Abstract
Humans are a highly mobile species that has colonized the entire globe in a few tens of thousands of years after it went out of Africa. There are still many unknowns about the routes followed by our ancestors during this expansion process, which has been influenced by various environmental, biological, and cultural factors, but these migrations have contributed to shape the genetic diversity of our species. A powerful approach to study the consequences of human dispersal on our genome is the modelling of complex evolutionary scenarios via computer simulation. In this chapter, we present three types of approaches used to simulate human dispersal in a geographic landscape. We focus on a spatially explicit method, simulating the demographic and migratory dynamic of populations forward in time and their resulting genetic diversity backward in time using the coalescent. We describe this approach and illustrate its interest with two important results: the process of gene surfing during population expansion and the genetic consequences of hybridization during species expansions. We show that a relatively simple scenario of global expansion of Homo sapiens from Africa, with rare hybridization events with archaic humans, such as Neanderthals or Denisovans, over a large geographic area reasonably explains the introgression pattern of archaic DNA in the genome of our species.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Homo sapiens is a highly mobile species. From its likely place of origin in Africa, it colonized the entire planet in less than 50,000 years. This early colonization phase was constrained by geographic and environmental boundaries (coastlines , mountains, forests), but also by the presence of other archaic Homo species (Neanderthals, Denisovans, H. erectus), who were present in most of the old world (Veeramah and Hammer 2014). Given the fossil record, Africa and the Near-East have been occupied by anatomically modern humans (AMH) before Asia, Europe, and the Americas (e.g. Henn et al. 2012). The most supported scenario for the origin of our own species is often termed “Recent African Origin” (RAO) and proposes an emergence of AMH in East Africa between 200 and 100 kya (Tattersall 2009), followed by a rapid spread toward the rest of the world starting around 60 kya (Henn et al. 2012), which led to a full replacement of all archaic human forms. The initial version of RAO has been challenged by the sequencing of the Neandertal and Denisovan genomes (Green et al. 2010; Reich et al. 2010; Prufer et al. 2014) and by statistical analyses of existing genomic data sets (Wall et al. 2013). These studies support the existence of a relatively low genetic contribution of archaic forms in current non-African humans, which probably arose through hybridization after the exit out of Africa. These new analyses changed the old RAO paradigm to a scenario one could call “RAO with hybridization” (Stringer 2014). However, very little is known on the exact extant of the interactions between AMH and Neanderthals (NE) and even less with other archaic populations.
Despite this general RAO scenario, many unknowns still exist on the migration routes followed by our ancestors. The exact dates of colonization of many regions of the world are also uncertain simply because old fossils are rare or absent in many regions, even though such an absence is not a proof that these regions have not been colonized by early humans. Consequently, several alternative hypotheses have been proposed to explain the colonization of different regions of the world (Veeramah and Hammer 2014). For instance, at least three main possible routes of migration have been proposed to explain the diffusion of AMH from Africa to East Asia and Oceania: a coastal “southern route” along the coast of the Pacific Ocean (Macaulay et al. 2005), a second more continental route in the south of Himalaya (Rasmussen et al. 2011), and another migration route North of the Himalaya (Di and Sanchez-Mazas 2011).
The dispersal of AMH out of Africa has been probably triggered by a combination of various factors such as new abilities or opportunities (due to technological advances or cultural changes), climatic variation, and demographic pressure (Powell et al. 2009; Eriksson et al. 2012; Lahr and Foley 1998). In any case, this initial global dispersal has been followed by many additional migrations, at various times and geographic scales. Climate variation played an important role in promoting those migrations through several mechanisms: first, the sea level was about 120 m below the current level during cold periods, which created many land bridges between continents and previously isolated islands (e.g. British Isles, New Guinea, Beringia); second, glaciers and ice caps extended over much larger areas during cold periods (Ray and Adams 2001) making them uninhabitable; third, vegetation cover has also significantly changed with temperature variations, displacing towards refugia (Hewitt 2000) most animals and plants that were food sources for humans (Banks et al. 2008). In addition, at the beginning of the Holocene, most human populations have passed through a major economic and cultural transition in different parts of the world, switching from a hunter-gatherer to a food production lifestyle (Bellwood 2001). This change corresponds to the appearance of plant and animal domestication, pottery, and of sedentary lifestyle and probably involved demographic increases and large-scale migrations (Zvelebil 2001). Finally, sedentarized human populations continued to exchange migrants until present time and additional waves of migration took place around the world during the historical period, but probably at a smaller scale (Sokal 1991).
Ongoing short range migrations have left traces in the genome of contemporary humans (Ray et al. 2005), which shows a strong concordance between geography and genetics in most parts of the world (e.g. Novembre et al. 2008). For these reasons, it seems necessary to consider the spatial constraints on the dispersal of modern humans when they colonized the planet. However, it is still very difficult to consider these population dynamics when analysing genomic data, as envisioned models quickly become very complex and are difficult to parameterize and test. Fortunately, modelling and computer simulations of complex evolutionary scenarios can be used to produce realistic models of human migration over time and space and to assess their impact on current genetic and genomic diversity. Thanks to a regular increase in computer power and to the development of efficient algorithms and realistic models, simulation approaches have become essential to study human evolution. In this review, we shall describe the main modelling approaches used to study the effect of human dispersal on genetic diversity at the global or continental scale, with a focus on spatially explicit simulations. We shall also present and comment their major contributions to our understanding of human evolution and discuss future developments.
2 Modelling and Simulating Human Dispersal
The modelling and simulation of evolutionary scenarios using computer programs consists of a combination of mathematical models defined by a series of parameters. Typically, three different approaches have been used to simulate human dispersal in a geographic landscape: Forward diffusion models, forward individual-based simulations, and forward demographic coupled with backward genetic simulations.
Diffusion approaches are based on a set of spatially explicit differential equations specifying density and diffusion (migration) parameters in a continuous space. They are extensions of Fisher’s approach to model the wave of advance of advantageous genes in one dimension (Fisher 1937). Fisher’s models have since been extended to model the colonization of different continents in two dimensions (e.g. Steele et al. 1998; Fort and Pujol 2008; Martino et al. 2007; Fort et al. 2004) or to the spread of cultures or technologies in already settled habitats (Ackland et al. 2007; Fort and Mendez 1999). These diffusion models have also been refined to take into account the heterogeneity of the habitat (see Steele 2009 for a review). Note that these models can mainly predict colonization times or the spread of beneficial variants in an already colonized habitat, and they have been coupled to the generation of genetic data only recently (Barton et al. 2013a, b).
Individual-based or agent-based simulations are the most flexible way to simulate genetic data, as they allow one to simulate very specific behaviour, such as sex-specific reproductive success or migration, age-specific mortality, or migration rates. They are usually performed in discrete demes positioned on a one-dimensional (e.g. Eswaran 2002; Eswaran et al. 2005; Ramachandran et al. 2005; Fix 1997) or two-dimensional lattice (Itan et al. 2009; Rasteiro et al. 2012; Deshpande et al. 2009; Rendine et al. 1986; Barbujani et al. 1995) corresponding to stepping-stone models, but they can be extended to 2D hexagonal demes (e.g. Eriksson et al. 2012) enabling a more realistic spatial diffusion process (Lavrentovich et al. 2013). Individual-based simulations also allow one to simulate loci under arbitrarily complex selection models (Itan et al. 2009; Currat et al. 2010; Fix 1996; Peischl et al. 2013) fully or partially linked with neutral loci (e.g. Chadeau-Hyam et al. 2008). However, this complexity has a cost in terms of computing power and time, implying that a small number of individuals and populations can be modelled, even though the rescaling of mutation, migration rates, and selection coefficients can allow one to model large populations with a smaller number of individuals (e.g. Chadeau-Hyam et al. 2008). Another drawback of forward approaches is that one needs to define starting conditions or to wait that some demo-genetic equilibrium has been reached before starting recording the simulations. Nevertheless, these models have been applied to very interesting problems in human evolution, ranging from the spread of Neolithic farmers into Europe and their interaction with local hunter-gatherers (Rasteiro et al. 2012; Rendine et al. 1986; Barbujani et al. 1995; Rasteiro and Chikhi 2013), the expansion out of Africa, either with (Eswaran 2002; Eswaran et al. 2005) or without (Ramachandran et al. 2005; Deshpande et al. 2009) hybridization, or to the estimation of the geographical origin of beneficial mutation in European populations (Itan et al. 2009).
Despite their great flexibility, forward individual-based simulations can sometimes be very slow and require a lot of memory, since they simulate all the individuals of many populations. A way to solve these problems is to use forward simulations to simulate the demography of the populations and then coalescent approaches to simulate their genetic diversity. These approaches have typically been implemented in the SPLATCHE program (see Currat et al. 2004; Ray et al. 2010, and details below) or in simpler 1 D serial founder effect models (DeGiorgio et al. 2011), and a similar approach has been used to simulate genetic data linked to a site under selection (Ewing and Hermisson 2010).
One usage of these modelling approaches is to generate genetic data for various alternative hypotheses (therefore various combinations of values assigned to the parameters), in order to evaluate which is the scenario most (or less) compatible with the observed genetic data and thus to perform model choice. They can also be used to describe and study evolutionary processes and to estimate demographic or genetic parameters. In the latter case, simulations can be coupled with Bayesian inference procedures such as the approximate Bayesian computation (ABC, Beaumont et al. 2002; Wegmann et al. 2010) to estimate demographic parameters under various evolutionary scenarios.
3 Realistic Simulation with Spatially Explicit Simulations
Here we focus on spatially explicit simulation of human dispersal and their genetic diversity. By spatially explicit, we mean that the simulation of populations or individuals considers their geographic position. The stepping-stone model (Kimura 1953), which consists of an array of cells in one or two dimensions is the most commonly used spatially explicit model. Here, the location of each population is defined by its coordinates and because migrants are only exchanged between neighbouring populations, the effective amount of gene flow between two populations is inversely related to their distance on the grid. This leads to genetic isolation by distance (IBD) (Wright 1943), a process that played an important role in human evolution (Morton 1977, 1982), and which largely explains the correspondence between geography and genetic variation in extent human populations (e.g. Novembre et al. 2008). Note that very detailed geographic (e.g. continental contours) and environmental (e.g. mountains, deserts) information can be used in spatially explicit simulations, which is facilitated using geographic information system (GIS). Note that spatially explicit simulations can also be done by specifying an arbitrary migration matrix between all pairs of populations in a model, as implemented in programs like SIMCOAL and FASTSIMCOAL (Excoffier et al. 2000; Excoffier and Foll 2011; Laval and Excoffier 2004) or in MS (Hudson 2002). However, in this case the migration matrix can become huge and difficult to set up for models with complex geographic features, and therefore we will thus not discuss these programs below. In Table 12.1, we give a list of other spatially explicit simulation models and programs that have been used or could potentially be used in the context of human dispersal. Since their exact implementation varies from one model to the other, we redirect the readers to the original papers to get details about their underlying methodology. However, we illustrate below the implementation of the main key elements of a spatially explicit simulation framework through the program SPLATCHE that we have developed.
4 SPLATCHE: An Example of Spatially Explicit Simulation Program
The program SPLATCHE is able to translate environmental information into genetic diversity (Currat et al. 2004; Ray et al. 2010). The simulations are done in two phases: during the first forward part, the demography and dispersal of a species can be simulated from one or several origins, considering environmental information. In a second phase, the molecular genetic diversity of one or several samples drawn from the population simulated during the first phase can be generated.
4.1 Spatial Structure
A digital map in ASCII raster format is used for the spatial specifications of the simulations. It represents the geographical contour of the region of interest with a geographic projection adapted to the representation of surfaces (Ray 2003). Additional information, such as vegetation cover, terrain, seacoast, or rivers, may also be incorporated in the program using the same file format. The map is divided into geographic cells that are characterized by their coordinates and possibly by environmental characteristics. The dimension of the cell (usually identical for all cells) can be of arbitrary size and represents a real geographic distribution. For studying human dispersal, cells of 50 × 50 km2 (e.g. Currat and Excoffier 2005) or 100 × 100 km2 are usually used (e.g. Currat and Excoffier 2011) depending on the desired resolution and on the computational constraints (more cells imply more computing time). Sizes of this magnitude seem to be adequate to represent subpopulations of hunter-gatherers (Anderson and Gillam 2000; Gronenborg 1999; Cavalli-Sforza and Hewlett 1982; Hewlett et al. 1982).
The environmental characteristics of each cell (vegetation, altitude, coastal area, river, etc.) may be directly used to compute two demographic variables: the carrying capacity K and the friction rate F (Fig. 12.1). It is possible to take into account the uncertainty of these parameters by performing multiple simulations with different parameter values and by doing sensitivity analyses (Ray et al. 2008). Each cell contains one deme, which represents a sub-unit of the whole populations (or species) under study, or alternatively, two demes representing two interacting species or populations (see Box 12.1 and Fig. 12.2 for details).
Schematic representation of the incorporation of environmental information in SPLATCHE. The influence of various environmental factors, such as vegetation, hydrography, topography, and coastlines, is translated into two demographic parameters: carrying capacity K which affects population densities; Friction F which affects migration
Schematic representation of a spatially explicit modelling of two interacting populations. The whole area under study is divided into geographic cells, which each contain two demes representing two interacting populations (for example, AMH and NE). Demes of the same population belonging to neighbouring cells can exchange migrants, while admixture (gene flow) can occur between demes within the same cell
Time
A single simulation consists of recording the density of all demes and the number of migrants exchanged between demes during a fixed number of generations (parameter t).
Dispersal and Migration
A migration probability (parameter m) for individual genes to move between (neighbouring) demes. This probability is constant in SPLATCHE but can be different during the dispersion phase and at demographic equilibrium in other programs (e.g. Eriksson et al. 2012; Deshpande et al. 2009). This basic migration scheme may be altered in different ways:
-
By using friction values (parameter F), which represent the difficulty of crossing a cell depending on its specific environmental characteristics. Higher F values assigned to a given cell generally imply less migrants entering it (Ray et al. 2008). This parameter can be used, for instance, to favour movements along coastlines or rivers, or to the opposite, to use rivers, hills, deserts, or mountains as barriers to gene flow.
-
By allowing for long-distance dispersals (Ray and Excoffier 2010), occurring at a given rate (parameter λ) and at various distance (parameter d).
-
By directing migration specifically into a given direction (Arenas et al. 2012), e.g. towards the South during glaciation or to the North during a post-glacial colonization.
Demographic Dynamics
Deme density is usually logistically regulated, reflecting intra-deme competition for resources. The increase in density is controlled by the growth rate (parameter r) and the maximum number of individuals that may be sustained by the cell (the carrying capacity, parameter K). This last parameter may reflect the influence of the environment (e.g. different vegetation types leading to different K) or the type of culture or economy (e.g. food production versus hunter-gathering techniques). Heterogeneous environments can also be considered in the dispersion model (Wegmann et al. 2006).
Generation of Genetic Data
SPLATCHE is using a coalescent approach (Hudson 1990; Kingman 1982) to reconstruct backward in time the genealogy of a series of sampled genes. For neutral loci, the virtual genetic diversity obtained at the end of a simulation is constrained by the demography of the simulated population and possibly by a mutation and recombination model that depends on the type of simulated genetic data (e.g. allele frequencies or molecular data).
The combination of the various elements described above allows one to construct and simulate realistic scenarios of human dispersal and testing those using genetic data. For instance, the genetic consequences of a range expansion (demographic increase linked to a geographical spread), bottleneck, population contraction to refuge area(s), interactions between populations (competition and admixture), or a combination of those processes, can be investigated using this general approach.
5 Main Results and Discussion
The simulation of human dispersal offers a powerful tool to study the evolution of our species, and it complements other methodological approaches. Most of the early models for the simulation of human dispersal have been developed in the context of the peopling history of Europe (Rendine et al. 1986; Barbujani et al. 1995; Currat and Excoffier 2005; Arenas et al. 2013), but we focus here only on simulations of human dispersal at the global scale. The first realistic attempts (Ray et al. 2005; Liu et al. 2006) to simulate worldwide dispersal showed an excellent fit between the predictions of the models and real data, confirming that models explicitly incorporating demography and geography were powerful to make inferences on human peopling history. This has been confirmed by a study incorporating Pleistocene climatic variation (Eriksson et al. 2012), which showed that climatic change had a significant impact on human dispersal and the establishment of current genetic diversity.
5.1 Genes Surfing the Waves of Expansion
An important result brought by the simulation of human dispersal was to explain the mechanisms by which genetic diversity progressively decreases with distance from Africa: a series of founder effects during the range expansion of human populations (Ramachandran et al. 2005; Deshpande et al. 2009). The same mechanism has also been proposed to explain clinical genetic patterns in Europe (Barbujani et al. 1995; Currat and Excoffier 2005). The process of decreasing diversity along a colonization route was extensively described and demonstrated theoretically by a series of spatially explicit simulations performed in a 2-dimensional stepping-stone (Deshpande et al. 2009). This effect results from a phenomenon called “allele surfing” (Edmonds et al. 2004; Klopfstein et al. 2006), which describes how the frequency of a neutral allele can dramatically increase during a population expansion due to pure neutral demographic effects. Surfing is a stochastic process that only applies to a relatively small number of alleles from the source population or to new mutations appearing during the expansion, but it has important evolutionary consequences (Excoffier and Ray 2008; Petit and Excoffier 2009). Simulations have shown that the frequency of surfing varies depending on the demographic parameters of the population (Klopfstein et al. 2006). The probability of surfing increases with the growth rate (parameter r), while it inversely decreases with the carrying capacity (K) and the migration rate (m). This gene surfing process has been identified theoretically by spatially explicit simulations before being confirmed by empirical studies, both in yeasts and bacteria (Hallatschek et al. 2007) and by a survey of the recent human colonization of the Saguenay Lac Saint-Jean in Quebec (Moreau et al. 2011). This finding shows that neutral evolutionary processes can produce patterns identical to those expected under the action of positive selection (i.e. an allele present at very high frequency in the final population). It thus challenged the view that large differences between populations such as those observed at some loci between Africans and non-Africans were due to ongoing selection (Currat et al. 2006).
5.2 Hybridization During Expansion
On their road out of Africa, AMH met various archaic human forms, such as Neanderthals (NE), Denisovans (DE), and probably others (Prufer et al. 2014). Very little is known about the exact nature of the interactions between AMH and NE and virtually nothing with DE. Simulation is thus an inestimable tool to assess, in a spatially dynamic context, the effects on genetic diversity of at least two kinds of interactions: admixture and competition. The main interest of spatially explicit simulation compared to previous mathematical models is that progressive admixture in time and space can be simulated between NE and AMH, instead of instantaneous merging of two panmictic populations (Nordborg 1998; Serre et al. 2004).
Spatially explicit simulations have shown that continuous interbreeding over space and time between NE and AMH during the spread out of Africa of the latter may well explain the current patterns of genetic introgression. First, it has been shown that the absence of Neanderthal type of mitochondrial DNA in contemporary human population is compatible with an extremely low interbreeding success rate (parameter ɣ in Box 12.1) between AMH and Neanderthal (Currat and Excoffier 2004). It was shown (Currat and Excoffier 2011) that less than 2% of interbreeding success rate (ɣ) is compatible with the observed presence of 2–3% of Neanderthal DNA in the genome of contemporary non-Africans (Green et al. 2010; Reich et al. 2010; Wall et al. 2013). Such low levels of introgression could be due to only about 200–300 successful hybridization events between NE and AMH over their whole cohabitation period (at least 10 Ky in Europe, even more in the Near-East) and over all their area of overlap. New estimations using a more symmetrical hybridization model (Excoffier et al. 2014) and considering in the estimation procedure that NE introgression is higher in Asia than in Europe (Wall et al. 2013) have produced similar results with an estimate ɣ of less than 3% (Box 12.2). These results thus demonstrate that the reported pattern of NE introgression in extent modern humans is compatible with a strong reproductive isolation between AMH and NE. In addition, spatially explicit simulations suggest that the hybridization between AMH and NE occurred over a large geographic zone covering Western and Central Asia and probably reaching southern Siberia. The analysis was not able to precisely delineate the Neanderthal occupation zone, but it suggested that it should be as big in Asia as in Europe at the time of the spread of AMH (Currat and Excoffier 2011).
5.3 Limitations and Future Developments
One sensitive point of any modelling approach is the choice of the parameter values, which may be difficult for some parameters. For instance, demographic parameters for prehistoric populations (densities, growth, and migration rates) are often difficult to evaluate and the mutation rate for the studied loci might not be known precisely (Gibbons 2012). To somehow circumvent this problem, a thorough examination of the literature is necessary to establish possible intervals of parameter values. For instance, density estimates may come from ethnographic comparisons (e.g. Pennington 2001) or long-term estimation (Biraben 1979), while growth and migration rates may be derived using estimates of colonization times in different continental areas (Ray et al. 2005; Currat and Excoffier 2005, 2011, 2004). The simulation approach then allows an extensive exploration of the parameter space and the validation of plausible values.
Compared to deterministic mathematical models, computer simulations have the advantage of considering stochastic processes in the analyses, which could play an important role in the evolution of humans, as in the case of the gene surfing phenomenon described above. A benefit of the simulation approach is that models can be improved step by step by adding new elements and new information brought by scientific discoveries (Currat and Silva 2013).
Another advantage of realistic simulations of human dispersal is their ability to integrate various sources of information, such as genetics, archaeology, and environment. This feature could be very useful in the near future as new types of data are regularly produced. For instance, in the last decades, it has been increasingly possible to extract DNA from fossil remains and this technical advance has been widely used in the study of human evolution. Computer simulations should allow one to analyse the increasing number of genomic data retrieved from ancient specimen (Sanchez-Quinto et al. 2012; Skoglund et al. 2014, 2012), which should be especially powerful when combined with modern DNA. Moreover, computer simulations could be used to check if selection has favoured the introgression of some genes of archaic origin (Caspermeyer 2014; Ding et al. 2014). Finally, computer simulation could be used to understand the relation between Denisovan, Southeast Asians, and Oceanians (Reich et al. 2011), but the lack of spatial information about the exact ancestral Denisovan range makes it a challenging task.
6 Conclusion
Computer simulations are a powerful tool to study the effects of population dynamics on the genetic diversity of populations. It has been shown that properly considering the spatial dynamics of populations can drastically change the interpretation of empirical data. For instance, a spatial expansion does not show the same typical bell-shaped mitochondrial mismatch distribution than a pure demographic expansion in a panmictic population, but rather a multimodal mismatch distribution when migration rates between demes are small to moderate (Ray et al. 2003; Excoffier 2004).
All in all, spatially explicit simulations support a relatively simple scenario of global human dispersal out of Africa with very rare interbreeding events with archaic humans over a large geographic range. They revealed that observed low levels of Neanderthal ancestry in Eurasians are compatible with a very low rate of interbreeding (<3%) and that those rare and distinct admixture events occurred also after the split of Europeans and Asian over a wide European and Asiatic range, well beyond the Middle East. A spatially explicit model of population expansion with continuous but limited interbreeding explains most observed patterns of human genomic diversity, such as: (1) a recent single origin (Stewart and Stringer 2012), (2) decreasing genetic diversity from Africa (Prugnolle et al. 2005); (3) limited and relatively uniform Neanderthal introgression in Eurasia (Green et al. 2010), larger in East Asia than in Europe (Wall et al. 2013), (4) introgression asymmetry between NE and AMH (Green et al. 2010), (5) lack of mitochondrial introgression (Reich et al. 2010; Serre et al. 2004), (6) introgression in area where Neanderthal never existed (Green et al. 2010), (7) more than one hybridization event with archaic humans (Wall et al. 2013). Spatially explicit simulation of human dispersal thus provides a simple but powerful framework for the interpretation of genomic data, with considerable room for improvements and extensions.
Box 12.1 Simulation of Interactions Between Two Populations
The program SPLATCHE offers the possibility to study the interactions between two populations (e.g. hunter-gatherers and Neolithic farmers) or two species (e.g. NE and AMH) in a spatially explicit framework (Ray et al. 2010). It simulates two demes per geographic cell, each of them representing one population (Fig. 12.2), and the simulated world can thus be seen as two superimposed layers of demes. SPLATCHE considers two kinds of interaction between interacting populations or species: competition and admixture (Fig. 12.2).
Competition is simulated using a classical Lotka–Volterra model (Lotka 1932), which is an extension of the logistic regulation model. The density of one species is directly constrained by the density of the other one, assuming that both are in competition for local resources (e.g. habitat, food). Competition coefficients (α) are used to reflect the intensity of this kind of interactions and can be different in the two populations, i.e. due to a competitive advantage of one over the other. The competition coefficients α can be fixed to a specific value, such as 1, which means that an individual of the rival population exerts as much competitive pressure as an individual belonging to the same population. Alternatively, α can be density dependent (Currat and Excoffier 2005), i.e. computed as αij = Nj/(Ni + Nj), where αij represents the effect of competition of an individual of population j on an individual of population i and Ni and Nj are the densities of both populations in the cell. In that case, the strength of competition between both populations evolves over time and the most numerous one has a competitive edge over the less numerous one. Under this model, if the carrying capacities (K) between the two populations are sufficiently different, as it is assumed for AMH over NE (Currat and Excoffier 2011, 2004), then the one with the lower K will eventually goes extinct due to competition.
Admixture is simulated by local gene flow between the two demes belonging to the same cell and it can be regulated by an interbreeding success rate (parameter ɣ). If ɣ is equal to 0, then there is no gene flow between the two populations. If ɣ is equal to 1, there is random mating between them. Lower values of ɣ imply the existence of barriers to gene flow between the two species, which can be either pre-zygotic (e.g. cultural avoidance, disassortative mating) or post-zygotic due to lower hybrid fitness or a combination of those various factors.
In previous studies, we implemented a model of density-dependent gene flow between AMH and NE (Currat and Excoffier 2011, 2004). A new admixture model has been implemented in SPLATCHE, which is fully symmetrical when both species are at demographic equilibrium and which is more accurate for the description of interspecific hybridization than the previous model (Excoffier et al. 2014). In this new model, each Ni′ newborn individuals in a population i have at least one parent belonging to population i. Then, assuming random mating the probability that the second parent originated from population j is simply computed as Nj/(Ni + Nj), where Ni and Nj are the densities of both populations in the previous generation. Thus, the expected number of gene flow events (introgressions) from population j to i at each generation is defined as:
This new admixture model gives results qualitatively and quantitatively very similar to the previous one (Excoffier et al. 2014).
Box 12.2 Estimation of Hybridization Between Neanderthals and AMH
To investigate hybridization between Neanderthals and AMH, we explored a series of scenarios of human dispersal out of Africa into Eurasia, with various demographic parameters, hybridization and competition zones and varying intensities of admixture. We redirect the readers to the original article for exact details on those alternative scenarios (Currat and Excoffier 2011). In all examined scenarios, Neanderthals is assumed to be at demographic equilibrium in its entire range at the beginning of the simulation, while AMH is expanding demographically and spatially (Fig. 12.3). Various sizes of Neanderthal occupation zone where hybridization with AMH occurred were tested, ranging from the Middle East only to a wider area extending to southern Siberia (Fig. 12.3). During the AMH expansion, admixture can occur, and Neanderthals disappear due to competition with AMH. At the end of a simulation, the proportion of Neanderthal ancestry is measured in modern human genomes in Europe and in East Asia and compared to the reported levels of 2–3% (Reich et al. 2010). For each of the 13 envisioned scenarios, we performed 10,000 coalescent simulations and we computed the proportion of those simulations that were compatible with the observation. In our new study, a simulation was declared compatible if it resulted in 2–3% of Neanderthal ancestry in both Europe and East Asia and if Neanderthal introgression was slightly larger in East Asia than in Europe (as shown in Wall et al. 2013). Figure 12.4 and Table 12.2 show the interbreeding rates obtained with this new series of simulations. They confirm previous results (Currat and Excoffier 2011; Excoffier et al. 2014), as very few successful hybridization events (ɣ < 3%) over a wide Eurasian range are sufficient to result in 2–3% of introgression in non-Africans. Such a low hybridization rate is sufficient to explain current Neanderthal introgression because the few Neanderthal genes that are incorporated continuously at the wavefront of the AMH expansion tend to be amplified by the surfing phenomenon (Currat et al. 2008). Indeed, a few introgression events occurring in an invading deme on the wavefront can result in many more introgressed copies, because these introgressions usually occur when the invading deme is still growing and has not reached its carrying capacity. Second, AMH pioneers are recruited at the front of the expansion and consequently have a higher probability to propagate their genes (including recently introgressed Neanderthal genes) further away in the expanding population. Rare but continuous interbreeding events during the expansion of AMH over a large Neanderthal Eurasian range are thus a simple and efficient model to explain patterns of Neanderthal ancestry in current genomes.
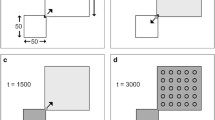
Example of the simulation of the dispersal of AMH out of Africa and the progressive disappearance of NE that inhabited an extended area in Eurasia. Black arrow represents the origin of AMH and pink arrows the current sampled locations. White represents empty cells, medium grey cells are occupied by NE only, black cells are occupied by AMH only, and dark grey represents the zone where NE and AMH coexist. Left panel shows the demographic forward phase and right panel shows the coalescent backward phase. Pink and red dots on the right panel represent at any time the location of lineages ancestral to the AMH sampled genes in AMH and NE demes, respectively. The parameters used for the simulations are those of scenario A in Table 12.2
Distribution of the proportion of simulations (among 10,000) resulting in Neanderthal introgression levels higher in the Chinese sample but still compatible with observations (1.9–3.1% in both French and Chinese samples). Each likelihood curve corresponds to a different demographic scenario described in Table 12.2. Results were obtained by assuming a deme area of 100 × 100 km2. Solid lines correspond to scenarios that are equally likely (within two AIC units from the scenario with the highest likelihood), whereas scenarios shown with a dotted line have an associated AIC more than two units larger and thus cannot be considered as equally well supported by the data (details of the estimation procedure may be found in Currat and Excoffier 2011)
References
Ackland GJ et al (2007) Cultural hitchhiking on the wave of advance of beneficial technologies. Proc Natl Acad Sci U S A 104(21):8714–8719
Anderson DG, Gillam C (2000) Paleoindian colonization of the Americas: implications from an examination of physiography, demography, and artifact distribution. Am Antiq 65(1):43–66
Arenas M et al (2012) Consequences of range contractions and range shifts on molecular diversity. Mol Biol Evol 29(1):207–218
Arenas M et al (2013) Influence of admixture and paleolithic range contractions on current European diversity gradients. Mol Biol Evol 30(1):57–61
Austerlitz F et al (1997) Evolution of coalescence times, genetic diversity and structure during colonization. Theor Popul Biol 51(2):148–164
Balloux F (2001) EASYPOP (version 1.7): a computer program for population genetics simulations. J Hered 92(3):301–302
Banks WE et al (2008) Human ecological niches and ranges during the LGM in Europe derived from an application of eco-cultural niche modeling. J Archaeol Sci 35(2):481–491
Barbujani G, Sokal RR, Oden NL (1995) Indo-European origins: a computer-simulation test of five hypotheses. Am J Phys Anthropol 96(2):109–132
Barton NH et al (2013a) Genetic hitchhiking in spatially extended populations. Theor Popul Biol 87:75–89
Barton NH, Etheridge AM, Veber A (2013b) Modelling evolution in a spatial continuum. J Stat Mech 2013:P01002
Beaumont MA, Zhang W, Balding DJ (2002) Approximate Bayesian computation in population genetics. Genetics 162(4):2025–2035
Bellwood P (2001) Early agriculturalist population. Annu Rev Anthropol 30:181–207
Biraben JN (1979) Essay on the evolution of numbers of mankind. Population 34(1):13–25
Brown JM, Savidge K, McTavish EJ (2011) DIM SUM: demography and individual migration simulated using a Markov chain. Mol Ecol Resour 11(2):358–363
Caspermeyer J (2014) Sunlight adaptation region of neanderthal genome found in up to 65% of modern East Asian populations. Mol Biol Evol 31(3):763
Cavalli-Sforza LL, Hewlett B (1982) Exploration and mating range in African Pygmies. Ann Hum Genet 46(Pt 3):257–270
Chadeau-Hyam M et al (2008) Fregene: simulation of realistic sequence-level data in populations and ascertained samples. BMC Bioinf 9:364
Currat M, Excoffier L (2004) Modern humans did not admix with Neanderthals during their range expansion into Europe. PLoS Biol 2(12):2264–2274
Currat M, Excoffier L (2005) The effect of the Neolithic expansion on European molecular diversity. Proc R Soc B 272(1564):679–688
Currat M, Excoffier L (2011) Strong reproductive isolation between humans and Neanderthals inferred from observed patterns of introgression. Proc Natl Acad Sci U S A 108(37):15129–15134
Currat M, Silva NM (2013) Investigating European genetic history through computer simulations. Hum Hered 76(3-4):142–153
Currat M, Ray N, Excoffier L (2004) SPLATCHE: a program to simulate genetic diversity taking into account environmental heterogeneity. Mol Ecol Notes 4(1):139–142
Currat M et al (2006) Comment on “Ongoing adaptive evolution of ASPM, a brain size determinant in homo sapiens” and “microcephalin, a gene regulating brain size, continues to evolve adaptively in humans”. Science 313:5784
Currat M et al (2008) The hidden side of invasions: massive introgression by local genes. Evolution 62(8):1908–1920
Currat M, Poloni ES, Sanchez-Mazas A (2010) Human genetic differentiation across the Strait of Gibraltar. BMC Evol Biol 10:237
DeGiorgio M, Degnan JH, Rosenberg NA (2011) Coalescence-time distributions in a serial founder model of human evolutionary history. Genetics 189(2):579–593
Dellicour S, Hardy OJ, Mardulyn P (2014) PhyloGeoSim 1.0. A program to simulate the evolution of DNA sequences under a spatially explicit model of coalescence. Mol Biol Evol 31(12):3359–3372
Deshpande O et al (2009) A serial founder effect model for human settlement out of Africa. Proc Biol Sci 276(1655):291–300
Di D, Sanchez-Mazas A (2011) Challenging views on the peopling history of East Asia: the story according to HLA markers. Am J Phys Anthropol 145(1):81–96
Ding Q et al (2014) Neanderthal introgression at chromosome 3p21.31 was under positive natural selection in East Asians. Mol Biol Evol 31(3):683–695
Edmonds CA, Lillie AS, Cavalli-Sforza LL (2004) Mutations arising in the wave front of an expanding population. Proc Natl Acad Sci U S A 101(4):975–979
Eriksson A, Manica A (2012) Effect of ancient population structure on the degree of polymorphism shared between modern human populations and ancient hominins. Proc Natl Acad Sci U S A 109(35):13956–13960
Eriksson A et al (2012) Late Pleistocene climate change and the global expansion of anatomically modern humans. Proc Natl Acad Sci U S A 109(40):16089–16094
Eswaran V (2002) A diffusion wave out of Africa. Curr Anthropol 43:749–764
Eswaran V, Harpending H, Rogers AR (2005) Genomics refutes an exclusively African origin of humans. J Hum Evol 49(1):1–18
Ewing G, Hermisson J (2010) MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics 26(16):2064–2065
Excoffier L (2004) Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol 13(4):853–864
Excoffier L, Foll M (2011) Fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics 27(9):1332–1334
Excoffier L, Ray N (2008) Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol Evol 23(7):347–351
Excoffier L, Novembre J, Schneider S (2000) SIMCOAL: a general coalescent program for the simulation of molecular data in interconnected populations with arbitrary demography. J Hered 91:506–510
Excoffier L, Quilodran CS, Currat M (2014) Models of hybridization during range expansions and their application to recent human evolution. In: Derevianko AP, Shunkov MV (eds) Cultural developments in the eurasian paleolithic and the origin of anatomically modern humans. Publishing Department of the Institute of Archaeology and Ethnography SB RAS, Novosibirsk, pp 122–137
Fisher RA (1937) The wave of advance of advantageous genes. Ann Eugenics 7:355–369
Fix AG (1996) Gene frequency clines in Europe: demic diffusion or natural selection? J Roy Anthropol Inst 2:625–643
Fix AG (1997) Gene frequency clines produced by kin-structured founder effects. Hum Biol 69(5):663–673
Fort J, Mendez V (1999) Reaction-diffusion waves of advance in the transition to agricultural economics. Phys Rev E 60(5 Pt B):5894–5901
Fort J, Pujol T (2008) Progress in front propagation research. Rep Prog Phys 71(8):086001
Fort J, Pujol T, Cavalli-Sforza LL (2004) Palaeolithic populations and waves of advance (human range expansions). Camb Archaeol J 14(1):53–61
Gibbons A (2012) Human evolution. Turning back the clock: slowing the pace of prehistory. Science 338(6104):189–191
Green RE et al (2010) A draft sequence of the neandertal genome. Science 328(5979):710–722
Gronenborg D (1999) A variation on a basic theme: the transition to farming in southern central Europe. J World Prehist 13(2):123–210
Guillaume F, Rougemont J (2006) Nemo: an evolutionary and population genetics programming framework. Bioinformatics 22(20):2556–2557
Hallatschek O et al (2007) Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci U S A 104(50):19926–19930
Henn BM, Cavalli-Sforza LL, Feldman MW (2012) The great human expansion. Proc Natl Acad Sci U S A 109(44):17758–17764
Hewitt GM (2000) The genetic legacy of the quartenary ice ages. Nature 405:907–913
Hewlett B, Van de Koppel JMH, Cavalli-Sforza L (1982) Exploration ranges of Aka pygmies of the Central African Republic. Man 17:418–430
Hudson RR (1990) Gene genealogies and the coalescent process. In: Futuyma D, Antonovics J (eds) Oxford surveys in evolutionary biology, vol 7. Oxford University Press, Oxford, p 300
Hudson RR (2002) Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18(2):337–338
Itan Y et al (2009) The origins of lactase persistence in Europe. PLoS Comput Biol 5(8):e1000491
Kimura M (1953) Stepping-stone model of population. Genetics 3:62–63
Kingman JFC (1982) The coalescent. Stoch Process Appl 13:235–248
Klopfstein S, Currat M, Excoffier L (2006) The fate of mutations surfing on the wave of a range expansion. Mol Biol Evol 23(3):482–490
Lahr MM, Foley RA (1998) Toward a theory of modern human origins: geography, demography, and diversity in recent human evolution. Yearb Phys Anthropol 41:137–176
Landguth EL, Cushman SA (2010) CDPOP: a spatially explicit cost distance population genetics program. Mol Ecol Resour 10(1):156–161
Laval G, Excoffier L (2004) SIMCOAL 2.0: a program to simulate genomic diversity over large recombining regions in a subdivided population with a complex history. Bioinformatics 20(15):1485–2487
Lavrentovich MO, Korolev KS, Nelson DR (2013) Radial Domany-Kinzel models with mutation and selection. Phys Rev E 87(1):012103
Liu H et al (2006) A geographically explicit genetic model of worldwide human-settlement history. Am J Hum Genet 79(2):230–237
Lotka AJ (1932) The growth of mixed populations: two species competing for a common food supply. J Wash Acad Sci 22:461–469
Macaulay V et al (2005) Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science 308(5724):1034–1036
Martino LA et al (2007) Fisher equation for anisotropic diffusion: simulating South American human dispersals. Phys Rev E 76(3):031923
Meirmans PG (2011) MARLIN, software to create, run, and analyse spatially realistic simulations. Mol Ecol Resour 11(1):146–150
Moreau C et al (2011) Deep human genealogies reveal a selective advantage to be on an expanding wave front. Science 334(6059):1148–1150
Morton NE (1977) Isolation by distance in human populations. Ann Hum Genet 40(3):361–365
Morton NE (1982) Estimation of demographic parameters from isolation by distance. Hum Hered 32:37–41
Neuenschwander S et al (2008) quantinemo: an individual-based program to simulate quantitative traits with explicit genetic architecture in a dynamic metapopulation. Bioinformatics 24(13):1552–1553
Nordborg M (1998) On the probability of Neanderthal ancestry. Am J Hum Genet 63(4):1237–1240
Novembre J et al (2008) Genes mirror geography within Europe. Nature 456(7218):98–101
Peischl S et al (2013) On the accumulation of deleterious mutations during range expansions. Mol Ecol 22(24):5972–5982
Pennington R (2001) Hunter-gatherer demography. In: Panter-Brick C, Layton RH, Rowley-Conwy P (eds) Hunter-gatherers: an interdisciplinary perspective. Cambridge University Press, Cambridge, pp 170–204
Petit RJ, Excoffier L (2009) Gene flow and species delimitation. Trends Ecol Evol 24(7):386–393
Powell A, Shennan S, Thomas MG (2009) Late pleistocene demography and the appearance of modern human behavior. Science 324(5932):1298–1301
Prufer K et al (2014) The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505(7481):43–49
Prugnolle F, Manica A, Balloux F (2005) Geography predicts neutral genetic diversity of human populations. Curr Biol 15(5):R159–R160
Ramachandran S et al (2005) Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci U S A 102(44):15942–15947
Rasmussen M et al (2011) An aboriginal Australian genome reveals separate human dispersals into Asia. Science 333(6052):94–98
Rasteiro R, Chikhi L (2013) Female and male perspectives on the neolithic transition in Europe: clues from ancient and modern genetic data. PLoS One 8(4):e0060944
Rasteiro R et al (2012) Investigating sex-biased migration during the Neolithic transition in Europe, using an explicit spatial simulation framework. Proc R Soc B 279(1737):2409–2416
Ray N (2003) Modélisation de la démographie des populations humaines préhistoriques à l'aide de données environnementales et génétiques. Université de Genève, Genève
Ray N, Adams JM (2001) A GIS-based vegetation map of the world at the last glacial maximum (25,000-15,000 BP). Internet Archaeol 11:319
Ray N, Excoffier L (2010) A first step towards inferring levels of long-distance dispersal during past expansions. Mol Ecol Resour 10(5):902–914
Ray N, Currat M, Excoffier L (2003) Intra-deme molecular diversity in spatially expanding populations. Mol Biol Evol 20(1):76–86
Ray N et al (2005) Recovering the geographic origin of early modern humans by realistic and spatially explicit simulations. Genome Res 15(8):1161–1167
Ray N, Currat M, Excoffier L (2008) Incorporating environmental heterogeneity in spatially-explicit simulations of human genetic diversity. In: Matsumura S, Forster P, Renfrew C (eds) Simulations, genetics and human prehistory. McDonald Institute for Archaeological Research, Cambridge, pp 103–117
Ray N et al (2010) SPLATCHE2: a spatially-explicit simulation framework for complex demography, genetic admixture and recombination. Bioinformatics 26(23):2993–2994
Rebaudo F et al (2013) SimAdapt: an individual-based genetic model for simulating landscape management impacts on populations. Methods Ecol Evol 4(6):595–600
Reich D et al (2010) Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468(7327):1053–1060
Reich D et al (2011) Denisova admixture and the first modern human dispersals into southeast Asia and Oceania. Am J Hum Genet 89(4):516–528
Rendine S, Piazza A, Cavalli-Sforza L (1986) Simulation and separation by principal components of multiple demic expansions in Europe. Am Nat 128(5):681–706
Sanchez-Quinto F et al (2012) Genomic affinities of two 7,000-year-old Iberian hunter-gatherers. Curr Biol 22(16):1494–1499
Serre D et al (2004) No evidence of neandertal mtDNA contribution to early modern humans. PLoS Biol 2(3):E57
Sgaramella-Zonta L, Cavalli-Sforza L (1973) A methode for the detection of a demic cline. In: Morton NE (ed) Genetic structure of population. University of Hawaii Press, Honolulu
Skoglund P et al (2012) Origins and genetic legacy of Neolithic farmers and Hunter-gatherers in Europe. Science 336:466–469
Skoglund P et al (2014) Genomic diversity and admixture differs for stone-age scandinavian foragers and farmers. Science 344(6185):747–750
Sokal RR (1991) Ancient movement patterns determine modern genetic variances in Europe. Hum Biol 63(5):589–606
Steele J (2009) Human dispersals: mathematical models and the archaeological record. Hum Biol 81(2-3):121–140
Steele J, Adams JM, Sluckin T (1998) Modeling Paleoindian dispersals. World Archaeol 30:286–305
Stewart JR, Stringer CB (2012) Human evolution out of Africa: the role of refugia and climate change. Science 335(6074):1317–1321
Stringer C (2014) Why we are not all multiregionalists now. Trends Ecol Evol 29(5):248–251
Sukumaran J, Holder MT (2011) Ginkgo: spatially-explicit simulator of complex phylogeographic histories. Mol Ecol Resour 11(2):364–369
Tattersall I (2009) Human origins: out of Africa. Proc Natl Acad Sci U S A 106(38):16018–16021
Veeramah KR, Hammer MF (2014) The impact of whole-genome sequencing on the reconstruction of human population history. Nat Rev Genet 15(3):149–162
Wall JD et al (2013) Higher levels of neanderthal ancestry in East Asians than in Europeans. Genetics 194(1):199
Wegmann D, Currat M, Excoffier L (2006) Molecular diversity after a range expansion in heterogeneous environments. Genetics 174(4):2009–2020
Wegmann D et al (2010) ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinf 11:116
Wright S (1943) Isolation by distance. Genetics 28:114–138
Zvelebil M (2001) The agricultural transition and the origins of Neolithic society in Europe. Documenta Praehistorica XXVlll. Neolithic Stud 8:1–26
Acknowledgments
This research was supported by a grant of the Center for Advanced Modelling Science (CADMOS) to MC and by Swiss NSF grants 31003A-143393 and 310030_188883 to LE and 31003A_182577 to MC. CSQ acknowledges support from CONICYT-Becas Chile and the Institute of Genetics and Genomics in Geneva (IGE3). All simulations were performed in the HPC cluster baobab.unige.ch.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Japan KK, part of Springer Nature
About this chapter
Cite this chapter
Currat, M., Quilodrán, C.S., Excoffier, L. (2021). Simulations of Human Dispersal and Genetic Diversity. In: Saitou, N. (eds) Evolution of the Human Genome II. Evolutionary Studies. Springer, Tokyo. https://doi.org/10.1007/978-4-431-56904-6_12
Download citation
DOI: https://doi.org/10.1007/978-4-431-56904-6_12
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-56902-2
Online ISBN: 978-4-431-56904-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)