Abstract
The phylum Ascomycota has been resolved into three major phylogenetic lineages: the subphyla Saccharomycotina (e.g., Saccharomyces, Pichia, Candida), Taphrinomycotina (e.g., Protomyces, Taphrina, Pneumocystis), and Pezizomycotina (e.g., Aspergillus, Neurospora, Peziza). We discuss the ecology, physiology, molecular biology, biotechnology, phylogeny, and systematics of Saccharomycotina and Taphrinomycotina, which represent the yeasts and yeastlike fungi of Ascomycota. Major changes in all aspects of our knowledge of these two subphyla have resulted from molecular studies, and the focus of the chapter is on these changes and their impact on present and future applications of the yeasts.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
I. Introduction
Yeasts are found in nearly all regions of the Earth, including hot deserts, polar areas, in freshwater, in salt water, and in the atmosphere, where they are commonly transported by prevailing winds. Though yeast growth is mainly saprotrophic, some yeasts are important pathogens of animals and plants. The term yeast has come to mean those fungi that divide by budding or fission and that have sexual states unenclosed in a fruiting body. Consequently, yeasts occur among the Ascomycota and the Basidiomycota . The focus of this chapter are those taxa assigned to Saccharomycotina and Taphrinomycotina of Ascomycota. As we will discuss, some members of these subphyla are among the economically most important fungi known.
II. Occurrence , Distribution, and Ecology
Although yeasts occur worldwide, some have restricted habitats, whereas others are found in many different environments. The key to understanding yeast ecology and the extent of habitat specificity is the accurate identification of species, which is now possible through DNA-based methods. Prior to the application of molecular methods, the identification of species from phenotype often resulted in misclassification, which rendered results from many ecological studies uncertain or misleading.
Yeasts are often associated with insects, and numerous studies have detailed these interactions (Phaff et al. 1956; van der Walt and Scott 1971; Vega and Blackwell 2005; Wickerham 1969). Contemporary studies have expanded on this earlier work and refined it through molecular-based species identification. Notable has been the work of H. J. Phaff and W. T. Starmer and colleagues (Phaff et al. 1987; Starmer et al. 1992), who examined yeast–Drosophila interactions among various species of cacti. In one of these studies, Pichia kluyveri was recognized to be comprised of three closely related and partially interfertile species that were found to show significant habitat differences.
Other yeasts are strongly associated with plants, including the well-known interaction of Saccharomyces cerevisiae with grapes (e.g., Mortimer and Polsinelli 1999). Ripe apples have significant surface populations of Saccharomyces , Torulaspora , Zygosaccharomyces, and other yeasts that are attributed to the transfer of soluble sugars onto the surface of the fruit, and many species of the genus Ogataea are found on leaves and decaying wood. Species of Ogataea utilize methanol as their sole source of carbon, which is present in the environment as a degradation product of lignin (de Koning and Harder 1992) and is formed in leaf respiratory processes (Fall and Benson 1996).
Some yeasts, such as Debaryomyces hansenii and Meyerozyma guilliermondii (anamorph: Candida guilliermondii), occur widely in nature and are common in water, plant debris, and soil. Both of the aforementioned species also represent opportunistic human pathogens. Although yeasts are commonly isolated from soil, few are believed to have soil as a primary habitat. Many Lipomyces species are an exception and have been isolated only from soil.
III. Importance
A. Food , Beverage, and Industrial Uses
Since ancient times, human societies worldwide have used beverages and foods fermented by yeasts (Legras et al. 2007). Archaeologists have found evidence that fermented beverages were consumed in Neolithic times (8500–4000 B.C.) in China, Iran, Egypt, and other areas of the world (Legras et al. 2007; McGovern et al. 2004), and the production of fermented beverages and foods seems to have paralleled the beginning of agriculture. Louis Pasteur provided the insight that fermentation was the result of microorganisms and noted that yeasts occurred on grapes, thereby providing a ready source of inoculum for wine (Dubos 1960; Mortimer and Polsinelli 1999). The commonplace processes of baking , brewing, and wine making are often taken for granted but represent major industries with a combined worldwide annual value that may exceed US$1 trillion (Hansen 2004; Verstrepen et al. 2006). Other food-related yeast processes include the natural fermentation of cocoa beans, coffee beans, pickles, olives, and similar products.
The industrial importance of yeasts has vastly expanded over the past few decades to include much more than bread making and brewing. Yarrowia lipolytica was initially used for the production of single-cell protein from hydrocarbons, but the species is now recognized as a major producer of citric acid, an important acidulant for industrial and food and beverage uses. Other industrially significant metabolites from ascomycetous yeasts include riboflavin from Eremothecium gossypii (≡Ashbya gossypii) (Wickerham et al. 1946), lactase from Kluyveromyces marxianus (Rubio-Texeira 2006), biosurfactants, such as sophorolipids, from members of the Starmerella clade (Kurtzman et al. 2010), and lipases from Y. lipolytica and Candida cylindracea (Gellissen et al. 2005). S. cerevisiae is widely used in the production of recombinant proteins for medical and other uses, but the methanol-assimilating yeasts , such as Komagataella pastoris (≡Pichia pastoris) and Ogataea polymorpha (≡Hansenula polymorpha), are also important in this role (Cregg and Madden 1988; Veenhuis et al. 1983).
The conversion of plant biomass to biofuels is of major interest, and the yeasts Pachysolen tannophilus , Scheffersomyces stipitis (≡Pichia stipitis), and Candida shehatae , which were discovered to ferment the d-xylose of biomass to ethanol, have been the mainstays in this effort (Slininger et al. 1987, and references therein). More recently, Spathaspora passalidarum and Candida jeffriesii were found to ferment d-xylose to ethanol (Nguyen et al. 2006).
B. Agriculturally Important Yeasts
1. Plant Pathogens
In this section, we discuss two important agricultural aspects of yeasts, those that are plant pathogens and those that are antagonists of pathogens. Taphrina and Protomyces , both members of Taphrinomycotina, are perhaps the best known of the yeastlike taxa that cause plant diseases . T. deformans, the cause of peach leaf curl, is worldwide in its distribution and the most economically devastating of the diseases caused by species of Taphrina (Fonseca and Rodrigues 2011; Mix 1949). Young leaves, stems, and fruit are often severely distorted when infected by T. deformans. Early application of fungicides usually controls peach leaf curl (Daughtrey et al. 2003), but failure to do so can result in significant crop losses. Other tree crops, such as almonds and pears, may be severely affected by Taphrina infections. Trees, such as alders and poplars, are also susceptible to various Taphrina species (Fonseca and Rodrigues 2011; Mix 1949). The effect of these infections on tree health is generally limited, but the appearance of infected leaves and flowers can be distressing when the trees are used in ornamental plantings.
Species of Protomyces cause symptoms similar to those seen from Taphrina infections. All known Protomyces species are plant parasitic and cause galls on stems, leaves, and fruits of Compositiae, Umbelliferae, and certain other plants (Tubaki 1957). Economic losses are seldom great, but P. macrosporus infection of coriander (Coriandrum sativum) was reported to have damaged up to 11 % of the crop during one growing season (Tripathi et al. 2003).
Several genera of Saccharomycotina also cause plant diseases. Most notable are species of Eremothecium , some of which were previously classified in the genera Ashbya, Nematospora, and Holleya, all of which are plant pathogens. E. ashbyi has a long history of causing cotton boll rot in various species of Gossypium and cankers on citrus fruit (Batra 1973). Similarly, E. gossypii causes staining and rot of cotton bolls and is pathogenic to coffee (Coffea spp.), soybean (Glycine max), and other crops. E. coryli can infect cotton, but it is also a pathogen of hazelnuts, tomatoes, and beans. Symptoms are generally disfigurement and disruption of the infected plant tissue. E. sinecaudum was discovered by Holley et al. (1984) to cause seed infection in oriental and yellow mustard in Saskatchewan, Canada. The remaining known species of Eremothecium, E. cymbalariae, appears uncommon but has been isolated as a pathogen of flax and other plants (Arnaud 1913). Other known pathogenic yeasts are Galactomyces candidus (anamorph Geotrichum candidum) and Galactomyces citri-aurantii (anamorph: Geotrichum citri-aurantii), which commonly cause sour rot of citrus, tomatoes, cantaloupes, peaches, lychee, and carrots (Butler et al. 1965; Wells 1977). Losses are sufficiently great to require treatment of the produce by a fungicide.
2. Biocontrol Yeasts
Yeasts assigned to Saccharomycotina as well to Basidiomycota are used in the biocontrol of plant diseases (Andrews 1992; Chalutz et al. 1991). Our focus will be on the ascomycetous yeasts used for this purpose. Yeasts are common on leaf surfaces and on fruits, the latter containing abundant, easily utilizable carbon sources. The finding that naturally occurring yeasts on apples can protect fruit against postharvest diseases (Janisiewicz 1987) markedly stimulated work on the utilization of yeasts as an alternative to chemical pesticides for the protection of fruits against storage diseases.
M. guilliermondii (anamorph: Candida guilliermondii) has been successfully used in a number of studies to control fruit rots. Guetsky et al. (2002) reported a 50 % reduction in Botrytis cinerea rot of strawberries following application of M. guilliermondii and suggested that the control mechanism is competition for nutrients. Similarly, M. guilliermondii was effective at controlling fungi that cause rot of citrus (Droby et al. 1993). M. guilliermondii is widespread in nature, but it is also an opportunistic human pathogen, raising concerns about its safety as a biocontrol agent on fruit and other produce that will be consumed without cooking. Candida oleophila , which is not a clinical yeast, shows good control of fruit storage rots and has been commercialized under the trade name Aspire for the protection of citrus from rots caused by species of Penicillium (Droby et al. 1998). Metschnikowia fructicola and Metschnikowia pulcherrima have also been tested extensively for the biocontrol of fruit rots (Janisiewicz et al. 2001; Karabulut et al. 2004). Additionally, Wickerhamomyces anomalus (≡Pichia anomala) has been effective in the biocontrol of mold-induced spoilage of ensiled maize (Passoth et al. 2006). A concern is that W. anomalus has been implicated in some human infections, and its suitability as a biocontrol agent is uncertain.
C. Food and Beverage Spoilage
The spoilage of foods and beverages by contaminating yeasts results in major economic losses worldwide (e.g., Fleet 1990). Yeasts responsible for food spoilage are not known to cause infection or food poisoning in humans, as do certain bacteria. The composition of foods and beverages often determines the species of yeasts that are likely to be found. Products with high sugar content (40–70 %) are commonly spoiled by Zygosaccharomyces spp., Torulaspora delbrueckii, Schizosaccharomyces octosporus, and Wickerhamomyces subpelliculosus (≡Pichia subpelliculosa). If salt (NaCl) is used as a preservative, other species can predominate. During the fermentation of cucumbers (10–16 % NaCl) and other products, such as soy sauce, Candida etchellsii and Candida versatilis are common. Species of Debaryomyces, especially D. hansenii, often overgrow aged cheeses, salami, and other salted meat products, imparting flavor and the potential for spoilage. Spoilage of fresh fruits may be caused by Candida stellata, P. kluyveri, Pichia fermentans, M. pulcherrima, and species of Hanseniaspora and its anamorph Kloeckera. Species of Brettanomyces and its teleomorph Dekkera are often responsible for turbidity and off-flavors in wine, beer, and soft drinks.
D. Human and Animal Pathogens
Candida albicans is the most common cause of candidiasis and is the species most often isolated from cases of oral infections and vaginitis (Bialkova and Subik 2006; Kaur et al. 2005). DNA sequence analysis of C. albicans strains resulted in the discovery of the closely related and phenotypically nearly identical species Candida dubliniensis (Sullivan et al. 2005). C. dubliniensis is mainly isolated from oral candidiasis in HIV-infected patients, but it is also isolated from healthy individuals, as is the case for C. albicans. Following C. albicans, the second most common cause of blood stream infections is Candida glabrata, and the increased frequency of infections by this species has been attributed to the larger population of immunocompromised individuals and the widespread use of antimycotics. DNA sequence analysis resulted in the discovery of the clinically important species Candida bracarensis and Candida nivariensis, both of which are phenotypically similar to C. glabrata (Alcoba-Flórez et al. 2005; Correia et al. 2006).
Numerous other ascomycetous yeasts are obtained as clinical isolates, but most are widespread in the environment and regarded as opportunistic pathogens. Among them are M. guilliermondii, Pichia kudriavzevii (anamorph: Candida krusei), Candida parapsilosis, Candida tropicalis, and even S. cerevisiae. Both animals and humans can develop yeast infections. Kazachstania pintolopesii has devastated colonies of laboratory mice (Kurtzman et al. 2005), and Macrorhabdus ornithogaster appears to be a cause of decline in birds such as budgerigars (Tomaszewski et al. 2003).
IV. Reproduction
A. Asexual
1. Budding, Fission, Endospores, Chlamydospores
Asexual reproduction , sometimes termed vegetative reproduction, occurs in ascomycetous yeasts by budding or by fission. The formation of pseudohyphae and septate (true) hyphae constitutes other forms of asexual reproduction that are discussed subsequently. Buds may arise either on yeast cells or on hyphal cells, and budding is initiated by the formation of a small evagination or outgrowth at some point on the surface of the cell. The parent (mother) cell remains more or less constant in size as the bud (blastoconidium) increases in size to form a new cell and then usually separates from the parent cell. Budding is termed holoblastic or enteroblastic, depending on how the bud is formed. All layers of the wall of the parent cell are involved in holoblastic budding and the bud separates, usually on a narrow base, leaving a scar through which no further budding occurs. Von Arx and Weijman (1979) considered holoblastic budding to be characteristic of Saccharomycotina. When budding is enteroblastic , the first bud arises through a rupture in the wall of the parent cell through which the innermost layer evaginates and ultimately grows out to form the outermost layer of the bud. The site of budding is eventually surrounded by a collarette owing to the recurrent formation and abscission of a succession of buds arising from the inner layer of the wall of the cell. Enteroblastic budding is characteristic of basidiomycetous yeasts and some members of Taphrinomycotina (Kurtzman and Sugiyama 2001; Sugiyama and Nishida 1995).
Budding is also classified on the basis of the position of the site where it occurs. Budding that is restricted to one pole of a cell is termed monopolar, and budding that occurs at both poles of a cell is termed bipolar . When the buds are abstricted on a rather broad base by the formation of a cross wall, the process is referred to as “budding on a broad base” and “bud fission” (Fig. 1.1). Recurrent budding leads to the formation of multiple scars or annellations at the poles of the cell (Streiblová 1971). Bipolar budding is characteristic of apiculate yeasts. Budding from various sites on a cell is termed multilateral or multipolar and is the most common type of budding among ascomycetous yeasts (Fig. 1.1). Budding is also described in terms of the way successive buds are produced. Sympodial budding occurs on a conidiophore that extends in growth by a succession of apices. A blastoconidium is produced at each apex, and the growth continues to the side of the apex; the result is a zigzag appearance, for example, Blastobotrys. Acropetal budding entails the formation of successive buds in a chain with the youngest at the apex. In basipetal budding, successive buds are formed, with the oldest at the apex.
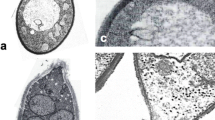
Various forms of asexual reproduction. (a) Multilateral budding (Pichia nakasei). (b) Bipolar budding on a wide base (Hanseniaspora osmophila). (c) Fission (Schizosaccharomyces pombe). (d) Pseudohyphae (Metschnikowia gruessii). (e) True (septate) hypha with side branches bearing blastoconidia (Candida ontarioensis). (Panels a–d, T. van Beest and T. Boekhout, CBS Web site; panel e, C.P. Kurtzman). Bars = 5 μm
Reproduction by fission is the division of an asexual cell by means of a septum growing inward from the cell wall to bisect the long axis of the cell. The newly formed fission cells, which are termed arthroconidia (arthrospores), elongate, and the process is repeated. Recurrent fission by a cell may give rise to transverse multiple scars or annellations (Streiblová 1971). This manner of reproduction is characteristic of Schizosaccharomyces and Dipodascus (Fig. 1.1).
Asexual cells may be globose, subglobose, ellipsoid, ovoid, obovoid, cylindrical, botuliform, bacilliform, elongate, apiculate, ogival, lunate, or triangular. Definitions and illustrations of the various possibilities can be found in Ainsworth & Bisby’s Dictionary of the Fungi (Kirk et al. 2008). The shape of the cell may reflect the mode of reproduction, and in some cases, it is characteristic of particular genera or species. Some examples include the lemon-shaped cells of the apiculate yeasts Hanseniaspora and Wickerhamia, the lunate cells of Metschnikowia lunata, and the triangular cells of Trigonopsis variabilis (Kurtzman and Robnett 2007).
Endospores are asexual cells that are formed within single cells and in hyphal cells and seem to arise by budding. Endospores are not commonly formed, but they have been observed in strains of Candida and a few other genera. No special media have been devised to stimulate the development of endospores (do Carmo-Sousa 1969).
Chlamydospores have been defined as thick-walled, nondeciduous, intercalary or terminal asexual spores formed by the rounding of a cell or cells (Ainsworth 1971; Hughes 1985; Stalpers 1987). The asexual nature of the chlamydospore distinguishes it from the teliospore of basidiomycetous yeasts from which the basidium is produced. Chlamydospores are generally rich in lipids and well adapted to maintain viability through periods of dormancy. In older cultures, chlamydospores shed their outer layers just before or during germination. Chlamydospores are characteristic of C. albicans, C. dubliniensis, and Metschnikowia species. Chlamydospores fulfill a dual function in Metschnikowia and either germinate by budding or are transformed into asci.
2. Pseudohyphae and True (Septate) Hyphae
Mature buds can either become detached as individual cells or remain attached to the parent cell and give rise to chains or clusters of cells. The tendency of some yeasts to form chains of cells results in the formation of pseudohyphae. A pseudohypha is defined as a filament composed of a chain of cells that has been formed by budding (Fig. 1.1). Pseudohyphae may be either rudimentary, in which case they consist of cells of similar size and shape, or they may be differentiated into elongated cells, each of which may produce blastoconidia. The form of a pseudohypha can be markedly affected by cultural conditions (van der Walt 1970). Some species of Dekkera form an unusual type of pseudohypha called a blastese, which is a slender, aseptate hypha that develops from germinating blastospores (Langeron and Guerra 1940).
Some yeasts produce true septate branching hyphae, which elongate by continuous growth of the hyphal tip followed by the formation of septa (Fig. 1.1). The fine structure of hyphal septa varies among taxa, but light microscopy does not reveal much detail except for the presence of large septal pore bodies in Ambrosiozyma . Hyphae may proliferate by simple branching, or they may produce blastoconidia on differentiated conidiogenous cells. The presence of blastoconidia on denticles is a characteristic of species of Trichomonascus (anamorph Blastobotrys), Hyphopichia, and certain other genera. Hyphae are sometimes joined by a process in which there is the fusion of branches of the same or different hyphae, and this is called anastomosis. The media most commonly used for detecting pseudohyphae and true hyphae are corn meal (maize) agar, morphology agar, 5 % malt extract agar, and potato-dextrose agar, but some clinical laboratories use rice agar, which will also promote the formation of chlamydospores by C. albicans.
B. Sexual Reproduction
Many yeasts reproduce sexually, resulting in an alternation of generations with the formation of characteristic cells in which reduction division takes place. In ascogenous yeasts, the site of meiosis is the ascus where the haploid generation of ascospores is formed by so-called free-cell formation, i.e., the process by which the cytoplasm surrounding the meiotic nuclei becomes enveloped by a wall. Ascogenous yeasts may be homothallic or heterothallic, and the asexual phase may be diploid or haploid, but sometimes both haploid and diploid cells are present in the same culture. Higher degrees of ploidy have also been reported for some species, such as S. cerevisiae and Lachancea kluyveri (≡Saccharomyces kluyveri) (Wickerham 1958).
For haploid homothallic yeasts, plasmogamy , karyogamy , and meiosis occur within the zygote, which is often formed by the conjugation of two separate budding cells or by conjugation between a cell and its bud (mother–daughter cell conjugation or bud meiosis). The diplophase is usually restricted to the diploid zygote within which the ascospores are formed. In the case of conjugation between a cell and its bud, the bud remains attached to the parent cell, which is converted into an ascus in which usually one to four ascospores are formed. Asci bearing such vestigial buds are found in Debaryomyces, Torulaspora, Pichia, and certain other genera. A process comparable to cell–bud conjugation appears to operate in the genus Nadsonia, where karyogamy is initiated by the fusion of the nuclei of a bud and its parent. The contents of the zygote then move into a bud at the opposite pole, which is abstricted by a septum and becomes the ascus. When diploidization occurs by the fusion of two independent haploid cells, the cells may form elongated conjugation tubes, which fuse to give a dumbbell shape, as is characteristic for Zygosaccharomyces, Kodamaea and certain other taxa (Fig. 1.2).
Various forms of ascospore formation. (a) Persistent, unconjugated asci with globose ascospores (Saccharomyces paradoxus). (b) Deliquescent, unconjugated asci with globose and hat-shaped ascospores (Pichia membranifaciens). (c) Asci with globose ascospores formed from conjugation of complementary mating types (Kodamaea ohmeri). (d) Persistent asci with globose ascospores formed by conjugation between cells and their buds (Schwanniomyces pseudopolymorphus). (e) Persistent asci with tapered bud-conjugants and globose ascospores (Torulaspora delbrueckii). (f) Persistent, unconjugated asci, each with a roughened, spherical ascospore (Citeromyces siamensis). (g) Deliquescent asci with bean-shaped ascospores (K. marxianus). (h) Hat-shaped ascospores released from deliquescent asci (Cyberlindnera veronae). (i) Saturn-shaped ascospores formed in deliquescent asci (Saturnispora ahearnii). (j) Elongated, needle-shaped ascospores in a persistent ascus (Metschnikowia hawaiiensis). (k) Elongated ascospores with a whiplike tail released from a deliquescent ascus (Eremothecium coryli). (l) Hat-shaped ascospores released from an ascus formed at the tip of an ascophore (Pachysolen tannophilus). (Panels a, c, d, g, j, T. van Beest and T. Boekhout, CBS Web site; panels b, e, f, h, i, k, l, C.P. Kurtzman). Bars = 5 μm
Heterothallic species may occur as haploid mating types or as diploid strains, which are normally heterozygous for the mating-type genes and are sometimes termed bisexual. Unisexual diploid strains are known (Wickerham 1958); asci from these strains are unconjugated, and unisexual haploid ascospores of both mating types are formed. Ascospores of opposite mating types either conjugate within the ascus, giving rise to the diplophase, as in Saccharomycodes, or the ascospores are released and germinate to give haploid asexual cells of opposite mating types. Normally, the diplophase can only be restored if conjugation of haploid cultures of opposite mating types occurs. Active cultures of mating types are not invariably stable and may revert to sporulating cultures as a result of mutation of the mating-type alleles (Takano and Oshima 1970). Some species, such as Pichia membranifaciens, seem to have both heterothallic and homothallic strains. Heterothallic strains of some species may show sexual agglutination, as in Wickerhamomyces canadensis (=Hansenula wingei) and Lachancea kluyveri (≡Saccharomyces kluyveri) (Wickerham 1958), in which cells of opposite mating types agglutinate when mixed. Agglutination in W. canadensis is mediated by complementary glycoproteins present on the surface of cells of the opposite mating types (Crandall and Brock 1968).
Ascospores vary in the number present in asci. Asci with either one or two ascospores are typical for Lodderomyces, Metschnikowia, and some species of Debaryomyces, whereas four spores are characteristic of some species of Pichia, Saccharomyces, and certain other genera. In contrast, species of Ascoidea and Vanderwaltozyma may form in excess of 100 ascospores in each ascus. The shape of ascospores varies widely and includes globose, ellipsoidal, cylindrical, reniform, crescentic, clavate, hat-shaped (galeate), cap-shaped, saturnoid, walnut-shaped, falcate, needle-shaped, and spindle-shaped with a whiplike appendage (Fig. 1.2). The surface may be smooth or rough, but surface ornamentation, including brims and ledges, may be reduced to such an extent that this morphology cannot be detected by light microscopy. Phylogenetic analyses have shown that ascospore shape is of uncertain value for assessing genus assignments and sometimes even varies within strains of a species (Kurtzman et al. 2008). An example of the latter is Kodamaea ohmeri, where both hat-shaped and globose ascospores have been found depending on the mating types paired (Wickerham and Burton 1954).
Ascosporulation may be induced under conditions that restrict asexual growth, but many strains sporulate without any special preparation. Some genera sporulate best on a particular medium. Acetate agar has been recommended for Saccharomyces (e.g., McClary et al. 1959) and dilute V8 agar for Metschnikowia (Pitt and Miller 1968). Many strains of Pichia and related genera sporulate on YM or malt agars. What follows represents a suggested start for ascospore detection. Strains placed on slant cultures of YM, 5 % malt extract, dilute V8, and RG agars should be incubated at 15 and 25 °C and examined weekly for 2 months [see Kurtzman et al. (2011) for the composition of culture media given in this chapter]. Some yeasts sporulate rapidly, i.e., within 24–48 h, especially when first isolated; others may require much longer, up to 6 weeks or more. If ascosporulation is not detected, then other media, such as Gorodkowa and acetate agars, can be tried. Strains that do not form ascospores may represent mating types and should be mixed to determine whether conjugation and ascosporulation occur. Conjugation often occurs within 24–48 h, but occasionally 1–2 weeks may be required. Ascospores can be stained with malachite green (Wickerham 1951), but visualization of unstained cells with either bright field or phase contrast microscopy is favored by many observers over staining.
V. Taxonomic Methods
A. Phenotypic Characterization
In addition to differentiation by cellular morphology, responses on fermentation and growth (assimilation) tests are still used in some laboratories to identify taxa. The physiological tests commonly used are fermentation of seven to eight carbohydrates, growth on various carbon and nitrogen sources, requirements for vitamins, growth at various temperatures, growth on media with a high content of sugar or sodium chloride, hydrolysis of urea, and resistance to antibiotics.
There is no known exception to the rule that when a yeast strain ferments a carbohydrate, it is also able to grow on it. However, the reverse does not hold true; many yeasts grow aerobically on sugars they cannot ferment. Yeasts vary in their ability to ferment sugars as measured by the production of carbon dioxide.
Various tests have been devised to detect the production of carbon dioxide from carbohydrates, but Durham tubes are the most useful method for the routine detection of carbohydrate fermentation (Kurtzman et al. 2011; Wickerham 1951). Durham tubes are test tubes with a small inverted tube inserted to collect any gas that may be produced. The fermentation of d-glucose, d-galactose, sucrose, maltose, lactose, raffinose, and trehalose is generally tested for routine identification; other compounds, such as inulin, starch, melibiose, cellobiose, and d-xylose, are sometimes used. The sugars are tested as 2 % (w/v) solutions, except for raffinose, where 4 % is usually used because some strains cleave and ferment only part of the molecule of this trisaccharide.
Assimilation tests determine the ability of a yeast to grow aerobically on a particular carbon compound supplied as the sole source of energy. The tests can be done either on solid media or in liquid media, but liquid media are believed by some taxonomists to give more reproducible results.
The size of growth tubes and the amount of medium used can vary widely among laboratories, but the method employing tubes of liquid media as described by Wickerham (1951) is commonly used. The results are improved by gently shaking the tubes during incubation. Some laboratories incubate the tests for a period of 3 weeks, others for 4 weeks. These long incubations allow the yeasts to adapt to utilize some compounds. Tests on solid media can be done in two ways. The first is the auxanographic method of Beijerinck (1889), in which the yeast is suspended in agar in pour plates and the test sugars are spotted at intervals around the circumference. The second method is to incorporate the test compound into a nutrient agar basal medium in petri dishes and inoculate the test yeast as either a streak or a point on the surface.
B. Genotypic Characterization
During the past 10–15 years, gene sequence analyses have been used for yeast identification, usually replacing the phenotypic methods described earlier. Nonetheless, the collection of phenotypic data provides important information about the biology of the strains under study and their potential biotechnological applications. DNA comparisons of yeasts have paralleled the increasing sophistication of methods for nucleic acid characterization. Initial studies were restricted to determining the mol% guanine + cytosine (G + C) content of DNA. From this work it was seen that ascomycetous yeasts had a nuclear DNA content of ca. 28–50 mol%, whereas basidiomycetous yeasts had a noticeably higher range of 50–70 mol% (Nakase and Komagata 1968; Price et al. 1978). These studies suggested that strains differing by 1–2 mol% were likely to represent different species, thereby providing a means for excluding strains incorrectly assigned to a particular species. Quantitation of gene sequence similarity between strains became possible with the development of DNA reassociation techniques that measure the extent of pairing of nucleotide sequences when DNA is made single-stranded and allowed to re-pair as a double strand. An interpretation of DNA reassociation data was provided by Martini and Phaff (1973) and Price et al. (1978), who suggested that, on the basis of shared phenotype, strains that showed 80 % or greater nuclear DNA relatedness are members of the same species. A limitation of DNA reassociation experiments has been that genetic resolution extends no further than to closely related species. In contrast, gene sequence comparisons offer the opportunity to resolve closely related species, as well as more distantly related taxa, and a database of sequences can be developed and expanded for further use.
The variable domain 2 (D2) from nuclear large subunit ribosomal RNA (LSU rRNA) was initially examined and found to resolve closely related species (Peterson and Kurtzman 1991). This work was expanded to include domains 1 and 2 (D1/D2) and applied to all described species of ascomycetous yeasts, resulting in a diagnostic database (barcode) for rapid species identification (Kurtzman and Robnett 1998). A comparison of nuclear DNA reassociation values suggested that conspecific strains differed by no more than 3 nucleotides among the 500–600 nucleotides of the D1/D2 domains , whereas differences of 6 or more nucleotides (1 %) indicated that the strains were different species. The preceding estimates of divergence were treated as a prediction (Kurtzman and Robnett 1998) because exceptions were known. Among them are hybrid species (Groth et al. 1999; Peterson and Kurtzman 1991; Vaughan-Martini and Kurtzman 1985) and certain DNA polymorphisms (Lachance et al. 2003).
A significant advantage to using rRNA gene sequences is that ribosomes have a common evolutionary history, and within the sequences are highly conserved regions between the variable regions that serve for pan-specific primer attachment for PCR amplification and sequencing. In contrast, protein coding genes tend to be variable across the entire gene, often making primer design difficult. Nonetheless, the gene sequences encoding several proteins have been examined for phylogenetically divergent groups of species. Daniel et al. (2001) compared Candida spp. from several clades and showed that phylogenetic trees generated from actin sequences were congruent with rRNA gene trees. Daniel and Meyer (2003) compared the resolution of closely related species from actin sequences and from D1/D2 LSU. As with D1/D2, actin did not always provide a clear separation of species, but in general, actin sequences had a greater number of substitutions, providing easier recognition of closely related species. Similar resolution was reported for the translation elongation factor-1α gene (Kurtzman et al. 2008) and the cytochrome oxidase II (COX II) gene (Belloch et al. 2000; Kurtzman and Robnett 2003).
In the examples presented, determination of whether strains are conspecific or members of separate species can be confused by hybridization events, by unexplained sequence polymorphisms, and by differences in nucleotide substitution rates. Multigene analyses offer a means for detecting these changes, which would be signalled by lack of congruence for a particular gene tree.
The multigene approach was recommended by Goodman (1976) for vertebrates, for bacteria by Dykhuizen and Green (1991), and for fungi by Taylor et al. (2000). The paper by Taylor et al. (2000) provides an inclusive review of species concepts, and the term genealogical concordance phylogenetic species recognition (GCPSR) was introduced to describe the concept of multigene analysis for species recognition (see Taylor and Berbee, Chap. 1, Vol. VII, Part A).
Comparison of strains from single gene sequences, such as D1/D2 LSU rRNA and the sequences of ITS, has provided a rapid means for species identification. However, the preceding discussion makes clear that some closely related species are not resolved by these sequences and that the occurrence of hybrids further complicates identification. Consequently, critical species identification requires a comparison of multiple genes, hence the increasingly widespread application of multilocus sequence typing (MLST) in strain identification.
VI. Phylogeny and Classification
A. Phylogeny
The relationship of ascomycetous yeasts with other members of the Ascomycota has been controversial for over 100 years. Because yeasts are morphologically simple, it was proposed that either they represent primitive forms of Ascomycota (e.g., Guilliermond 1912) or that they represent morphologically reduced forms of more evolved taxa (Cain 1972; Redhead and Malloch 1977; von Arx and van der Walt 1987).
The issue of relationships within the Ascomycota remained uncertain until the use of gene sequence analysis to estimate phylogeny. Walker (1985) sequenced 5S rRNA for selected ascomycetes, and the analysis of this data set divided the Ascomycota into three groups: (1) Schizosaccharomyces and Protomyces, (2) budding yeasts, and (3) so-called filamentous fungi. Berbee and Taylor (1993) analyzed a larger group of species from nuclear SSU rRNA gene sequences, which showed the same three major ascomycete lineages and that yeasts and filamentous fungi are sister taxa, whereas Schizosaccharomyces and relatives diverged prior to these two clades. Kurtzman and Robnett (1994) showed from partial LSU and SSU rRNA sequences that all currently accepted ascomycetous yeast genera were members of a single clade, which was separate from Schizosaccharomyces and members of the filamentous fungi. The finding from single-gene analyses that Ascomycota is comprised of three separate lineages was supported by multigene sequence analyses (Fitzpatrick et al. 2006; James et al. 2006; Kuramae et al. 2006). The phylogenetic trees generated from sequence analyses have significantly changed classification within the fungi, and these changes will be discussed. The outline of ascomycetous yeast classification is given in Table 1.1 and is based on the classification of the kingdom Fungi presented by Hibbett et al. (2007) and from multigene analyses reported by Kurtzman (2003), Kurtzman and Robnett (2003, 2007, 2010, 2013), Kurtzman and Suzuki (2010), and Kurtzman et al. (2007, 2008). Figure 1.3 shows the phylogenetic relationship among taxa of Saccharomycotina, Taphrinomycotina, and Pezizomycotina and was determined from multigene sequence analysis (Sugiyama et al. 2006). Support for early diverging lineages in many multigene trees is often weak, and assignment of genera to families is tentative for many of the taxa. The analyses presented in Figs. 1.3 and 1.4a, b are examples of this issue, and the uncertain placement of the anamorphic genus Saitoella has been indicated in Fig. 1.3, whereas Fig. 1.4b suggests that Taphrinomycotina may consist of two clades. However, other multigene analyses have supported the monophyly of Taphrinomycotina (Liu et al. 2009; Schoch et al. 2009).
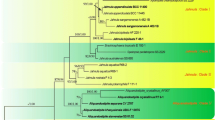
Phylogenetic resolution of members of the Ascomycota into three major clades (Pezizomycotina, Saccharomycotina, Taphrinomycotina) from a multigene analysis based on the 50 % majority rule consensus of 18,000 Bayesian MCMCMC generated trees. In this analysis, the alternate positions for Saitoella complicata provide an example of the effect that weak basal lineages have on taxon placement. Numbers on branches are Bayesian posterior probability. Numbers separated by a slash (/) indicate bootstrap values based on parsimony analysis with third codon position/parsimony analysis without third codon position/neighbor joining (NJ) analysis with third codon position/NJ analysis without third codon position. An asterisk (*) indicates lack of bootstrap support. Branches supported by ≥95 % posterior probability and ≥70 % bootstrap value in all analyses (parsimony vs. NJ, third codon included vs. excluded) are indicated by thick lines. Dotted arrows indicate alternative groupings found with ≥70 % bootstrap support: I = parsimony analysis with third codon position; II = both parsimony and NJ analyses with third codon position; III = NJ analysis without third codon position, which demonstrates the impact of data set composition on the outcome of the analysis. For details and GenBank accession numbers of the four genes (SSU rRNA, D1/D2 LSU rRNA, RNA polymerase 2, and beta-tubulin) used in this analysis, see Sugiyama et al. (2006). Additionally, see Assembling the Fungal Tree of Life Web site (http://aftol.org/data.php) and the GenBank homepage (http://www.ncbi.nih.gov/Genbank/index.html). Modified from Sugiyama et al. (2006); reproduced with permission from Mycologia. @The Mycological Society of America
(a) Phylogenetic relationships among type species of ascomycetous yeast genera and reference taxa determined from maximum-likelihood analysis using concatenated gene sequences for LSU rRNA, SSU rRNA, EF-1α, RPB1, and RPB2. Filobasidiella neoformans was the designated outgroup species in the analysis. Names in bold font are type species of currently recognized genera, whereas names in standard font are not type species. Pneumocystis is represented by the type species, but not the type strain. Protomyces and Taphrina are not represented by type species. Bootstrap values (1,000 replicates) >50 % are given at branch nodes. Strain accession numbers are NRRL unless otherwise indicated. Designations in brackets indicate the coenzyme Q value for each species. Y and YB prefixes are NRRL strain numbers. Type species of the genera Ascobotryozyma (anamorph, Botryozyma), Coccidiascus, Endomyces, Helicogonium, Phialoascus, Macrorhabdus, and Schizoblastosporon (teleomorph, Nadsonia), several of which are not known from culture, were not included in the analysis (Kurtzman and Robnett 2013). Basal lineages of some taxa are not well resolved, which demonstrates the need to include additional gene sequences in analyses. Because of this, many family assignments are tentative. Clade 1, Saccharomycetaceae and Saccharomycodaceae (Saccharomycodes and Hanseniaspora, which appear more closely related in some analyses); Clade 2, Phaffomycetaceae; Clade 3, Komagataella; Clade 4, Saccharomycopsis (Saccharomycopsidaceae) and Ascoidea (Ascoideaceae); Clade 5, Pichiaceae and closely related genera. (b) Phylogenetic relationships among type species of ascomycetous yeast genera and reference taxa determined from maximum likelihood analysis using concatenated gene sequences for LSU rRNA, SSU rRNA, EF-1α, RPB1, and RPB2. Clade 6, Debaryomycetaceae, Metschnikowiaceae, and other related taxa; Clade 7, Alloascoidea, an Ascoidea-like new genus; Clade 8, Sporopachydermia; Clade 9, Dipodascaceae and Trichomonascaceae; Clade 10, Trigonopsis and Botryozyma; Clade 11, Lipomycetaceae; Clade 12, Taphrinomycotina, which in this analysis shows a dichotomy but in some other analyses all genera share a common lineage
B. Classification
Two events are having a profound impact on the classification of fungi. The first of these, as discussed earlier, is gene sequence analyses, which have changed our understanding of relationships among members of Ascomycota. The genera accepted in Saccharomycotina and Taphrinomycotina are listed in Table 1.1, and most have now been circumscribed from multigene phylogenetic analyses. Phylogenetic relationships among the genera are shown in Fig. 1.4a, b. This analysis is based on five gene sequences, but bootstrap support for placement of some genera in the phylogenetic tree is weak and their assignment to families is tentative.
The second event that is having a major impact on the classification of fungi is the result of recent changes in the International Code of Nomenclature for algae, fungi and plants (Melbourne Code, e.g., Hawksworth 2012; Knapp et al. 2011; Norvell 2011). The genera of fungi may now include species with known sexual states (teleomorphs) as well as species for which sexual states have not been discovered (anamorphs). With this change, genera will be phylogenetically circumscribed to include related species whether or not sexual states are known, and the terms anamorph and teleomorph will have no status in genus descriptions. Because this change in rules for classification is so new, few yeast genera have been recircumscribed to include both sexual and asexual species. For this reason, the terms anamorph and teleomorph are being used in this chapter to provide a reference to the classification of yeasts presented in the recently published fifth edition of The Yeasts, A Taxonomic Study (Kurtzman et al. 2011), which was prepared before the new code was adopted.
1. Saccharomycotina
Many of the teleomorphic clades of Sacccharomycotina include species of Candida. Under the old code, the genus Candida was a “dumping ground” for budding yeasts that do not form ascospores, and it represents a polyphyletic group of species. With the introduction of sequence analysis, many species of Candida are seen to be members of teleomorphic clades and, because of the aforementioned changes in the code, will be transferred to those genera. However, other Candida species are in clades with no known ascosporic state and will be classified in new genera. What has become apparent from recent research is that much of the future classification of yeasts will rest on the phylogenetic analysis of gene sequences rather than on the phenotypic characters that we observe on a petri dish or under the microscope.
2. Taphrinomycotina
On the basis of phylogenetic analyses of various gene sequences, members of the subphylum Taphrinomycotina are composed of the early diverging and biologically diverse members of Ascomycota (e.g., Kurtzman and Robnett 1998; Kurtzman and Sugiyama 2001; Sugiyama et al. 2006). The number of known taxa assigned to Taphrinomycotina is surprisingly small considering that the more recently evolved Saccharomycotina and Pezizomycotina are much more species rich. Whether this reflects an absence of species diversity or ineffective isolation strategies is unknown, but the recent description of Archaeorhizomyces (the new class Archaeorhizomycetes) (Rosling et al. 2011), an early diverging and widely distributed genus, suggests that more effective isolation methods will reveal Taphrinomycotina to be a larger group of species than previously thought.
Taphrinomycotina are characterized by ascosporic states that lack ascogenous hyphae. Asexual reproduction is by budding or fission. With the exception of Neolecta , an apothecial ascomycete genus (Landvik 1996; Landvik et al. 1993), neither ascomata nor conidiomata are formed. Saccharomycotina have cell walls that show two layers and undergo holoblastic conidiogenesis, in contrast to basidiomycetous yeasts, which characteristically have multilayered cell walls and enteroblastic, repetitive percurrent conidiogenesis (Moore 1987). However, Saitoella complicata and Taphrina wiesneri are exceptional because, although they have two-layer cell walls, they show enteroblastic budding (Goto et al. 1987; Sjamsuridzal et al. 1997).
Originally, it was thought that Taphrina and Schizosaccharomyces lacked chitin in their cell walls (e.g., Cavalier-Smith 1987), thereby separating them from other fungi, but Sietsma and Wessels (1990) reported glucosaminoglycan in Schizosaccharomyces pombe. Bowen et al. (1992) determined the DNA sequence of a chitin synthase fragment from S. pombe, and Nishida and Sugiyama (1994a) demonstrated DNA sequences for chitin synthase fragments in Taphrina wiesneri, Protomyces inouyii, and Saitoella complicata. Furthermore, Garner et al. (1991) reported that chitin is an integral part of the cell wall of Pneumocystis carinii trophozoites and cysts. The carbohydrate composition of cell walls from Taphrina, Protomyces, and Saitoella is characterized by rhamnose, glucose, and mannose (Prillinger et al. 1990; Sugiyama et al. 1985), but Schizosaccharomyces does not contain rhamnose (Prillinger et al. 1990). Members of Taphrinomycotina show a negative diazonium blue B (DBB) reaction, as is typical of Saccharomycotina (Kurtzman and Sugiyama 2001; Kurtzman et al. 2011; Sugiyama and Nishida 1995).
Order Pneumocystidales. Pneumocystis carinii is a principal causal agent of pneumonia in patients with HIV/AIDS, and for many years this organism was considered to be a protozoan. Edman et al. (1988) showed P. carinii to be a fungus based on 18S sequence comparisons. Watanabe et al. (1989) suggested from 5S rRNA analysis that Pneumocystis is closely related to Zygomycotina, but ultrastructural studies showed cell division by fission (Yoshida 1989). Taylor et al. (1994) and Sugiyama and Nishida (1995) suggested that P. carinii and S. pombe have a similar life cycle, and Nishida and Sugiyama (1994b) placed Pneumocystis in Taphrinomycotina. Subsequently, Hibbett et al. (2007) accommodated the genus in the class Pneumocystidiomycetes in the subphylum Taphrinomycotina.
Pneumocystis was included for the first time in The Yeasts, A Taxonomic Study, 5th Edition, in which Cushion and Keely (2011) fully redescribed and accepted five species, i.e., the type species P. carinii and four other species.
According to the genus diagnosis (Cushion and Keely 2011), “no species of Pneumocystis has been continuously cultivated outside the mammalian lung,” asexual reproduction is by binary fission, and eight ascospores are produced within an ascus as a result of meiosis. The correct name for the taxon called Pneumocystis carinii from human lungs is Pneumocystis jirovecii, whereas the name P. carinii is only found in the lungs of immunosuppressed rats (Cushion and Keely 2011).
Order Schizosaccharomycetales. Schizosaccharomyces is saprotrophic and undergoes asexual reproduction by fission. The genus is comprised of four species: S. japonicus, S. octosporus, S. pombe, and S. cryophilus (Vaughan-Martini and Martini 2011). Yamada and Banno (1987) reassigned S. octosporus and S. japonicus to other genera on the basis of differences in ascospore morphology, ubiquinone type, and cellular linoleic acid content, but this separation was not supported by rRNA gene sequence analyses (Kurtzman and Robnett 1991, 1998; Naehring et al. 1995). Strains of Schizosaccharomyces are isolated from high sugar substrates, such as fruit juices, honey, dried fruits, and tree fluxes. Schizosaccharomyces is widely used in studies of molecular biology.
Order Neolectales. The genus Neolecta is characterized by clavate, stalked apothecia and cylindrical aparaphysate eight-spored asci. Korf (1973) placed the genus in Helotiales of the Discomycetes, whereas Redhead (1977) created the new family Neolectaceae and tentatively placed it in Lecanorales. Landvik et al. (1993) and Landvik (1996) proposed the new order Neolectales because their SSU rRNA gene sequence analyses placed Neolecta outside of Pezizomycotina. Because of these and other comparisons, Eriksson and Hawksworth (1995) and Sjamsuridzal et al. (1997) placed the genus Neolecta in what is now Taphrinomycotina. According to Redhead’s (1977) description and illustrations, asci of Neolecta vitellina occasionally are filled with numerous conidia, which has also been observed in Taphrina (cf. Sugiyama 1998).
Order Taphrinales. Included in this order are two families, Protomycetaceae and Taphrinaceae (Table 1.1). Protomycetaceae includes six genera, Burenia, Protomyces, Protomycopsis, Saitoella, Taphridium, and Volkartia. Some of these genera are well studied, others are not, and we will discuss three of the better studied genera of the order. Protomyces is parasitic on plants mainly of Apiaceae (Umbelliferae) and Asteraceae (Compositae). Protomyces inouyei, for example, attacks the composite Youngia japonica in Japan, causing a gall on stems (Tubaki 1957). The thick-walled resting spores that are formed in infected plant tissue germinate, giving an ascuslike tube in which spores develop (Fig. 8 in Kurtzman and Sugiyama 2001). Protomyces species generally grow at lower temperatures (15–25 °C) and produce pigmented colonies, much as is seen for Taphrina and Saitoella, which also tend to grow at lower temperatures.
The genus Taphrina, which may include as many as 95 species, is parasitic on a wide variety of vascular plants, primarily ferns, the Rosales, and the Fagales (Kramer 1973; Mix 1949). The morphology and life cycles of Taphrina spp. are unique. Species are dimorphic with a saprotrophic haploid, uninucleate state, and a parasitic ascogenous binucleate mycelial state that develops in the host tissue. Moore (1990) described the genus Lalaria for the anamorphic states of Taphrina, some of which are occasionally isolated from the environment as a yeast state. Taphrina deformans, which causes peach leaf curl, is especially well studied (Kramer 1960; Martin 1940; Syrop and Beckett 1976). Another well-studied species is T. wiesneri (=T. cerasi), which attacks the Japanese cherry tree (Cerasus yedoensis), causing witches’ broom (Fig. 9 in Kurtzman and Sugiyama 2001; Tubaki 1978). Ascospores of T. wiesneri bud within asci on the leaves of the host plant (Fig. 10 in Kurtzman and Sugiyama 2001). Characteristic of most other Taphrina spp., the budding haploid phase of T. wiesneri appears as pigmented yeastlike colonies on agar media. The color is due to the formation of carotenoid pigments.
The yeast states of Taphrina (Fonseca and Rodrigues 2011), Protomyces (Kurtzman 2011), and Saitoella (Sugiyama and Hamamoto 2011) are similar culturally, biochemically, and chemotaxonomically. Especially noteworthy is the fact that the cell walls of the three genera are two-layered and typical of ascomycetous yeasts, whereas conidiogenesis (budding) is enteroblastic and typical of basidiomycetous yeasts (Goto et al. 1987; Sjamsuridzal et al. 1997).
Phylogenetic analysis of SSU rRNA gene sequences from 14 species of Taphrina and 4 species of Protomyces verified the presence of two genera and demonstrated that they are monophyletic (Sjamsuridzal et al. 1997). On the basis of rRNA sequence analyses (Kurtzman 1993), Protomycetales appear to be synonymous with Taphrinales, hence placement of the preceding genera in a single order, Taphrinales. This treatment has been supported by the multigene phylogenetic analysis of Hibbett et al. (2007).
The history of studies on the anamorphic genus Saitoella was fully described by Sugiyama et al. (1993). The first known species of this genus, S. complicata, was initially identified as Rhodotorula glutinis (Goto and Sugiyama 1970), but later Goto et al. (1987) proposed that S. complicata was closely related to Taphrinales. This supposition, based on phenotypic comparisons, was verified from analyses of SSU rRNA gene sequences (Nishida and Sugiyama 1993, 1994b; Nishida et al. 1993; Sugiyama et al. 1993). On the basis of multigene phylogenetic analysis, Saitoella appears to be a member of Taphrinomycotina (Fig. 2 in Sugiyama et al. 2006). A second species of Saitoella was reported and described as S. coloradoensis (Kurtzman and Robnett 2012). This new species was isolated from insect frass occurring in an Engelmann spruce (Picea engelmannii) growing in the state of Colorado in the USA, and multigene analysis showed that S. complicata and S. coloradoensis are closely related. The teleomorph of Saitoella is unknown, and the evolutionary relationships between Saitoella and other members within Taphrinomycotina remain uncertain. At present, the genus Saitoella has two assigned species described from three strains (Kurtzman and Robnett 2012; Sugiyama and Hamamoto 2011), but the genus may be larger. Allison et al. (2010) cloned DNA from soil sampled in an Alaskan (USA) boreal forest. Of the 433 fungal sequences obtained, 3 (GU212336, GQ892426, GQ892425) were identical to the D1/D2 LSU rRNA gene sequence of S. complicata.
New major lineages within Ascomycota. Several studies on fungal diversity in soils using a total environmental DNA sampling have revealed potentially new early diverging ascomycete lineages independent of Taphrinomycotina (Blackwell 2011; Jumpponen and Johnson 2005; Schadt et al. 2003; Sugiyama et al. 2006; Vandenkoornhuyse et al. 2002). Subsequently, Porter et al. (2008) suggested a major new clade as Soil Clone Group I (SCGI) of Ascomycota equivalent to a subphylum. Currently the SCGI clade is known only from sequence data. As discussed earlier, the genus Archaeorhizomyces (Rosling et al. 2011), which was assigned to Archeorhizomycetes, appears to be a member of the Taphrinomycotina. Both molecular- and cultivation-based methods for isolation of novel ascomycete taxa from environments are needed to elucidate the whole phylogenetic picture of Ascomycota.
VII. Isolation , Maintenance, and Culture Availability
A. Isolation
Yeasts are recovered from a wide range of aquatic, marine, and terrestrial habitats, as well as from the atmosphere. Many yeasts are widely distributed, whereas others appear to be confined to specific habitats. Yeasts seldom occur in the absence of either molds or bacteria. Consequently, selective techniques are often used for the recovery of yeasts. The composition of selective media is determined by the fact that yeasts are generally capable of developing at pH levels and water activities that reduce or inhibit the growth of bacteria. Antibiotics may also be used to suppress bacteria. Fungistatic agents for the suppression of molds can be used, but these compounds may also inhibit yeasts. In addition to the methods discussed in this chapter, the publications of Beech and Davenport (1971), Deak (2003), and Kurtzman et al. (2011) discuss the isolation of yeasts from natural habitats, and the publications of Buckley (1971) and Staib et al. (1989) provide methods for isolating clinical yeasts. When yeasts are present in high numbers, they may be isolated by directly plating the material, or suspensions of the material, on acidified agar media that may also contain antibiotics or have other selective formulations. Dilution plate techniques can be used for quantitative studies.
Temperatures for isolation and growth . Cultures are usually incubated at 20–25 °C because most yeasts are mesophilic; however, temperatures between 4 and 15 °C are essential for psychrophilic taxa. Higher temperatures, in the range of 30–37 °C, are often required for yeasts that are strictly associated with warm-blooded sources. Among these species are human and animal pathogens assigned to Candida, Kazachstania, Macrorhabdus, and Cyniclomyces. The latter two genera have exceptional nutritional requirements (Kurtzman et al. 2011). Incubation temperatures can also serve to selectively isolate particular groups of species.
Acidified media (pH 3.5–5). Acidified media provide selective isolation, and hydrochloric and phosphoric acids are often used to acidify the media. Organic acids, such as acetic acid, are not recommended because they are only slightly dissociated at pH 3.5–5, and high concentrations of undissociated acids have an inhibitory effect on many yeasts. Exceptions include Zygosaccharomyces bailii, Z. bisporus, and some strains of Pichia membranifaciens (Yarrow 1998).
Agar in media with a low pH is hydrolyzed when autoclaved. To resolve this problem, sterilized molten agar is cooled to approximately 45 °C, and a predetermined volume of acid is added. The medium and acid are quickly but gently mixed to avoid air bubbles and immediately poured into petri dishes. The addition of approximately 0.7 % (v/v) 1 N hydrochloric acid to YM agar or glucose–peptone–yeast extract agar usually gives the desired pH of 3.7–3.8. Many yeasts can be recovered at pH 3.7, but some species, such as those of the genus Schizosaccharomyces, are inhibited by high acid media, and moderately acidic media with a pH in the range 4.5–5.0 will give a higher recovery.
When yeasts are present in low numbers, population size can be increased by incubating the sample in a liquid medium at a pH of 3.7–3.8. The use of antibiotics or high or low temperatures can provide further selection. The development of molds can be restricted by excluding air from the culture by pouring sterile paraffin oil over the surface of the medium to form a layer about 1 cm deep. This procedure favors the development of fermentative strains but may fail to recover aerobic strains. Another method for restricting mold development involves incubating flasks of isolation media on a rotary shaker (Wickerham 1951). Molds in shaken flasks often do not conidiate and tend to grow as pellets that are outgrown by yeasts. The yeasts may be separated from the molds either by allowing the pellets of mold to settle for a few minutes and then streaking the suspension of yeasts onto agar in petri dishes or by removing suspended pellets by filtering through sterile glass wool. These pregrowth methods cannot be used for the quantitation of the initial population size in the sample tested.
Osmotic media . Yeasts can often grow on media with concentrations of sugar that are high enough to inhibit the development of many bacteria. A medium such as glucose–peptone–yeast extract agar or YM agar containing glucose at a concentration of 30–50 % is suitable for recovering osmophilic and osmotolerant yeasts from foodstuffs and juice concentrates of low water activity. The selective action of these media can be enhanced by lowering the pH to around 4.5. Osmotolerant yeasts recovered in this way can usually be successfully subcultured on media containing successively decreasing amounts of sugar, for example, 30, 10, 4, and 2 %.
Antibiotics and other selective compounds. Several media containing antibiotics have been described (Bills and Foster 2004) that can be used to suppress co-occurring microorganisms. Antibacterial antibiotics, which are often used, include tetracycline at 50 mg/L or a combination of penicillin G and streptomycin sulfate, each at a concentration of 150–500 mg/L. Many of these antibiotics are heat labile and must be added after the medium has been autoclaved and is cool to the touch. In contrast, chloramphenicol (actidione) is heat stable and can be added to the medium prior to sterilization. Antifungal antibiotics, which suppress filamentous fungi, may also suppress yeasts. Commonly used antifungals include cycloheximide (100–500 mg/L), cyclosporin A (4–10 mg/L), and pimaricin (5–100 mg/L).
Selective media can be used to isolate genera, species, or groups of species with a particular property. Van der Walt and van Kerken (1961) isolated species of Dekkera using media containing cycloheximide and sorbic acid at pH 4.8. Van Dijken and Harder (1974) used a medium containing methanol, cycloserine, and penicillin G for the isolation of methanol-assimilating yeasts of biotechnological importance. Various species of the Lipomycetaceae can be recovered from soil and insect frass by a procedure that depends on the utilization of thymine as a nitrogen source and resistance to cycloheximide.
Isolation using membrane filters . Yeasts can be recovered from liquid substrates by passing the liquid through a 0.45 μm membrane filter (e.g., Mulvany 1969). Solid substrates, such as soils, can be washed to suspend the yeast cells prior to passing the wash solution through the membrane. The filters are placed face up on the surface of a selective agar medium, followed by incubation at a temperature appropriate for the target organisms. Plates should be inspected daily. This technique is particularly useful for recovering yeasts when they are present in low concentrations, and the method can serve as a means to quantitate the densities of yeast communities.
Purification of cultures . Isolates are obtained in pure culture from natural materials or from enriched cultures by streaking on a suitable medium, such as glucose–peptone–yeast extract agar or YM agar (Kurtzman et al. 2011). Persistent bacterial contamination can often be eliminated by acidifying the media or by adding antibiotics. Single, well-separated colonies of each form are selected and streaked again. Twice is generally sufficient to obtain pure cultures. When two or more morphologically distinct colonies persistently appear after replating of a single colony, these may represent morphological or sexual variants of a single species. Because of this, it is useful to initially save multiple colonies from isolations because some may represent mating types.
B. Maintenance
The maintenance of yeast cultures on a medium that contains glucose as the only carbon source reduces the risk of changes in growth and fermentative patterns due to the selection of mutants (Scheda 1966). The majority of yeasts may be stored at temperatures between 4 and 12 °C and subcultured at intervals of 6–8 months. Some yeasts, such as certain Kazachstania spp., need to be subcultured every month. Strains of Dekkera and Brettanomyces produce excessive amounts of acetic acid, and inclusion of 1–2 % calcium carbonate in the medium prolongs viability. Nevertheless, these yeasts still need to be subcultured every 2 months, or sometimes more frequently.
Some yeasts lose their ability to produce ascospores when maintained by serial cultivation on laboratory media, whereas other isolates still sporulate after 50 or more years in cultivation. However, for many strains, the ability to sporulate is either impaired or lost within a period that varies from a few weeks to several years. For ascosporogenous heterothallic species, one mating type may selectively predominate in laboratory culture, which results in the loss of the sexual state. For these reasons, it is important to preserve nomenclatural types and reference strains with one of the more permanent conservation techniques as soon as possible after acquisition. Suitable techniques are lyophilization (Kirsop and Kurtzman 1988), L-drying (Mikata and Banno 1989), and freezing in either liquid nitrogen vapor or a mechanical freezer at temperatures between −60 and −135 °C, although liquid nitrogen is preferred.
The procedure of lyophilization freeze-dries cultures to an inactive, low-moisture state. The process starts with suspending actively growing cultures in a cryoprotectant, such as sterile bovine serum or skim milk. The volume of cryoprotectant depends on the size of the ampoule used, and ampoules used in the ARS Culture Collection (NRRL) are made from 6 mm outside diameter pyrex glass tubing. Lyophilized strains generally survive for many decades, though there are exceptions, and the cultures should be checked periodically for viability. Lyophilized cultures are usually stored in a refrigerator at 4–5 °C.
In the ARS process, ca. 0.1 mL of suspension containing cells and bovine serum is added aseptically to the sterile, labeled lyophil tubes, which have a small cotton plug (Kurtzman et al. 2011). After filling, tubes are inserted into rubber connectors on the manifold of the lyophil machine. The manifold is lowered so that the ends of the tubes are immersed in a 50 % ethylene glycol bath cooled to ca. −30 °C with dry ice. Once the contents of the tubes are frozen, the bath temperature is raised to ca. −20 to −25 °C with fresh ethylene glycol, and the vacuum pump is started. Drying takes ca. 2 h and is finalized by raising the manifold and allowing further drying at room temperature. Ampoules are sealed with a gas-oxygen torch. Some lyophil machines use larger ampoules that may contain 1 mL of cell suspension. These larger tubes may be initially frozen but do not need immersion in a freezing bath because evaporation of this larger volume is adequate to keep the preparations frozen.
A process termed L-drying has been successfully used by several large culture collections in Japan (Mikata and Banno 1989). The process is similar to lyophilization, with the exception that cells and cryoprotectant are dried more slowly under vacuum and do not freeze. L-dried ampoules are sealed in the same manner as lyophilized preparations.
Frozen storage at low temperatures is a common alternative for preserving microorganisms that do not survive lyophilization or L-drying. Storage in −80 and −125 °C mechanical freezers is often satisfactory, but storage at still lower temperatures is preferable. The choice is the vapor phase of liquid nitrogen, which is ca. −180 °C. Liquid nitrogen at atmospheric pressure has a slightly lower temperature of −196 °C (−320 °F), but a concern with immersion storage is that ampoules occasionally leak, and when thawed, the trapped liquid nitrogen will expand to gas so rapidly that the ampoules explode.
The general procedure for liquid nitrogen storage of microorganisms is to suspend cells from an actively growing culture in a cryoprotectant such as 10–15 % glycerol. Usually 1 mL of suspension is placed in a 2 mL cryoampoule. Ordinarily, the ampoules are transferred to a storage box in the liquid nitrogen freezer and allowed to undergo uncontrolled freezing. Occasionally, the freeze rate needs to be controlled to ensure high cell viability, which can be done using commercially available devices that have a programmable freezing rate (see Verkley et al., Chap. 8, this volume).
C. Culture Availability and Distribution
The shipment of cultures is governed by a variety of national and international laws and regulations, and some of these regulations have developed in recent years because of the threat of global bioterrorism. Within the USA, shipment of plant and animal pathogens requires a permit from the USDA Animal and Plant Health Inspection Service (APHIS) (APHIS 526 for plant pathogens, APHIS VS 16-3 for animal pathogens). A permit is required from the U.S. Public Health Service when a pathogen is imported into the USA or when a foreign pathogen is redistributed within the USA. Other countries often have their own specific regulations. Failure to comply with requirements can result in the closure of laboratories, large fines, or even incarceration. The shipment of cultures may be governed by the Convention on Biodiversity, also known as the Rio Treaty. Here the concern is that part of a country’s national heritage, i.e., unique microbial germplasm, will be exploited by outside parties without due compensation. Consequently, before novel or other strains are shipped between countries, the requirements of the Rio Treaty must be known. Culture collections that maintain a wide diversity of yeasts are listed in Table 1.2.
VIII. Future Directions
Yeasts are central to many present-day agricultural, medical, and industrial processes, and with the development of recombinant DNA technologies, as well as new societal needs, the role of yeasts in human advancement can be expected to increase. Rapid DNA sequencing technologies have made whole genome sequencing relatively inexpensive, thereby permitting large-scale comparative genomics for yeasts, which will lead to enormous advances in understanding the genetics of metabolic pathways, reproductive processes, and bioengineering. Molecular systematics will play a key role in future uses. The application of molecular methods for strain identification will markedly assist medical diagnostics as well as define industrially and agriculturally important species, including those important in food spoilage and the biocontrol of pests and pathogens. Molecular identification methods will lead to the discovery of numerous new species, which will give us a far better understanding of biodiversity.
References
Ainsworth GC (1971) Ainsworth & Bisby’s dictionary of the fungi, 6th edn. Commonwealth Mycological Institute, Kew
Alcoba-Flórez J, Méndez-Álvarez S, Cano J, Guarro J, Pérez-Roth E, Arévalo M (2005) Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J Clin Microbiol 43:4107–4111
Allison SD, McGuire KL, Treseder KK (2010) Resistance of microbial and soil properties to warming treatment seven years after boreal fire. Soil Biol Biochem 42:1872–1878
Arnaud G (1913) Sur le genre Eremothecium Borzi. Bull Trim Soc Mycol Fr 29:572–576
Andrews JH (1992) Biological control in the phyllosphere. Annu Rev Phytopathol 30:603–635
Batra LR (1973) Nematosporaceae (Hemiascomycetidiae): taxonomy, pathogenicity, distribution and vector relations. Tech Bull No 1469, US Dept of Agriculture, Washington, DC
Beech FW, Davenport RR (1971) Isolation, purification and maintenance of yeasts. In: Morris JR, Ribbons DW (eds) Methods in microbiology, vol 4. Academic, New York, pp 153–182
Beijerinck MW (1889) L’auxanographie, ou la méthode de l’hydrodiffusion dans la gélatine appliquée aux recherchesmicrobiologiques. Arch Néerl Sci Exactes Nat 23:367–372
Belloch C, Querol A, Garcia MD, Barrio E (2000) Phylogeny of the genus Kluyveromyces inferred from the mitochondrial cytochrome-c oxidase II gene. Int J Syst Evol Microbiol 50:405–416
Berbee ML, Taylor JW (1993) Ascomycete relationships: dating the origin of asexual lineages with 18S ribosomal RNA gene sequence data. In: Reynolds DR, Taylor JW (eds) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, pp 67–78
Bialkova A, Subik J (2006) Biology of the pathogenic yeast Candida glabrata. Folia Microbiol (Praha) 51:3–20
Bills GF, Foster MS (2004) Formulae for selected materials used to isolate and study fungi and fungalales. In: Bills GF, Fostser MS, Mueller GM (eds) Biodiversity of fungi. Inventory and monitoring methods. Elsevier, Amsterdam, pp 595–618
Blackwell M (2011) The fungus: 1, 2, 3…5.1 million species? Am J Bot 98:426–438
Bowen AR, Chen-Wu JL, Momany M, Young R, Szaniszlo PJ, Robbins PW (1992) Classification of fungal chitin synthases. Proc Natl Acad Sci U S A 89:519–523
Buckley HR (1971) Fungi pathogenic for man and animals: 2. The subcutaneous and deep-seated mycoses. In: Norris JR, Ribbons DW (eds) Methods in microbiology, vol 4. Academic, New York, pp 461–478
Butler EE, Webster RK, Eckert JW (1965) Taxonomy, pathogenicity and physiological properties of the fungus causing sour rot of citrus. Phytopathology 55:1262–1268
Cain RF (1972) Evolution of the fungi. Mycologia 64:1–14
Cavalier-Smith T (1987) The origin of fungi and pseudofungi. In: Rayner ADM, Brasier CM, Moore D (eds) Evolutionary biology of the fungi. Cambridge University Press, Cambridge, pp 339–353
Chalutz E, Droby S, Cohen L, Weiss B, Barkai-Golan R, Daus A, Fuchs Y, Wilson CL (1991) Biological control of Botrytis, Rhizopus, and Alternaria rots of tomato fruit by Pichia guilliermondii. In: Wilson CL, Chalutz E (eds) Biological control of postharvest diseases of fruits and vegetables. Workshop Proc. US GPO, Washington, DC, pp 71–85
Correia A, Sampaio P, James S, Pais C (2006) Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int J Syst Evol Microbiol 56:313–317
Crandall MA, Brock TD (1968) Molecular basis of mating in the yeast Hansenula wingei. Bacteriol Rev 32:139–163
Cregg JM, Madden KR (1988) Development of the methylotrophic yeast, Pichia pastoris, as a host system for the production of foreign proteins. Dev Ind Microbiol 29:33–41
Cushion MT, Keely SP (2011) Pneumocystis Delanoë & Delanoë. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam, pp 709–717
Daniel H-M, Meyer W (2003) Evaluation of ribosomal RNA and actin gene sequences for the identification of ascomycetous yeasts. Int J Food Microbiol 86:71–78
Daniel H-M, Sorrell TC, Meyer W (2001) Partial sequence analysis of the actin gene and its potential for studying the phylogeny of Candida species and their teleomorphs. Int J Syst Evol Microbiol 51:1593–1606
Daughtrey ML, Hodge KT, Shishkoff N (2003) Archiascomycete and Hemiascomycete pathogens. In: Trigiano RN, Windham MT, Windham AS (eds) Plant pathology: concepts and laboratory exercises. CRC, Boca Raton, FL, pp 111–116
Deak T (2003) Detection, enumeration and isolation of yeasts. In: Boekhout T, Robert V (eds) Yeasts in food: beneficial and detrimental aspects. Behr’s Verlag, Hamburg, pp 39–68
de Koning W, Harder W (1992) Methanol-utilizing yeasts. In: Murell JC, Dalton H (eds) Methane and methanol utilizers. Plenum, New York, pp 207–244
do Carmo-Sousa L (1969) Endospore formation in the genus Trichosporon. In: Kocková-Kratochvílová A (ed) Proc 2nd symp on yeasts, 1966, Bratislava, pp 87–92
Droby S, Hofstein R, Wilson CL, Wisniewski M, Fridlender B, Cohen L, Weiss B, Daus A, Timar D, Chalutz E (1993) Pilot testing of Pichia guilliermondii: a biocontrol agent of postharvest diseases of citrus fruit. Biol Control 3:47–52
Droby S, Cohen L, Daus A, Weiss B, Horev B, Chalutz E, Katz H, Keren-Tzur M, Shachnai A (1998) Commercial testing of Aspire: a yeast preparation for the biological control of postharvest decay of citrus. Biol Control 12:97–101
Dubos RJ (1960) Louis Pasteur, free lance of science. De Capo, New York
Dykhuizen DE, Green L (1991) Recombination in Escherichia coli and the definition of biological species. J Bacteriol 173:7257–7268
Edman JC, Kovacs JA, Masur H, Santi DV, Elwood HJ, Sogin ML (1988) Ribosomal RNA sequence shows Pneumocystis carinii to be a member of the fungi. Nature 334:519–522
Eriksson OE, Hawksworth DL (1995) Notes on ascomycete systematics—nos. 1885–2023. Syst Ascomyc 14:41–77
Fall R, Benson A (1996) Leaf methanol—the simplest natural product from plants. Trends Plant Sci 1:296–301
Fitzpatrick DA, Logue ME, Stajich JE, Butler G (2006) A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol 6:99–113
Fleet GH (1990) Food spoilage yeasts. In: Spencer JFT, Spencer DM (eds) Yeast technology. Springer, Berlin, pp 124–166
Fonseca Á, Rodrigues MG (2011) Taphrina Fries (1832). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam, pp 823–858
Garner R, Walker AN, Horst MN (1991) Morphologic and biochemical studies of chitin expression in Pneumocystis carinii. J Protozool 38:125–145
Gellissen G, Kunze G, Gaillardin C, Cregg JM, Berardi E, Veenhuis EM, van der Klei I (2005) New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and on dimorphic Arxula adeninivorans and Yarrowia lipolytica—a comparison. FEMS Yeast Res 5:1079–1096
Goodman M (1976) Protein sequences in phylogeny. In: Ayala FJ (ed) Molecular evolution. Sinauer Associates, Sunderland, MA, pp 141–159
Goto S, Sugiyama J (1970) Studies on Himalayan yeasts and molds (IV). Several asporogenous yeasts, including two new taxa of Cryptococcus. Can J Bot 48:2097–2101
Goto S, Sugiyama J, Hamamoto M, Komagata K (1987) Saitoella, a new anamorphic genus in the Cryptococcaceae to accommodate two Himalayan yeast isolates formerly identified as Rhodotorula glutinis. J Gen Appl Microbiol 33:75–85
Groth C, Hansen J, Piskur J (1999) A natural chimeric yeast containing genetic material from three species. Int J Syst Bacteriol 49:1933–1938
Guetsky R, Shtienberg D, Elad Y, Fischer E, Dinoor A (2002) Improving biological control by combining biocontrol agents each with several mechanisms of disease suppression. Phytopathology 92:976–985
Guilliermond A (1912) Les Levures. Encyclopédie scientifique. O Doin et Fils, Paris
Hansen EB (2004) Microorganisms. In: Hui YH, Meunier-Goddik L, Hansen AS, Josephsen J, Nip W-K, Stanfield PS, Toldrá F (eds) Handbook of food and beverage fermentation technology. Marcel Dekker, New York, pp 9–22
Hawksworth DL (2012) Managing and coping with name of pleomorphic fungi in a period of transition. IMA Fungus 3:15–24
Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lucking R, Thorsten Lumbsch H, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Koljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo JM, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schussler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao YJ, Zhang N (2007) A higher-level phylogenetic classification of the Fungi. Mycol Res 111:509–547
Holley RA, Allan-Wojtas P, Phipps-Todd BE (1984) Nematospora sinecauda sp. nov., a yeast pathogen of mustard seeds. Antonie Van Leeuwenhoek 50:305–320
Hughes SJ (1985) The term chlamydospore. In: Arai T, Kuga T, Terao K, Yamazaki M, Miyaji M, Unemoto T (eds) Filamentous microorganisms: biomedical aspects. Japan Scientific Societies Press, Tokyo, pp 1–20
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüssler A, Longcore JE, O'Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R (2006) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443:818–822
Janisiewicz WJ (1987) Postharvest biological control of blue mold on apples. Phytopathology 77:481–485
Janisiewicz WJ, Tworkoski TJ, Kurtzman CP (2001) Biocontrol potential of Metschnikowia pulcherrima strains against blue mold of apple. Phytopathology 91:1098–1108
Jumpponen A, Johnson LC (2005) Can rDNA analyses of diverse fungal communities in soil and roots detect effects of environmental manipulations—a case study from tallgrass prairie. Mycologia 97:1177–1194
Karabulut OA, Tezcan H, Daus A, Cohen L, Wiess B, Droby S (2004) Control of preharvest and postharvest fruit rot in Strawberry by Metschnikowia fructicola. Biocontrol Sci Technol 14:513–521
Kaur R, Domergue R, Zupancic ML, Cormack BP (2005) A yeast by any other name: Candida glabrata and its interaction with the host. Curr Opin Microbiol 8:378–384
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Ainsworth & Bisby’s dictionary of the fungi, 10th edn. CAB International, Wallingford
Kirsop BE, Kurtzman CP (eds) (1988) Living resources for biotechnology. Yeasts. Cambridge University Press, Cambridge
Knapp S, McNeill J, Turland NJ (2011) Changes to publication requirements made at the XVIII International Botanical Congress in Melbourne—what does e-publication mean for you? BMC Evol Biol 11:251–254
Korf RP (1973) Discomycetes and Tuberales. In: Ainsworth GC, Sparrow FK, Sussman AS (eds) The Fungi, an advanced treatise, vol IV A. Academic, New York, pp 249–319
Kramer CL (1960) Morphological development and nuclear behavior in the genus Taphrina. Mycologia 52:295–320
Kramer CL (1973) Protomycetales and taphrinales. In: Ainsworth GC, Sparrow FK, Sussman AS (eds) The fungi—an advanced treatise, vol. IVA. Academic, New York, pp 33–41
Kuramae EE, Robert V, Snel B, Weiss M, Boekhout T (2006) Phylogenomics reveal a robust fungal tree of life. FEMS Yeast Res 6:1213–1220
Kurtzman CP (1993) Systematics of the ascomycetous yeasts assessed from ribosomal RNA sequence divergence. Antonie Van Leeuwenhoek 63:165–174
Kurtzman CP (2003) Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces. Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res 4:233–245
Kurtzman CP (2011) Protomyces Unger (1833). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam, pp 725–731
Kurtzman CP, Robnett CJ (1991) Phylogenetic relationships among species of Saccharomyces, Schizosaccharomyces, Debaryomyces and Schwanniomyces determined from partial ribosomal RNA sequences. Yeast 7:61–72
Kurtzman CP, Robnett CJ (1994) Orders and families of ascosporogenous yeasts and yeast-like taxa compared from ribosomal RNA sequence similarities. In: Hawksworth DL (ed) Ascomycete systematics: problems and perspectives in the nineties. Plenum, New York, pp 249–258
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371
Kurtzman CP, Robnett CJ (2003) Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res 3:417–432
Kurtzman CP, Robnett CJ (2007) Multigene phylogenetic analysis of the Trichomonascus, Wickerhamiella and Zygoascus yeast clades, and the proposal of Sugiyamaella gen.nov. and 14 new species combinations. FEMS Yeast Res 7:141–151
Kurtzman CP, Robnett CJ (2010) Systematics of methanol assimilating yeasts and neighboring taxa from multigene sequence analysis and the proposal of Peterozyma gen. nov., a new member of the Saccharomycetales. FEMS Yeast Res 10:353–361
Kurtzman CP, Robnett CJ (2012) Saitoella coloradoensis sp. nov., a new species of the Ascomycota, subphylum Taphrinomycotina. Antonie Van Leeuwenhoek 101:795–802
Kurtzman CP, Robnett CJ (2013) Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Res 13(1):23–33
Kurtzman CP, Sugiyama J (2001) Ascomycetous yeasts and yeastlike taxa. In: McLaughlin DJ, McLaughlin EG, Lemke PA (eds) The mycota, vol VII (Systematics and evolution), Part A. Springer, Berlin, pp 179–200
Kurtzman CP, Suzuki M (2010) Phylogenetic analysis of ascomycete yeasts that form coenzyme Q-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces and Scheffersomyces. Mycoscience 51:2–14
Kurtzman CP, Robnett CJ, Ward JN, Brayton C, Gorelick P, Walsh TJ (2005) Multigenic phylogenetic analysis of pathogenic Candida species in the Kazachstania (Arxiozyma) telluris complex and description of their ascosporic states as Kazachstania bovina sp. nov., K. heterogenica sp. nov., K. pintolopesii sp. nov. and K. slooffiae sp.nov. J Clin Microbiol 43:101–111
Kurtzman CP, Albertyn J, Basehoar-Powers E (2007) Multigene phylogenetic analysis of the Lipomycetaceae and proposed transfer of Zygozyma species to Lipomyces and Babjevia anomala to Dipodascopsis. FEMS Yeast Res 7:1027–1034
Kurtzman CP, Robnett CJ, Basehoar-Powers E (2008) Relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene phylogenetic analysis and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Res 8:939–954
Kurtzman CP, Price NP, Ray KJ, Kuo TM (2010) Production of sophorolipids biosurfactants by multiple species of the Starmerella (Candida) bombicola yeast clade. FEMS Microbiol Lett 311:140–146
Kurtzman CP, Fell JW, Boekhout T, Robert V (2011) Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam, pp 87–110
Lachance M-A, Daniel H-M, Meyer W, Prasad GS, Gautam SP, Boundy-Mills K (2003) The D1/D2 domain of the large-subunit rDNA of the yeast species Clavispora lusitaniae is unusually polymorphic. FEMS Yeast Res 4:253–258
Landvik S (1996) Neolecta, a fruit-body-producing genus of the basal ascomycetes, as shown by SSU and LSU rRNA sequences. Mycol Res 100:199–202
Landvik S, Eriksson OE, Gargas A, Gustafsson P (1993) Relationships of the genus Neolecta (Neolectales ordo nov., Ascomycotina) inferred from 18S rDNA sequences. Syst Ascomyc 11:107–118
Langeron M, Guerra P (1940) Valeur et nature des variations et dissociations de colonies chez les champignons levuriformes. Ann Parasitol Hum Comp 17:447–469
Legras J-L, Medinoglu D, Cornuet J-M, Karst F (2007) Bread beer and wine: Saccharomyces diversity reflects human history. Mol Ecol 16:2091–2102
Liu Y, Leigh JW, Brinkmann H, Cushion MT (2009) Phylogenomic analyses support the monophyly of Taphrinomycotina, including Schizosaccharomyces fission yeasts. Mol Biol Evol 26:27–34
Martin EM (1940) The morphology and cytology of Taphrina deformans. Am J Bot 27:743–751
Martini A, Phaff HJ (1973) The optical determination of DNA–DNA homologies in yeasts. Ann Microbiol 23:59–68
McClary DO, Nulty WL, Miller GR (1959) Effect of potassium versus sodium in the sporulation of Saccharomyces. J Bacteriol 78:362–368
McGovern PE, Zhang J, Tang J, Zhang Z, Hall GR, Moreau RA, Nuñez A, Dutyrm ED, Richards MP, Wang C-S, Cheng G, Zhao Z, Wang C (2004) Fermented beverages of pre-proto-historic China. Proc Natl Acad Sci U S A 101:17593–17598
Mikata K, Banno I (1989) Preservation of yeast cultures by L-drying: viability after 5 years of storage at 5 °C. Inst Ferment Osaka Res Commun 14:80–103
Mix AJ (1949) A monograph of the genus Taphrina. Univ Kansas Sci Bull 23:1–167
Moore RT (1987) Micromorphology of yeasts and yeast-like fungi and its taxonomic implications. In: de Hoog GS, Smith MT, Weijman ACM (eds) The expanding realm of yeast-like fungi. Elsevier, Amsterdam, pp 203–226
Moore RT (1990) The genus Lalaria gen. nov.: Taphrinales anamorphosum. Mycotaxon 38:315–330
Mortimer R, Polsinelli M (1999) On the origins of wine yeast. Res Microbiol 150:199–204
Mulvany JG (1969) Membrane filter techniques in microbiology. In: Norris JR, Ribbons DW (eds) Methods in microbiology, vol 1. Academic, New York, pp 205–253
Naehring J, Kiefer S, Wolf K (1995) Nucleotide sequence of the Schizosaccharomyces japonicus var. versatilis ribosomal RNA gene cluster and its phylogenetic implications. Curr Genet 28:353–359
Nakase T, Komagata K (1968) Taxonomic significance of base composition of yeast DNA. J Gen Appl Microbiol 14:345–357
Nguyen NH, Suh SO, Marshall CJ, Blackwell M (2006) Morphological and ecological similarities: wood-boring beetles associated with novel xylose-fermenting yeasts, Spathaspora passalidarum gen. sp. nov. and Candida jeffriesii sp. nov. Mycol Res 110:1232–1241
Nishida H, Sugiyama J (1993) Phylogenetic relationships among Taphrina, Saitoella, and other higher fungi. Mol Biol Evol 10:431–436
Nishida H, Sugiyama J (1994a) Phylogeny and molecular evolution among higher fungi. Nippon Nogeikagaku Kaishi 68:54–57 (in Japanese)
Nishida H, Sugiyama J (1994b) Archiascomycetes: detection of a major new lineage within the Ascomycota. Mycoscience 35:361–366
Nishida H, Sugiyama J (1995) A common group I intron between a plant parasitic fungus and its host. Mol Biol Evol 12:883–886
Nishida H, Blanz PA, Sugiyama J (1993) The higher fungus Protomyces inouyei has two group I introns in the 18S rRNA gene. J Mol Evol 37:25–28
Nishida H, Tajiri Y, Sugiyama J (1998) Multiple origins of fungal group I introns located in the same position of nuclear SSU rRNA gene. J Mol Evol 46:442–448
Nishida H, Ogura A, Yokota A, Yamaguchi I, Sugiyama J (2000) Group I intron located in PR protein homologue gene in Youngia japonica. Biosci Biotechnol Biochem 64:606–609
Norvell LL (2011) Melbourne approves a new Code. Mycotaxon 116:481–490
Passoth V, Fredlund E, Druvefors UA, Schnurer J (2006) Biotechnology, physiology and genetics of the yeast Pichia anomala. FEMS Yeast Res 6:3–13
Peterson SW, Kurtzman CP (1991) Ribosomal RNA sequence divergence among sibling species of yeasts. Syst Appl Microbiol 14:124–129
Phaff HJ, Miller MW, Shifrine M (1956) The taxonomy of yeasts isolated from Drosophila in the Yosemite region of California. Antonie Van Leeuwenhoek 22:145–161
Phaff HJ, Starmer WT, Tredick-Kline J (1987) Pichia kluyveri sensu lato—a proposal for two new varieties and a new anamorph. In: de Hoog GS, Smith MT, Weijman ACM (eds) The expanding realm of yeast-like fungi. Elsevier, Amsterdam, pp 403–414
Pitt JI, Miller MW (1968) Sporulation in Candida pulcherrima, Candida reukaufii and Chlamydozyma species: their relationships with Metschnikowia. Mycologia 60:663–685
Porter TM, Schadt CW, Rizvi L, Martin AP, Schmidt SK, Scott-Denton L, Vilgalys R, Moncalvo JM (2008) Widespread occurrence and phylogenetic placement of a soil clone group adds a prominent new branch to the fungal tree of life. Mol Phylogenet Evol 46:635–644
Price CW, Fuson GB, Phaff HJ (1978) Genome comparison in yeast systematics: delimitation of species within the genera Schwanniomyces, Saccharomyces, Debaryomyces and Pichia. Microbiol Rev 42:161–193
Prillinger H, Dörfler C, Laaser G, Eckerlein B, Lehle L (1990) Ein Beitrag zur Systematik und Entwicklungsbiologie höherer Pilze: Hefe-Typen der Basidiomyceten. Teil I: Schizosaccharomycetales, Protomyces-Typ. Z Mykol 56:219–250
Redhead SA (1977) The genus Neolecta (Neolectaceae fam. nov., Lecanolales, Ascomycetes) in Canada. Can J Bot 55:301–306
Redhead SA, Malloch D (1977) The Endomycetaceae: new concepts, new taxa. Can J Bot 55:1701–1711
Rosling A, Cox F, Cruz-Martinez K, Ihrmark K, Grelet GA, Lindahl BD, Menkis A, James TY (2011) Archaeorhizomycetes: unearthing an ancient class of ubiquitous soil fungi. Science 333:876–879
Rubio-Texeira M (2006) Endless versatility in the biotechnological applications of Kluyveromyces LAC genes. Biotechnol Adv 24:210–223
Schadt CW, Martin AP, Lipson DA, Schmidt SK (2003) Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301:1359–1361
Scheda R (1966) Merkmalsveränderungen bei Hefen der Gattung Saccharomyces. Monatsschr Brau 19:256–258
Schoch CL, Sung G-H, López-Giráldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny PB, Kauff F, Wang Z, Gueidan C, Andrie RM, Trippe K, Ciufetti LM, Wynns A, Fraker E, Hodkinson BP, Bonito G, Yahr R, Groenewald JZ, Arzanlou M, de Hoog GS, Crous PW, Hewitt D, Pfister DH, Peterson K, Gryzenhout M, Wingfield MJ, Aptroot A, Suh S-O, Blackwell M, Hillis DM, Griffith GW, Castlebury LA, Rossman AY, Lumbsch HT, Lücking R, Büdel B, Rauhut A, Diederich P, Ertz D, Geiser DM, Hosaka K, Inderbitzin P, Kohlmeyer J, Volkmann-Kohlmeyer B, Mostert L, O’Donnell K, Sipman H, Rogers JD, Shoemaker RA, Sugiyama J, Summerbell RC, Untereiner W, Johnston P, Stenroos S, Zuccaro A, Dyer P, Crittenden P, Cole MS, Hansen K, Trappe JM, Lutzoni F, Spatafora JW (2009) The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol 58:224–239
Sietsma JH, Wessels JGH (1990) Occurrence of glucosaminoglycan in the wall of Schizosaccharomyces pombe. J Gen Microbiol 136:2261–2265
Sjamsuridzal W, Tajiri Y, Nishida H, Thuan T, Kawasaki H, Hirata A, Yokota A, Sujiyama J (1997) Evolutionary relationships of members of the genera Taphrina, Protomyces, Schizosaccharomyces, and related taxa within the Archiascomycetes: integrated analysis of genotypic and phenotypic characters. Mycoscience 38:267–280
Slininger PJ, Bolen PL, Kurtzman CP (1987) Pachysolen tannophilus properties and process considerations for ethanol production from D-xylose. Enzyme Microb Technol 9:5–15
Staib F, Seibold M, Antweiler E, Frölich B (1989) Staib agar supplemented with a triple antibiotic combination for the detection of Cryptococcus neoformans in clinical specimens. Mycoses 32:448–452
Stalpers JA (1987) Pleomorphy in holobasidiomycetes. In: Sugiyama J (ed) Pleomorphic fungi: the diversity and its taxonomic implications. Kodansha, Tokyo, pp 201–220
Starmer WT, Ganter PF, Aberdeen V (1992) Geographic distribution and genetics of killer phenotypes for the yeast Pichia kluyveri across the United States. Appl Environ Microbiol 58:990–997
Streiblová E (1971) Cell division in yeasts. In: Pérez-Miravette A, Peláez D (eds) Recent advances in microbiology. Proc. 10th int congress on microbiology, Mexico City, Mexico, pp 131–140
Sugiyama J (1998) Relatedness, phylogeny, and evolution of the fungi. Mycoscience 39:487–511
Sugiyama J, Hamamoto M (2011) Saitoella S. Goto, Sugiyama, Hamamoto & Komagata (1987). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam, pp 1313–1315
Sugiyama J, Nishida H (1995) The higher fungi: their evolutionary relationships and implications for fungal systematics. In: Arai R, Kato M, Doi Y (eds) Biodiversity and evolution. National Science Museum Foundation, Tokyo, pp 177–195
Sugiyama J, Fukagawa M, Chiu S-W, Komagata K (1985) Cellular carbohydrate composition, DNA base composition, ubiquinone systems and diazonium blue B color test in the genera Rhodosporidium, Leucosporidium, Rhodotorula and related basidiomycetous yeasts. J Gen Appl Microbiol 31:519–550
Sugiyama J, Hosaka K, Suh SO (2006) Early diverging Ascomycota: phylogenetic divergence and related evolutionary enigmas. Mycologia 98:996–1005
Sugiyama J, Nishida H, Suh S-O (1993) The paradigm of fungal diagnoses and descriptions in the era of molecular systematics: Saitoella complicata as an example. In: Reynolds DR, Taylor JW (eds) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, pp 261–269
Sullivan DJ, Moran GP, Coleman DC (2005) Candida dubliniensis: ten years on. FEMS Microbiol Lett 253:9–17
Syrop M, Beckett S (1976) Leaf curl disease of almond caused by Taphrina deformans. III. Ultrastructural cytology of the pathogen. Can J Bot 54:293–305
Takano I, Oshima Y (1970) Mutational nature of an allele-specific conversion of the mating type by the homothallic gene HOα in Saccharomyces. Genetics 65:421–427
Taylor JW, Swann EC, Berbee ML (1994) Molecular evolution of ascomycete fungi: phylogeny and conflict. In: Hawksworth DL (ed) Ascomycete systematics: problems and perspectives in the nineties. Plenum, New York, pp 201–212
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32
Tomaszewski EK, Logan KS, Snowden KF, Kurtzman CP, Phalen DN (2003) Phylogenetic analysis identifies the ‘megabacterium’ of birds as a novel anamorphic ascomycetous yeast, Macrorhabdus ornithogaster gen. nov., sp. nov. Int J Syst Evol Microbiol 53:1201–1205
Tripathi AK, Chauhan RKS, Bartaria AM, Chauhan S (2003) Quantitative and qualitative loss in coriander due to Protomyces macrosporus. Indian Phytopathol 56:451–452
Tubaki K (1957) Biological and cultural studies of three species of Protomyces. Mycologia 49:44–54
Tubaki K (1978) Taphrina wiesneri (Rathay) Mix. In: Udagawa S, Tubaki K, Horie Y, Miura K, Minoura K, Yamazaki M, Yokoyama T, Watanabe S (eds) Kinruizukan (‘Compendium of Fungi’), part 1. Kodansha, Tokyo, pp 329–330 (in Japanese)
Vandenkoornhuyse P, Baldauf SL, Leyval C, Straczek J, Young JPW (2002) Extensive fungal diversity in plant roots. Science 295:2051
van der Walt JP (1970) Criteria and methods used in classification. In: Lodder J (ed) The yeasts, a taxonomic study, 2nd edn. North-Holland, Amsterdam, pp 34–113
van der Walt JP, Scott DB (1971) Saccharomycopsis synnaedendra, a new yeast from South African insect sources. Mycopathol Mycol Appl 44:101–106
van der Walt JP, van Kerken AE (1961) The wine yeasts of the Cape. Part V. Studies on the occurrence of Brettanomyces intermedius and Brettanomyces schanderlii. Antonie Van Leeuwenhoek 27:81–90
van Dijken JP, Harder W (1974) Optimal conditions for the enrichment and isolation of methanol-assimilating yeasts. J Gen Microbiol 84:409–411
Vaughan-Martini A, Kurtzman CP (1985) Deoxyribonucleic acid relatedness among species of the genus Saccharomyces sensu stricto. Int J Syst Bacteriol 35:508–511
Vaughan-Martini A, Martini A (2011) Schizosaccharomyces Lindner (1893). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam, pp 779–784
Veenhuis M, van Dijken JP, Harder W (1983) The significance of peroxisomes in the metabolism of one-carbon compounds in yeasts. Adv Microb Physiol 24:1–82
Vega FE, Blackwell M (2005) Insect–fungal associations: ecology and evolution. Oxford University Press, Oxford
Verstrepen KJ, Chambers PJ, Pretorius IS (2006) The development of superior yeast strains for the food and beverage industries: challenges, opportunities, and potential benefits. In: Querol A, Fleet GH (eds) Yeasts in foods and beverages. Springer, Berlin, pp 399–444
von Arx JA, van der Walt JP (1987) Ophiostomatales and endomycetales. In: de Hoog GS, Smith MT, Weijman ACM (eds) The expanding realm of yeast-like fungi. Elsevier, Amsterdam, pp 167–176
von Arx JA, Weijman ACM (1979) Conidiation and carbohydrate composition in some Candida and Torulopsis species. Antonie Van Leeuwenhoek 45:547–555
Walker WF (1985) 5S ribosomal RNA sequences from ascomycetes and evolutionary implications. Syst Appl Microbiol 6:48–53
Watanabe J, Hori H, Tanabe K, Nakamura Y (1989) Phylogenetic association of Pneumocystis carinii with the “Rhizopoda/Myxomycota/Zygomycota group” indicated by comparison of 5S ribosomal RNA sequences. Mol Biochem Parasitol 32:163–168
Wells JM (1977) Sour rot of peaches caused by Monilia implicata and Geotrichum candidum. Phytopathology 67:404–408
Wickerham LJ (1951) Taxonomy of yeasts. Tech Bull 1029, U.S. Department of Agriculture, Washington, DC
Wickerham LJ (1958) Sexual agglutination of heterothallic yeasts in diverse taxonomic areas. Science 128:1504–1505
Wickerham LJ (1969) New homothallic taxa of Hansenula. Mycopathol Mycol Appl 37:15–32
Wickerham LJ, Burton KA (1954) A clarification of the relationships of Candida guilliermondii to other yeasts by a study of their mating types. J Bacteriol 68:594–597
Wickerham LJ, Flickinger MH, Johnston RM (1946) The production of riboflavin by Ashbya gossypii. Arch Biochem 9:95–98
Yamada Y, Banno I (1987) Hasegawaea gen. nov., an ascosporogenous yeast genus for the organism whose ascospores have smooth surfaces without papillae and which are characterised by the absence of coenzyme Q and by the presence of linoleic acid in cellular fatty acid composition. J Gen Appl Microbiol 33:295–298
Yarrow D (1998) Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam, pp 77–100
Yoshida Y (1989) Ultrastructural studies of Pneumocystis carinii. J Protozool 36:53–60
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Kurtzman, C.P., Sugiyama, J. (2015). 1 Saccharomycotina and Taphrinomycotina: The Yeasts and Yeastlike Fungi of the Ascomycota. In: McLaughlin, D., Spatafora, J. (eds) Systematics and Evolution. The Mycota, vol 7B. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-662-46011-5_1
Download citation
DOI: https://doi.org/10.1007/978-3-662-46011-5_1
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-662-46010-8
Online ISBN: 978-3-662-46011-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)