clinical problem and a tractable disease to explore. The combined availability of both primary tissue and cell lines has meant that breast cancer researchers have acted as “early adopters” of profiling technologies, and breast cancer has often served as a test case for new technology implementation. A number of factors are now leading to increased use of lipidomics techniques in the study of breast cancer, beyond technological developments within the lipidomics field itself. It is now clear that obesity is a major environmental cause of cancer, which contributes both directly and indirectly to breast cancer incidence. The importance of obesity in driving common cancers is leading to increased recognition of the fact that lipid metabolism is also greatly altered in cancer relative to normal cells, although whether and how these phenomena are linked need further investigation. Alterations in cancer lipid metabolism have been shown through direct investigations and indirectly through genomics, transcriptomics, and proteomics approaches, which highlight alterations in gene copy number, expression, or protein levels of key regulators of lipid metabolism. While molecular therapies for cancer continue to represent a major area of drug development, there is increased recognition that drugs developed for metabolic conditions such as obesity may also be applied for cancer therapy [85].
Lipidomics faces some particular challenges not shared by other “omics” fields, such as genomics, transcriptomics, and proteomics. Sequencing the human genome deduced the gene set available to build both transcripts and proteins and therefore defined the theoretical boundaries of molecules relevant to these fields. In contrast, the number of biological lipids has not been defined and at present, cannot be predicted [99]. The full identification of all lipid species is rendered further challenging as some are likely to be present at low abundance [99], and if these cannot be predicted, they are less likely to be identified. Furthermore, lipids exert their functions through interactions with proteins, RNA, and other molecules within cells, and these interactions are only beginning to be analyzed and defined [121].
Despite such challenges, the lipidome is likely to present a wealth of opportunities in terms of cancer diagnosis and treatment. Identification of lipid classes and their structures opens new possibilities for exploration of lipid alterations in cancer, providing novel biomarkers and the basis to develop novel therapeutics strategies. The metabolome has been described as the amplified output of a biological system, with small changes in individual enzymes potentially leading to large outputs that can be robustly quantified [122]. The immense structural diversity of lipids, while currently a major challenge, also provides opportunities to define highly specific biomarkers in disease states such as cancer. Just as enzymatic regulators of lipid metabolism have been proposed as therapeutic targets in breast and other cancers, disease-restricted lipids themselves may prove to be therapeutic targets, which may be less susceptible to the development of drug resistance through individual gene mutations. Given the immense biological and clinical relevance of lipids to many human diseases, ongoing efforts to identify and classify biological lipids, and more frequent integration of lipidomics with other experimental approaches, we may see lipidomics grow to rival other more well-established “omics” fields within the next 10 years.
References
Jönsson G, Staaf J, Olsson E, Heidenblad M, Vallon-Christersson J, Osoegawa K, et al. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosomes Cancer. 2007;46:543–58.
Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146.
Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumors. Nature. 2000;406:747–52.
Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumors. Nature. 2012;490:61–70.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumors reveals novel subgroups. Nature. 2012;486:346–52.
Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33.
Brasaemle DL. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48:2547–59.
Escribá PV, González-Ros JM, Goñi FM, Kinnunen PK, Vigh L, Sánchez-Magraner L, et al. Membranes: a meeting point for lipids, proteins and therapies. J Cell Mol Med. 2008;12:829–75.
Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–90.
Resh MD. Use of analogs and inhibitors to study the functional significance of protein palmitoylation. Methods. 2006;40:191–7.
Suzuki M, Shinohara Y, Ohsaki Y, Fujimoto T. Lipid droplets: size matters. J Electron Microsc. 2011;60(S1):S101–16.
Harvatine KJ, Boisclair YR, Bauman DE. Recent advances in the regulation of milk fat synthesis. Animal. 2009;3:40–54.
Neville MC, Anderson SM, McManaman JL, Badger TM, Bunik M, Contractor N, et al. Lactational and neonatal nutrition: defining and refining the critical questions. J Mammary Gland Biol Neoplasia. 2012;17:167–88.
McManaman JL, Reyland ME, Thrower EC. Secretion and fluid transport mechanisms in the mammary gland: comparisons with the exocrine pancreas and the salivary gland. J Mammary Gland Biol Neoplasia. 2006;11:249–68.
Greenstein JP. Biochemistry of cancer. New York: Academic; 1954.
Medes G, Thomas A, Weinhouse S. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 1953;13:27–9.
Ookhtens M, Kannan R, Lyon I, Baker N. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am J Physiol. 1984;247:R146–53.
Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–77.
Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010;70:8117–26.
Rees ED, Shuck AE, Ackermann H. Lipid composition of rat mammary carcinomas, mammary glands, and related tissues: endocrine influences. J Lipid Res. 1966;7:396–402.
Hilf R, Goldenberg H, Michel I, Orlando RA, Archer FL. Enzymes, nucleic acids, and lipids in human breast cancer and normal breast tissue. Cancer Res. 1970;30:1874–82.
Sakai K, Okuyama H, Yura J, Takeyama H, Shinagawa N, Tsuruga N, et al. Composition and turnover of phospholipids and neutral lipids in human breast cancer and reference tissues. Carcinogenesis. 1992;13:579–84.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Kridel SJ, Lowther WT, Pemble 4th CW. Fatty acid synthase inhibitors: new directions for oncology. Expert Opin Investig Drugs. 2007;16:1817–29.
Takahiro T, Shinichi K, Toshimitsu S. Expression of fatty acid synthase as a prognostic indicator in soft tissue sarcomas. Clin Cancer Res. 2003;9:2204–12.
Camassei FD, Cozza R, Acquaviva A, Jenkner A, Ravà L, Gareri R, et al. Expression of the lipogenic enzyme fatty acid synthase (FAS) in retinoblastoma and its correlation with tumor aggressiveness. Invest Ophthalmol Vis Sci. 2003;44:2399–403.
Kuhajda FP. Fatty acid synthase and cancer: new application of an old pathway. Cancer Res. 2006;66:5977–80.
Graner E, Tang D, Rossi S, Baron A, Migita T, Weinstein LJ, et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5:253–61.
Shah US, Dhir R, Gollin SM, Chandran UR, Lewis D, Acquafondata M, et al. Fatty acid synthase gene overexpression and copy number gain in prostate adenocarcinoma. Hum Pathol. 2006;37:401–9.
Epstein JI, Carmichael M, Partin AW. OA-519 (fatty acid synthase) as an independent predictor of pathologic state in adenocarcinoma of the prostate. Urology. 1995;45:81–6.
Alo’ PL, Visca P, Marci A, Mangoni A, Botti C, Di Tondo U. Expression of fatty acid synthase (FAS) as a predictor of recurrence in stage I breast carcinoma patients. Cancer. 1996;77:474–82.
Visca P, Sebastiani V, Botti C, Diodoro MG, Lasagni RP, Romagnoli F, et al. Fatty acid synthase (FAS) is a marker of increased risk of recurrence in lung carcinoma. Anticancer Res. 2004;24:4169–73.
Sebastiani V, Visca P, Botti C, Santeusanio G, Galati GM, Piccini V, et al. Fatty acid synthase is a marker of increased risk of recurrence in endometrial carcinoma. Gynecol Oncol. 2004;92:101–5.
Chirala SS, Chang H, Matzuk M, Abu-Elheiga L, Mao J, Mahon K, et al. Fatty acid synthesis is essential in embryonic development: fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc Natl Acad Sci U S A. 2003;100:6358–63.
Chakravarthy MV, Pan Z, Zhu Y, Tordjman K, Schneider JG, Coleman T, et al. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005;1:309–22.
Swinnen JV, Van Veldhoven PP, Timmermans L, De Schrijver E, Brusselmans K, Vanderhoydonc F, et al. Fatty acid synthase drives the synthesis of phospholipids partitioning into detergent-resistant membrane microdomains. Biochem Biophys Res Commun. 2003;302:898–903.
Baron A, Migita T, Tang D, Loda M. Fatty acid synthase: a metabolic oncogene in prostate cancer? J Cell Biochem. 2004;91:47–53.
Menendez JA. Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: molecular mechanisms and therapeutic perspectives. Biochim Biophys Acta. 1801;2010:381–91.
Bertucci F, Borie N, Ginestier C, Groulet A, Charafe-Jauffret E, Adélaïde J, et al. Identification and validation of an ERBB2 gene expression signature in breast cancers. Oncogene. 2004;23:2564–75.
Isola J, Chu L, DeVries S, Matsumura K, Chew K, Ljung BM, et al. Genetic alterations in ERBB2-amplified breast carcinomas. Clin Cancer Res. 1999;5:4140–5.
Pegram MD, Konecny G, Slamon DJ. The molecular and cellular biology of HER2/neu gene amplification/overexpression and the clinical development of herceptin (trastuzumab) therapy for breast cancer. Cancer Treat Res. 2000;103:57–75.
Slamon DJ, Clark GM, Wong SG. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82.
Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer. 2012;12:553–63.
Kumar-Sinha C, Ignatoski KW, Lippman ME, Ethier SP, Chinnaiyan AM. Transcriptome analysis of HER2 reveals a molecular connection to fatty acid synthesis. Cancer Res. 2003;63:132–9.
Menendez JA, Ropero S, Mehmi I, Atlas E, Colomer R, Lupu R. Overexpression and hyperactivity of breast cancer-associated fatty acid synthase (oncogenic antigen-519) is insensitive to normal arachidonic fatty acid-induced suppression in lipogenic tissues but it is selectively inhibited by tumoricidal alpha-linolenic and gamma-linolenic fatty acids: a novel mechanism by which dietary fat can alter mammary tumorigenesis. Int J Oncol. 2004;24:1369–83.
Zhang D, Tai LK, Wong LL, Chiu LL, Sethi SK, Koay ES. Proteomic study reveals that proteins involved in metabolic and detoxification pathways are highly expressed in HER-2/neu-positive breast cancer. Mol Cell Proteomics. 2005;4:1686–96.
Menendez JA, Mehmi I, Verma VA, Teng PK, Lupu R. Pharmacological inhibition of fatty acid synthase (FAS): a novel therapeutic approach for breast cancer chemoprevention through its ability to suppress Her-2/neu (erbB-2) oncogene-induced malignant transformation. Mol Carcinog. 2004;41:164–78.
Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–75.
Yang Y, Morin PJ, Han WF. Regulation of fatty acid synthase expression in breast cancer by sterol regulatory element binding protein-1c. Exp Cell Res. 2003;282:132–7.
Menendez JA, Decker JP, Lupu R. In support of fatty acid synthase (FAS) as a metabolic oncogene: extracellular acidosis acts in an epigenetic fashion activating FAS gene expression in cancer cells. J Cell Biochem. 2005;94:1–4.
Yoon S, Lee MY, Park SW, Moon JS, Koh YK, Ahn YH, et al. Up-regulation of acetyl-CoA carboxylase alpha and fatty acid synthase by human epidermal growth factor receptor 2 at the translational level in breast cancer cells. J Biol Chem. 2007;282:26122–31.
Kallioniemi A, Kallioniemi O-P, Piper J, Tanner M, Stokke T, Chen L, et al. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci U S A. 1994;91:2156–60.
Diamandis EP, Hudson T, Kallioniemi O, Liu ET, López-Otín C. Cancer genomes. Clin Chem. 2010;56:1660–4.
Hillmer L, Yao F, Inaki K, Lee WH, Ariyaratne PN, Teo AS, et al. Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes. Genome Res. 2011;21:665–75.
Teschendorff AE, Caldas C. The breast cancer somatic ‘muta-ome’: tackling the complexity. Breast Cancer Res. 2009;11:301.
Brunet J, Vazquez-Martin A, Colomer R, Graña-Suarez B, Martin-Castillo B, Menendez JA. BRCA1 and acetyl-CoA carboxylase: the metabolic syndrome of breast cancer. Mol Carcinog. 2008;47:157–63.
Menendez JA, Vellon L, Lupu R. DNA topoisomerase IIalpha (TOP2A) inhibitors up-regulate fatty acid synthase gene expression in SK-Br3 breast cancer cells: in vitro evidence for a ‘functional amplicon’ involving FAS, Her-2/neu and TOP2A genes. Int J Mol Med. 2006;18:1081–7.
Pitel F, Fillon V, Heimel C, Le Fur N, el Khadir-Mounier C, Douaire M, et al. Mapping of FASN and ACACA on two chicken microchromosomes disrupts the human 17q syntenic group well conserved in mammals. Mamm Genome. 1998;9:297–300.
Kauraniemi P, Kallioniemi A. Activation of multiple cancer-associated genes at the ERBB2 amplicon in breast cancer. Endocr Relat Cancer. 2006;13:39–49.
Kourtidis A, Srinivasaiah R, Carkner RD, Brosnan MJ, Conklin DS. Peroxisome proliferator-activated receptor-gamma protects ERBB2-positive breast cancer cells from palmitate toxicity. Breast Cancer Res. 2009;11:R16.
Kourtidis A, Jain R, Carkner RD, Effert C, Brosnan MJ, Conklin DS. An RNA interference screen identifies metabolic regulators NR1D1 and PBP as novel survival factors for breast cancer cells with the ERBB2 signature. Cancer Res. 2010;70:1783–92.
Fontaine C, Dubois G, Duguay Y, Helledie T, Vu-Dac N, Gervois P, et al. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipocyte differentiation. J Biol Chem. 2003;278:37672–80.
Jia Y, Qi C, Kashireddi P, Surapureddi S, Zhu YJ, Rao MS, et al. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver. J Biol Chem. 2004;279:24427–34.
Laitinen S, Fontaine C, Fruchart JC, Staels B. The role of the orphan nuclear receptor Rev-Erb alpha in adipocyte differentiation and function. Biochimie. 2005;87:21–5.
Misra P, Owuor ED, Li W, Yu S, Qi C, Meyer K, et al. Phosphorylation of transcriptional coactivator peroxisome proliferator-activated receptor (PPAR)-binding protein (PBP). Stimulation of transcriptional regulation by mitogen-activated protein kinase. J Biol Chem. 2002;277:48745–54.
Wang J, Lazar MA. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol Cell Biol. 2008;28:2213–20.
Zhu Y, Kan L, Qi C, Kanwar YS, Yeldandi AV, Rao MS, et al. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J Biol Chem. 2000;275:13510–6.
Moncur JT, Park JP, Memoli VA, Mohandas TK, Kinlaw WB. The “Spot 14” gene resides on the telomeric end of the 11q13 amplicon and is expressed in lipogenic breast cancers: implications for control of tumor metabolism. Proc Natl Acad Sci U S A. 1998;95:6989–94.
Wells WA, Schwartz GN, Morganelli PM, Cole BF, Gibson JJ, Kinlaw WB. Expression of “Spot 14” (THRSP) predicts disease free survival in invasive breast cancer: immunohistochemical analysis of a new molecular marker. Breast Cancer Res Treat. 2006;98:231–40.
Parkin DM, Boyd L, Walker LC. 16. The fraction of cancer attributable to lifestyle and environmental factors in the UK in 2010. Br J Cancer. 2011;105:S77–81.
Cleary MP, Grossmann ME. Minireview: obesity and breast cancer: the estrogen connection. Endocrinology. 2009;150:2537–42.
Gayde C, Goolam I, Bangash HK, Tresham J, Fritschi L, Wylie E. Outcome of mammography in women in large breasts. Breast. 2012;21:493–8.
Park J, Euhus DM, Scherer PE. Paracrine and endocrine effects of adipose tissue on cancer development and progression. Endocr Rev. 2011;32:550–70.
Giovannucci E, Rimm EB, Colditz GA, Stampfer MJ, Ascherio A, Chute CG, et al. A prospective study of dietary fat and risk of prostate cancer. J Natl Cancer Inst. 1993;85:1571–9.
Hursting SD, Thornquist M, Henderson MM. Types of dietary fat and the incidence of cancer at five sites. Prev Med. 1990;19:242–53.
Saadatian-Elahi M, Norat T, Goudable J, Riboli E. Biomarkers of dietary fatty acid intake and the risk of breast cancer: a meta-analysis. Int J Cancer. 2004;111:584–91.
Bougnoux P, Hajjaji N, Couet C. The lipidome as a composite biomarker of the modifiable part of the risk of breast cancer. Prostaglandins Leukot Essent Fatty Acids. 2008;79:93–6.
Gal D, MacDonald PC, Porter JC, Simpson ER. Cholesterol metabolism in cancer cells in monolayer culture. III. Low-density lipoprotein metabolism. Int J Cancer. 1981;28:315–9.
Kuemmerle NB, Rysman E, Lombardo PS, Flanagan AJ, Lipe BC, Wells WA, et al. Lipoprotein lipase links dietary fat to solid tumor cell proliferation. Mol Cancer Ther. 2011;10:427–36.
Ahn J, Schatzkin A, Lacey Jr JV, Albanes D, Ballard-Barbash R, Adams KF, et al. Adiposity, adult weight change, and postmenopausal breast cancer risk. Arch Intern Med. 2007;167:2091–102.
Pichard C, Plu-Bureau G, Neves ECM, Castro M, Gompel A. Insulin resistance, obesity and breast cancer risk. Maturitas. 2008;60:19–30.
Subbaramaiah K, Howe LR, Bhardwaj P, Du B, Gravaghi C, Yantiss RK, et al. Obesity is associated with inflammation and elevated aromatase expression in the mouse mammary gland. Cancer Prev Res (Phila). 2011;4:329–46.
Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–503.
Abramson HN. The lipogenesis pathway as a cancer target. J Med Chem. 2011;54:5615–38.
Puig T, Vázquez-Martín A, Relat J, Pétriz J, Menéndez JA, Porta R, et al. Fatty acid metabolism in breast cancer cells: differential inhibitory effects of epigallocatechin gallate (EGCG) and C75. Breast Cancer Res Treat. 2008;109:471–9.
Puig T, Aguilar H, Cufí S, Oliveras G, Turrado C, Ortega-Gutiérrez S, et al. A novel inhibitor of fatty acid synthase shows activity against HER2+ breast cancer xenografts and is active in anti-HER2 drug-resistant cell lines. Breast Cancer Res. 2011;13:R131.
Shi Y, Burn P. Lipid metabolic enzymes: emerging targets for the treatment of obesity. Nat Rev Drug Discov. 2004;3:695–710.
Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov. 2008;7:489–503.
Straub BK, Herpel E, Singer S, Zimbelmann R, Breuhahn K, Macher-Goeppinger S, et al. Lipid droplet-associated PAT-proteins show frequent and differential expression in neoplastic steatogenesis. Mod Pathol. 2010;23:480–92.
Ruggieri S. Separation of the methyl esters of fatty acids by thin layer chromatography. Nature. 1962;193:1282–3.
Bennett RD, Heftmann E. Thin-layer chromatography of sterols. J Chromatogr. 1962;9:359–62.
Li M, Zhou Z, Nie H, Bai Y, Liu H. Recent advances of chromatography and mass spectrometry in lipidomics. Anal Bioanal Chem. 2011;399:243–9.
Hauff S, Vetter W. Quantification of fatty acids as methyl esters and phospholipids in cheese samples after separation of triacylglycerides and phospholipids. Anal Chim Acta. 2009;636:229–35.
Gawrisch K, Eldho NV, Polozov IV. Novel NMR tools to study structure and dynamics of biomembranes. Chem Phys Lipids. 2002;116:135–51.
Marsh D, Páli T. The protein-lipid interface: perspectives from magnetic resonance and crystal structures. Biochim Biophys Acta. 2004;1666:118–41.
Fahy E, Cotter D, Sud M, Subramaniam S. Lipid classification, structures and tools. Biochim Biophys Acta. 1811;2011:637–47.
Han X, Gross RW. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc Natl Acad Sci U S A. 1994;91:10635–9.
Hou W, Zhou H, Elisma F, Bennett SA, Figeys D. Technological developments in lipidomics. Brief Funct Genomic Proteomic. 2008;7:395–409.
Postle AD. Lipidomics. Curr Opin Clin Nutr Metab Care. 2012;15:127–33.
Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610.
Hartler J, Tharakan R, Köfeler HC, Graham DR, Thallinger GG. Bioinformatics tools and challenges in structural analysis of lipidomics MS/MS data. Brief Bioinform. 2013;14:375–90.
Ivanova PT, Milne SB, Myers DS, Brown HA. Lipidomics: a mass spectrometry based systems level analysis of cellular lipids. Curr Opin Chem Biol. 2009;13:526–31.
Taguchi R, Houjou T, Nakanishi H, Yamazaki T, Ishida M, Imagawa M, et al. Focused lipidomics by tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;823:26–36.
Taguchi R, Nishijima M, Shimizu T. Basic analytical systems for lipidomics by mass spectrometry in Japan. Methods Enzymol. 2007;432:185–211.
Nakanishi H, Ogiso H, Taguchi R. Qualitative and quantitative analyses of phospholipids by LC-MS for lipidomics. Methods Mol Biol. 2009;579:287–313.
Haynes CA, Allegood JC, Sims K, Wang EW, Sullards MC, Merrill Jr AH. Quantitation of fatty acyl-coenzyme as in mammalian cells by liquid chromatography-electrospray ionization tandem mass spectrometry. J Lipid Res. 2008;49:1113–25.
Dória ML, Cotrim Z, Macedo B, Simões C, Domingues P, Helguero L, et al. Lipidomic approach to identify patterns in phospholipid profiles and define class differences in mammary epithelial and breast cancer cells. Breast Cancer Res Treat. 2012;133:635–48.
Dória ML, Cotrim CZ, Simões C, Macedo B, Domingues P, Domingues MR, et al. Lipidomic analysis of phospholipids from human mammary epithelial and breast cancer cell lines. J Cell Physiol. 2013;228:457–68.
Milne S, Ivanova P, Forrester J, Alex Brown H. Lipidomics: an analysis of cellular lipids by ESI-MS. Methods. 2006;39:92–103.
Lv W, Yang T. Identification of possible biomarkers for breast cancer from free fatty acid profiles determined by GC-MS and multivariate statistical analysis. Clin Biochem. 2012;45:127–33.
Pollack JR, Sørlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963–8.
Adélaïde J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–75.
Staaf J, Jönsson G, Ringnér M, Vallon-Christersson J, Grabau D, Arason A, et al. High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer. Breast Cancer Res. 2010;12:R25.
Harvell DM, Richer JK, Singh M, Spoelstra N, Finlayson C, Borges VF, et al. Estrogen regulated gene expression in response to neoadjuvant endocrine therapy of breast cancers: tamoxifen agonist effects dominate in the presence of an aromatase inhibitor. Breast Cancer Res Treat. 2008;112:489–501.
Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41.
Buczynski MW, Dumlao DS, Dennis EA. An integrated omics analysis of eicosanoid biology. J Lipid Res. 2009;50:1015–38.
Hilvo M, Denkert C, Lehtinen L, Müller B, Brockmöller S, Seppänen-Laakso T, et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011;71:3236–45.
Kilpinen S, Autio R, Ojala K, Iljin K, Bucher E, Sara H, et al. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol. 2008;9:R139.
Brockmöller SF, Bucher E, Müller BM, Budczies J, Hilvo M, Griffin JL, et al. Integration of metabolomics and expression of glycerol-3-phosphate acyltransferase (GPAM) in breast cancer-link to patient survival, hormone receptor status, and metabolic profiling. J Proteome Res. 2012;11:850–60.
Feng L. Probing lipid-protein interactions using lipid microarrays. Prostaglandins Other Lipid Mediat. 2005;77:158–67.
Denkert C, Bucher E, Hilvo M, Salek R, Orešič M, Griffin J, et al. Metabolomics of human breast cancer: new approaches for tumor typing and biomarker discovery. Genome Med. 2012;4:37.
Acknowledgments
We wish to thank Sarah Frost and Sara Jaffer for critical reading of the manuscript and past and present group members and external collaborators for their support.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer India
About this chapter
Cite this chapter
Kamili, A., Byrne, J.A. (2014). Lipidomics in Breast Cancer. In: Barh, D. (eds) Omics Approaches in Breast Cancer. Springer, New Delhi. https://doi.org/10.1007/978-81-322-0843-3_11
Download citation
DOI: https://doi.org/10.1007/978-81-322-0843-3_11
Published:
Publisher Name: Springer, New Delhi
Print ISBN: 978-81-322-0842-6
Online ISBN: 978-81-322-0843-3
eBook Packages: MedicineMedicine (R0)